3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(13):2885-2891. doi:10.7150/jca.31167 This issue Cite
Review
Regulatory mechanisms and clinical perspectives of circRNA in digestive system neoplasms
1. Department of Anorectal Surgery, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan 450052, China
2. Department of gastrointestinal surgery, The people's hospital of Dezhou, Dezhou, Shandong 253014, China
3. Department of Pathology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan 450052, China
4. Department of Pathology, School of Basic Medicine, Zhengzhou University, Zhengzhou, Henan 450052, China
#Equal contributors
Received 2018-11-2; Accepted 2019-4-14; Published 2019-6-2
Abstract
A new star, circular RNA (circRNA), is a class of noncoding RNA with a stable cyclic structure. Exonic circRNA mainly exists in the eukaryotic cytoplasm. Intronic circRNAs (ciRNA) and exonic circRNAs with introns (EIciRNA) are found in the nucleus. Recent evidences showed the functional diversity of circRNAs, which could be microRNA (miRNA) sponges, interact with protein or translate into small peptide. Due to the change of human eating habits, digestive cancer remains one of the most common cancers worldwide and it is prone to metastasis. Increasing studies have found a number of circRNAs using RNA sequencing technology and displayed double roles of circRNA in digestive cancer. In this review, we surveyed the biogenesis and regulation of circRNAs, discussed circRNA functions and clinical applications (especially circRNAs in exosome) in digestive cancers, which implied that circRNAs could be as potential biomarkers in diagnosis and treatment of digestive cancers in the future.
Keywords: CircRNA, Digestive cancers, Biogenesis, Regulation, Clinical applications
Introduction
In the 1970s, circRNAs were first reported in plant viroids [1]. Through observation using an electron microscope, circRNAs were proven to exist in the eukaryotic cytoplasm [2]. Moreover, ciRNA and EIciRNA were subsequently detected in the nucleus of eukaryotes [3-5]. CircRNAs were found as a class of noncoding RNA without 5' caps and 3' ploy (A) tails [6]. Different from linear noncoding RNAs, such as long noncoding RNA (lncRNA) and miRNAs, circRNAs have a covalently closed ring shape and are insensitive to exonuclease R, which leads to the stabilization of the structure [7].
With the development of bioinformatics, most studies showed a variety of circRNA played an important role in various diseases. To explore the circRNA role in diabetes and depression, Jiang et al. found 183 upregulated hsa-circRNAs and 64 downregulated hsa-circRNAs in the treatment group. KEGG results showed that potential circRNAs might be involved in the thyroid hormone, Wnt, ErbB, and mitogen-activated protein kinase signaling pathways [8]. Moreover, dysregulated circRNAs were implicated in osteogenesis, diabetes mellitus and brain dysfunction, especially cancer [9-11]. Recently, Huang et al. recognized several differentially expressed circRNAs using a circRNA microarray. Furthermore, circRNA_100338, one of the upregulated circRNAs, promoted invasive potential of hepatitis B-related hepatocellular carcinoma [12]. CircFAT1 (e2), as a novel circular RNA, has been reported to inhibit gastric cancer progression [13]. One intronic circRNA derived from CTNNB1 (circ-CTNNB1) was upregulated in several cancer including gastric cancer, prostate cancer and colon cancer. Function identification showed that circ-CTNNB1 promoted β-catenin activation, growth, invasion, and metastasis in these cancer cells [14]. These studies suggested that circRNAs might be key regulators in cancer progression.
Due to the change of human eating habits, digestive cancers are very common in the world, such as colorectal cancer, gastric cancer and esophageal squamous cell carcinoma [15-17]. Therefore, it is essential for increasing the survival rate of digestive cancer patients via early diagnosis and therapy [18]. In this study, we summarized a series of studies on circRNA biogenesis and its regulation. We also discussed circRNA functions, mechanisms and potential clinical significance in digestive cancers.
Biogenesis and regulations of circRNA
Increasing circRNAs were identified by microarray or high-throughput sequencing, but the biogenesis of circRNA is still elusive. Recently, some researchers have found that intron pairing, exon skipping, and RNA binding protein (RBP) pairing were the main reasons that drove the circularization of RNA (Figure 1) [7, 19-22]. In 2012, Jeck et al. identified a number of “backsplices” by high-throughput sequencing. The features of the circularized exons were driven by intron-pairing, which contained complementary ALU repeats using bioinformatic analysis [23]. Additional evidence strengthened the present theory that reverse complementary matches (RCMs) in circRNA flanking introns are the key step of circRNA biogenesis, which was diminished by enzyme adenosine deaminase 1 (ADAR1) [22]. Exon-skipping events are another mainstream hypothesis for circularization. A study reported that exon-skipping promoted the shaping of a spliced lariat containing the circularized exon [20]. Moreover, RNA binding protein showed a potential induction in circRNA formation. Quaking is an RNA binding protein that affects the circRNA abundance related to the intronic Quaking binding motifs [21]. As a result, the flanking complementary sequences could promote back-splicing, further shaping circRNAs. Recently, Xiang et al. reported that the immune factors, NF90/NF110, enhanced the formation of back-splicing in the nucleus, which suggested that NF90/NF110 was involved in circRNA biogenesis [24]. Recently, the processing of circRNA was reported in eukaryotes. Ling et al. showed that circRNAs in eukaryotes were from pre-mRNA reverse splicing, and its productions were correlated with the extension rate of Pol II [25].
Based on the adequate biogenesis studies of circRNA, increasing studies have focused on its regulation (Figure 1). Currently, circRNAs play an important role via sponging miRNAs, interacting with proteins or being translated into functional small peptide [26-28].
Biogenesis and regulations of circRNAs. Three modes of circRNA biogenesis are shown in the ring. (a) Flanking intron pairing, which contains ALU repeats, drives the formation of backsplicing. (b) Lariat-driven circularization shapes a lariat. The lariat is spliced. 5'-GU-rich and 3'-C-rich regions assist intron circularization and shape ciRNAs. (c) RBP is involved in the circularization. Based on this, the regulation of circRNA is shown in A, B and C.
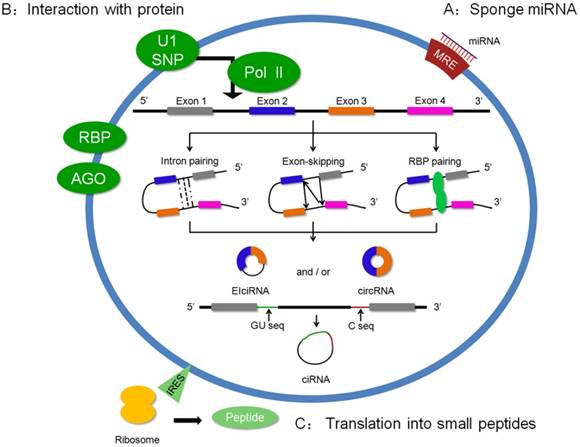
miRNA sponge
It is well known that lncRNAs could be endogenous sponges that bind to their target miRNAs because of the presence of the microRNA response element (MRE) in lncRNA sequences [29]. Subsequently, Thomas B et al. proved that circRNAs are implicated in the ceRNA mechanism as an miRNA sponge. A previous study showed a high level of circRNA expression in mouse and human brains [28]. Further study found that a circRNA has more than 70 certain miRNA seed match segments and could be a sponge of miR-7 [27]. A world of circRNA functions was opened and increasing evidences had shown a wide range of regulation mechanisms by the circRNA-miRNA axis.
An exonic circRNA, ciRS-7, was found to have 63 conserved miRNA target sites of miR-7 and was bound by AGO complexes [30]. Monika et al. verified that ciRS-7 could be bound by AGO miRNA complexes with miR-7 and miR-671. Deficiency of the ciRS-7 gene led to the dysfunction of draining, including sensorimotor gating damage and abnormal synaptic transmission. A miR-7 target, the Fos gene, is upregulated in the ciRS-7 deficient group and showed a potential relationship with the behavioral phenotype in mouse [11]. Knockdown ciRS-7 or miR-7 showed that ciRS-7 was proposed as an anti-miRNA circRNA, which is crucial to normal brain function. Similarly, Kun et al. identified a heart-related circRNA, HRCR, which could be bound by miR-223. ARC was verified as a downstream target of miR-223 by RNAhybrid prediction and target protector technology. Overexpressed miR-223 promoted hypertrophy in the mouse heart and cardiomyocytes. Interestingly, enhanced HRCR expression showed a contrasting effect in vivo and in vitro, which implied that HRCR could restrain the development of heart disease [31]. In cancer, accumulated studies verified that circRNAs could function similar to ceRNA, regulating tumor progression. Upregulated circUBAP2 in human osteosarcoma indicated a poor patient prognosis. It accelerated tumor growth and suppressed apoptosis by sponging miR-143 and releasing the miR-143 direct target, anti-apoptotic Bcl-2 [32, 33]. Resembling circUBAP2, circ_0001982 presented a tumor suppressor effect by sponging miR-143 in breast cancer cells [34]. CircRNA_100290 was another identified circRNA in oral cancer by microarray technology. Through competitively binding miR-29b, silencing of circRNA_100290 markedly inhibited the expression level of CDK6. As a target of miR-29b, CDK6 combined with cyclin promoted the cell cycle from G1 phase to S phase [35-37]. There are many studies of circRNA-miRNA regulation. These studies suggested that circRNAs sponging miRNAs showed complex regulation networks in cancer and disease, and more ceRNAs of circRNAs will be found in the future.
Interaction with protein
Previous studies have revealed that circRNAs could influence pre-mRNA and regulate parental gene expression. Ashwal-Fluss et al. found a conserved muscle-blind binding site in the flanking sequences of circRNA, circMbI. Interestingly, intensive research showed that MBL could specifically bind with circMbI and showed a forceful influence in the biosynthesis of circMbI. This study illustrated a function of circRNA in transcription via competing with pre-mRNA splicing [38]. The others of circRNA in regulating transcription display an effect on the parental gene. Cir-ITCH is one of the circRNAs that modulates the parental gene through sponging miRNA. Based on common miRNA binding sites in circITCH and the ITCH sequence, circITCH competitively bound with miR-7, miR-17, and miR-214, indirectly increasing ITCH expression [39]. As we showed, the sequence of EIciRNAs contained the exon and its flanking intron. A classical study reported that EIciRNAs, such as circEIF3J and circPIAP2 in the nucleus, promoted parental gene transcription via shaping EIciRNAs-U1 snRNP complexes and interacting with Pol II, which suggested that EIciRNAs could bind with the protein and had an effect on the regulation of transcription of the parental gene [5]. In addition, exonic circular RNAs, as regulators, play identical roles in combining with proteins. Upregulated circ-Foxo3 in non-cancer cells was correlated with the cell cycle. An interesting result showed that circ-Foxo3 bound to CDK2 and p21, forming an RNA-protein complex to arrest the cell cycle [40].
Translation into small peptides
Recent studies indicated that circRNA could be translated into the protein. Additionally, a non-circle structure, internal ribosome entry site (IRES), is the key aspect of circRNA translation into a protein. Nagarjuna et al. found several translating circRNAs in the fly head. These circRNAs not only retained conservative termination codons in evolution but could also be bound to the ribosome utilizing the start codon of its host mRNA. This translation course was possibly regulated by starvation and FOXO [41]. Similar to the fly head, anendogenous circular RNA, FBXW7, contained an IRES, which could be encoded by a 21 kDa protein in the human brain. A functional study displayed the suppressor role of the FBXW7 protein in malignant phenotypes of human glioblastoma [42]. These studies strongly proved that endogenous circRNAs had a translation capacity, which exploited a new field of circRNA function.
Dysregulated circRNAs in digestive cancers
CircRNAs are a new member of the non-coding RNA family. Genome-wide statistical analysis promoted the discovery of circRNAs in multiple cancer tissues [12, 43]. Through microarray technology, 125 circRNAs were upregulated and 76 circRNAs were identified in paired colorectal cancer tissues and normal tissues [44]. Similarly, compared with the normal tissues, 522 downregulated and 191upregulated circRNAs were identified in gastric cancer. Further online functional analysis showed that these dysregulated circRNAs were involved in carcinogenesis [45]. Radiotherapy is a primary method of oncotherapy. Acquired radioresistance led to a decline in patient survival. Recently, a study showed that dysregulated circRNAs were implicated in radioresistant esophageal cancer cells. Most of their target genes were enriched in the Wnt signaling pathway, which implied that these circRNAs exert their regulation in resistance to esophageal cancer through the Wnt signaling pathway [46]. As we showed, dysregulated circRNAs exist in digestive cancers and may play important roles in digestive cancers (Table 1).
Dysregulated circRNA in digestive cancers.
| circRNA | Location | Gene symbol | Cancer | Dysregulation | Ref |
|---|---|---|---|---|---|
| hsa_circ_0074362 | chr5:142264862-142311690 | ARHGAP26 | Gastric cancer | Downregulated | [47] |
| hsa_circ_0000520 | chr14:20811436-20811559 | RPPH1 | Gastric cancer | Downregulated | [48] |
| hsa_circ_0047905 | chr18:61156579-61172318 | SERPINB5 | Gastric cancer | Upregulated | [49] |
| hsa_circ_0138960 | chr9:74817486-74828907 | GDA | |||
| hsa_circ_0000745 | chr17:20107645-20109225 | SPECC1 | Gastric cancer | Downregulated | [50] |
| hsa_circ_0067934 | chr3:170013698-170015181 | PRKCI | Esophageal squamous cell carcinoma | Upregulated | [51] |
| hsa_circ_0000677 | chr16:16101672-16162159 | ABCC1 | Colorectal cancer | Upregulated | [52] |
| hsa_circ_0003159 | chr7:81689743-81746489 | CACNA2D1 | Gastric cancer | Downregulated | [53] |
| hsa_circ_0000190 | chr1:224553580-224559125 | CNIH4 | Gastric cancer | Downregulated | [54] |
| hsa_circ_0000069 | chr1:47745912-47748131 | STIL | Colorectal cancer | Upregulated | [55] |
| hsa_circ_0001724 | chr7:92462409-92463134 | CDK6 | Colorectal cancer | Upregulated | [56] |
| hsa_circ_0026344 | chr12:52314542-52317145 | ACVRL1 | Colorectal cancer | Downregulated | [57] |
Role of circular RNA in digestive cancers
Recently, most studies showed that circRNAs are important regulators involved in cancer progression. Moreover, these circRNAs play double roles which could excert different functions through different regulations in digestive cancers (Figure 2).
Roles of circRNAs in digestive cancers. Green rectangles indicate onco-circRNAs. Red rectangles represent circRNA suppressors.
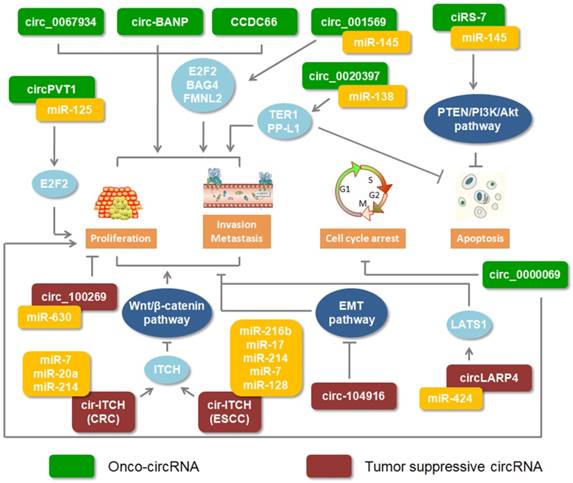
Onco-circRNA
The expression of circular BANP was reported to be upregulated in colorectal cancer tissues compared to its adjacent normal tissues. Through cellular localization analysis, circ-BANP was shown to primarily exist in the cytoplasm. Silencing circ-BANP expression inhibited colorectal cell proliferation, which implied that circ-BANP was an oncogene in colorectal cancer [58]. High expression levels of circ_0000069 in colorectal cancer performed a similar function in promoting cell proliferation, migration, and invasion [55]. In esophageal squamous cell carcinoma, increased circ_0067934 was identified compared with adjacent normal tissues. The results of knocking down circ_0067934 showed inhibiting effect on cancer cell proliferation and migration [51]. Increasing miRNAs were implicated in the regulation of circRNA. Based on RNA-Seq and functional analysis, circPVT1 was identified as an oncogene in gastric cancer. Moreover, miR-125 families could be bound by circPVT1 [59]. Another positive circRNA, circ_001569 assisted the gene expression of miR-145 targets, E2F5, BAG4 and FMNL2 [52]. High expression level of circHIPK3 was identified in colorectal cancer and promoted the cancer growth and metastasis by sponging miR-7 [60]. In addition, circRNAs were involved in signaling pathways regulated by miRNA. Recently, the PTEN/PI3K/Akt signaling pathway was frequently discussed in cancer. Inhibiting PTEN, which attenuates the activity of Akt or increasesPI3K and Akt, could block tumor cell apoptosis [61, 62]. Overexpression of ciRS-7 in gastric cancer could relieve the braking effect of miRNAs on regulating the PTEN/PI3K/Akt signaling pathway [63]. These evidences suggested that circRNAs could be as positive regulators to promote digestive cancer progression.
Anti-oncogene circRNA
Adversely, few studies illustrated that certain circRNAs, as anti-oncogene, were involved in digestive cancers. There was low expression of circ-104916 in gastric cancer tissues. Overexpressed circ-104916 hindered cell proliferation, migration and invasion via regulating key factors of the EMT process [64]. By sponging onco-miRNA, upregulated circRNA_100269 and circ-LARP4 inhibited gastric cancer cell proliferation and invasion in vitro [65, 66]. As we previously discussed, circ-ITCH implemented its function by modulating its parental genes via sponging miRNA. In colorectal cancer and esophageal squamous cell carcinoma, circ-ITCH played a suppressive role and bound to the miRNA, impeding the Wnt/β-Catenin pathway [39, 67]. CircITGA7 was found to suppress cell growth and metastasis via inhibiting the Ras signalling pathway and promoting the transcription of its host gene ITGA7 in colorectal cancer [68]. Those results implied that circRNA could be as anti-oncogene to be involved in digestive cancer progression.
Clinical application in digestive cancers
In recent years, digestive cancer has high morbidity and high mortality. Although most tumor markers have been found and applied in clinical detection, their sensitivity and veracity are still barely satisfactory. CircRNAs are novel noncoding RNAs with a number of observable and stable ectopic-circRNAs present in digestive cancer [30]. Yan et al. identified 48 dysregulated circRNAs and illustrated a tight link with four circRNAs and early recurrence in gastric cancer patients with stage III cancer after radical surgery [69]. Downregulated circ_0000190 in gastric cancer indicated a correlation with tumor diameter, lymphatic metastasis, distal metastasis, TNM stage, and CA19-9 levels [54]. In colorectal cancer, circ_001988 was reported as a new biomarker due to its significant clinical behavior [70]. These studies illustrate that circRNAs are promising biomarkers in the diagnosis and treatment digestive cancers based on their functions.
Exosomes is extracellular vesicles which secrected by many cell type. Then they release into the variety of body fluids, such as serum, saliva, urine, amniotic fluid and cerebrospinal fluid for intercellular communication [71-73]. Exosomes derived from cancer cells were found to have great clinical applications in cancer because they could be drivers containing biomacromolecules. Recently, a study reported that enriched and stable circRNAs were identified in the serum exosomes derived from colorectal cancer patient. The process of how circRNAs move from the cancer cell to the body fluid is assisted by the exosome [74]. It was a breakthrough of circRNA potentially applying clinical transformation.
Conclusion
From trash to treasure, our understanding of circRNAs is gradually increasing in recent years. To date, exon skipping, intron pairing and RBP pairing were the main reasons of RNA circularization [7, 19-22]. In addition, Ling et al. thought that there verse splicing of pre-mRNAs was the main source for circRNAs in eukaryotes [25]. Through sponging the miRNA, interacting with protein, and translating into small peptides, circRNAs showed diverse regulatory mechanisms. Digestive cancers are the most common cancers in the world. Using RNA sequencing technology provided us more circRNAs that were identified from esophageal squamous cell carcinoma, gastric cancer, and colorectal cancer. These circRNAs played double roles in the development of digestive cancer, implying potential applications in clinical diagnosis and therapy. Where do circRNAs come from? How are they regulated? What is their role in digestive cancers? These three questions are indispensable to be solved by further studies. Therefore, there are still other unknown circRNA functions and regulatory mechanisms that need to be investigated in the future.
Acknowledgements
Sun Zhenqiang received funding from The National Natural Science Foundation of China (81560385), The Medical Scientific and Technological Research Project of Henan Province (201702027), Youth Innovation Fund Project of The First Affiliated Hospital of Zhengzhou University (YNQN2017035), and The China Postdoctoral Science Foundation (2017M610462).
Authors' contributions
ZQS, CC and SHY wrote the manuscript and created the figures. QBZ, GXW, ZL, JS and ZYZ collected the related paper. ZQS, WTY and JBL provided guidance and revised this manuscript. All authors have read and approved the final version.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Sanger HL, Klotz G, Riesner D. et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proceedings of the National Academy of Sciences of the United States of America. 1976;73:3852-6
2. Hsu M, Cocaprados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339-40
3. Zhang Y, Zhang XO, Chen T. et al. Circular Intronic Long Noncoding RNAs. Molecular Cell. 2013;51:792-806
4. Chen L, Huang C, Wang XL. et al. Circular RNAs in Eukaryotic Cells. Current Genomics. 2015;16:312-18
5. Li ZY, Huang C, Bao C. et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nature Structural & Molecular Biology. 2015;22:256-64
6. Danan M, Schwartz S, Edelheit S. et al. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Research. 2012;40:3131-42
7. Chen LL, Yang L. Regulation of circRNA biogenesis. Rna Biology. 2015;12:381-8
8. Jiang G, Ma Y, An T. et al. Relationships of circular RNA with diabetes and depression. Scientific Reports. 2017;7:7285
9. Xiaoyun Lia BP, Xiaofeng Zhu, Panpan Wang, Yingquan Xiong, Hengrui Liu, Kehuan Sun, Haixia Wang, Ling Ou, Zhidi Wu, Xiaoguang Liu, Haibin He, Shu Mo, Xunqian Peng, Ya Tian, Ronghua Zhang, Li Yang. Changes in related circular RNAs following ERβ knockdown and the relationship to rBMSC osteogenesis. Biochemical and Biophysical Research Communications. 2017;493:100-7
10. Shan K, Liu C, Liu BH. et al. Circular Non-Coding RNA HIPK3 Mediates Retinal Vascular Dysfunction in Diabetes Mellitus. Circulation. 2017;136:1629-42
11. Piwecka M, Glažar P, Hernandez-Miranda LR. et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357:eaam8526
12. Huang XY, Huang ZL, Xu YH. et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep. 2017;7:5428
13. Fang J, Hong H, Xue X. et al. A novel circular RNA, circFAT1(e2), inhibits gastric cancer progression by targeting miR-548g in the cytoplasm and interacting with YBX1 in the nucleus. Cancer letters. 2019;442:222-32
14. Yang F, Fang E, Mei H. et al. Cis-acting circ-CTNNB1 promotes beta-catenin signaling and cancer progression via DDX3-mediated transactivation of YY1. Cancer research. 2018;79:557-71
15. Chen WQ, Zheng RS, Baade PD. et al. Cancer Statistics in China, 2015. CA-Cancer J Clin. 2016;66:115-32
16. Zhu JZ, Tan ZQ, Hollis-Hansen K. et al. Epidemiological Trends in Colorectal Cancer in China: An Ecological Study. Dig Dis Sci. 2017;62:235-43
17. Jemal A, Bray F, Center MM. et al. Global cancer statistics. Ca A Cancer Journal for Clinicians. 2011;61:69-90
18. Imai H, Sawada K, Sato A. et al. Complete Resection of Liver Metastases of Colorectal Cancer after High Efficacy Bevacizumab, S-1, and CPT -11 Combination Chemotherapy. Gan to Kagaku Ryoho. 2015;42:101-4
19. Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes & Development. 2014;28:2233-47
20. Kelly S, Greenman C, Cook PR. et al. Exon Skipping Is Correlated with Exon Circularization. Journal of Molecular Biology. 2015;427:2414-7
21. Conn SJ, Pillman KA, Toubia J. et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125-34
22. Ivanov A, Memczak S, Wyler E. et al. Analysis of Intron Sequences Reveals Hallmarks of Circular RNA Biogenesis in Animals. Cell Reports. 2015;10:170-7
23. Jeck WR, Sorrentino JA, Wang K. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna-a Publication of the Rna Society. 2013;19:141-57
24. Li X, Liu CX, Xue W. et al. Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Molecular Cell. 2017;67:214-27
25. Zhang Y, Xue W, Li X. et al. The Biogenesis of Nascent Circular RNAs. Cell Reports. 2016;15:611-24
26. Chao CW, Al E. The mouse formin (Fmn) gene: abundant circular RNA transcripts and gene-targeted deletion analysis. Molecular Medicine. 1998;4:614-28
27. Hansen TB, Jensen TI, Clausen BH. et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-8
28. Hansen TB, Wiklund ED, Bramsen JB. et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. Embo Journal. 2011;30:4414-22
29. Cesana M, Cacchiarelli D, Legnini I. et al. A Long Noncoding RNA Controls Muscle Differentiation by Functioning as a Competing Endogenous RNA. Cell. 2011;147:358-69
30. Memczak S, Jens M, Elefsinioti A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-8
31. Wang K, Long B, Liu F. et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. European Heart Journal. 2016;37:2602-11
32. Zhang H, Wang GC, Ding C. et al. Increased circular RNA UBAP2 acts as a sponge of miR-143 to promote osteosarcoma progression. Oncotarget. 2017;8:61687-97
33. Oltersdorf T, Elmore SW, Shoemaker AR. et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677-81
34. Tang YY ZP, Zou TN, Duan JJ, Zhi R, Yang SY, Yang DC, Wang XL. Circular RNA hsa_circ_0001982 Promotes Breast Cancer Cell Carcinogenesis Through Decreasing miR-143. DNA and cell biology. 2017;36:901-8
35. Zhu KG, Liu L, Zhang JL. et al. MiR-29b suppresses the proliferation and migration of osteosarcoma cells by targeting CDK6. Protein & Cell. 2016;7:434-44
36. Cole AM, Myant K, Reed KR. et al. Cyclin D2-Cyclin-Dependent Kinase 4/6 Is Required for Efficient Proliferation and Tumorigenesis following Apc Loss. Cancer Research. 2010;70:8149-58
37. Asghar U, Witkiewicz AK, Turner NC. et al. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nature Reviews Drug Discovery. 2015;14:130-46
38. Ashwal-Fluss R, Meyer M, Pamudurti NR. et al. circRNA Biogenesis Competes with Pre-mRNA Splicing. Molecular Cell. 2014;56:55-66
39. Li F, Zhang LY, Li W. et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6:6001-13
40. Du WW, Yang W, Liu E. et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Research. 2016;44:2846-58
41. Pamudurti NR, Bartok O, Jens M. et al. Translation of CircRNAs. Molecular Cell. 2017;66:9-21
42. Yang Y, Gao X, Zhang M. et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. JNCI: Journal of the National Cancer Institute. 2018;110:435
43. Peng NC, Shi LX, Zhang Q. et al. Microarray profiling of circular RNAs in human papillary thyroid carcinoma. Plos One. 2017;12:e0170287
44. Zhang PL, Zuo ZG, Shang WJ. et al. Identification of differentially expressed circular RNAs in human colorectal cancer. Tumor Biol. 2017;39:1010428317694546
45. Dang Y, Ouyang XJ, Zhang F. et al. Circular RNAs expression profiles in human gastric cancer. Scientific Reports. 2017;7:9060
46. Su HF, Lin FQ, Deng X. et al. Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. J Transl Med. 2016;14:225
47. Xie Y, Shao Y, Sun W. et al. Downregulated expression of hsa_circ_0074362 in gastric cancer and its potential diagnostic values. Biomarkers in Medicine. 2017;12:11-20
48. Sun H, Tang W, Rong D. et al. Hsa_circ_0000520, a potential new circular RNA biomarker, is involved in gastric carcinoma. Cancer Biomarkers. 2017:1-8
49. Lai Z, Yang Y, Yan Y. et al. Analysis of co-expression networks for circular RNAs and mRNAs reveals that circular RNAs hsa_circ_0047905, hsa_circ_0138960 and has-circRNA7690-15 are candidate oncogenes in gastric cancer. Cell Cycle. 2017:1-11
50. Huang M, He YR, Liang LC. et al. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World Journal of Gastroenterology. 2017;23:6330-8
51. Xia W, Qiu M, Chen R. et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Scientific Reports. 2016;6:35576
52. Xie H, Ren X, Xin S. et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7:26680-91
53. Tian M, Chen R, Li T. et al. Reduced expression of circRNA hsa_circ_0003159 in gastric cancer and its clinical significance. Journal of Clinical Laboratory Analysis. 2017;32:e22281
54. Chen S, Li T, Zhao Q. et al. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clinica chimica acta. 2017;466:167-71
55. Guo J, Jin L, Zhu C. et al. Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. Oncotargets & Therapy. 2016;9:7451-8
56. Xu XW, Zheng BA, Hu ZM. et al. Circular RNA hsa_circ_000984 promotes colon cancer growth and metastasis by sponging miR-106b. Oncotarget. 2017;8:91674-83
57. Yuan Y, Liu W, Zhang Y. et al. CircRNA circ_0026344 as a prognostic biomarker suppresses colorectal cancer progression via microRNA-21 and microRNA-31. Biochemical and Biophysical Research Communications. 2018;503:870-5
58. Zhu MC, Xu YJ, Chen Y. et al. Circular BANP, an upregulated circular RNA that modulates cell proliferation in colorectal cancer. Biomedicine & Pharmacotherapy. 2017;88:138-44
59. Chen J, Li Y, Zheng QP. et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Letters. 2017;388:208-19
60. Zeng K, Chen X, Xu M. et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death & Disease. 2018:9
61. Sansal I, Sellers WR. The biology and clinical relevance of the PTEN tumor suppressor pathway. J Clin Oncol. 2004;22:2954-63
62. Osaki M, Oshimura M, Ito H. PI3K-Akt pathway: Its functions and alterations in human cancer. Apoptosis. 2004;9:667-76
63. Pan H, Li T, Jiang Y. et al. Overexpression of circular RNA ciRS-7 abrogates the tumor suppressive effect of miR-7 on Gastric cancer via PTEN/PI3K/AKT signaling pathway. Journal of Cellular Biochemistry. 2017;119:440-46
64. Jin L, Li Z, Yan Z. et al. Circ-104916 is downregulated in gastric cancer and suppresses migration and invasion of gastric cancer cells. Oncotargets & Therapy. 2017;10:3521-29
65. Zhang Y, Liu H, Li WD. et al. CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630. Aging-Us. 2017;9:1585-94
66. Zhang J, Liu H, Hou LD. et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Molecular Cancer. 2017;16:151
67. Huang G, Hua Z, Shi Y. et al. cir-ITCH Plays an Inhibitory Role in Colorectal Cancer by Regulating the Wnt/β-Catenin Pathway. Plos One. 2015;10:e0131225
68. Li X, Wang J, Zhang C. et al. Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating transcription of its host gene ITGA7. Journal Of Pathology. 2018;246:166-79
69. Zhang Y, Li J, Yu J. et al. Circular RNAs signature predicts the early recurrence of stage III gastric cancer after radical surgery. Oncotarget. 2017;8:22936-43
70. Wang XN, Zhang Y, Huang L. et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. International Journal of Clinical and Experimental Pathology. 2015;8:16020-5
71. Hu C, Chen M, Jiang R. et al. Exosome-related tumor microenvironment. Journal Of Cancer. 2018;9:3084-92
72. Keller S, Ridinger J, Rupp A-K. et al. Body fluid derived exosomes as a novel template for clinical diagnostics. J Transl Med. 2011:9
73. Simpson RJ, Lim JWE, Moritz RL. et al. Exosomes: proteomic insights and diagnostic potential. Expert Review Of Proteomics. 2009;6:267-83
74. Li Y, Zheng QP, Bao CY. et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Research. 2015;25:981-4
Author contact
![]() Corresponding authors: Email: zqsun82edu.cn (Zhenqiang Sun), E-mail: yuanweitangedu.cn (Weitang Yuan) and Email: 1999liujbcom (Jinbo Liu). Tel. +86-037167967141.
Corresponding authors: Email: zqsun82edu.cn (Zhenqiang Sun), E-mail: yuanweitangedu.cn (Weitang Yuan) and Email: 1999liujbcom (Jinbo Liu). Tel. +86-037167967141.

 Global reach, higher impact
Global reach, higher impact