Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(16):3582-3592. doi:10.7150/jca.30342 This issue Cite
Research Paper
EMP2 acts as a suppressor of melanoma and is negatively regulated by mTOR-mediated autophagy
1. State Key Laboratory of Oral Diseases, National Clinical Research Center for Oral Disease, Department of Orthodontics and Paediatrics, West China Hospital of Stomatology, Sichuan University, Chengdu, 610041, PR China
2. Department of Radiation Oncology, Sichuan Cancer Hospital & Institute, Sichuan Cancer Center, School of Medicine, University of Electronic Science and Technology of China, Radiation Oncology Key Laboratory of Sichuan Province, Chengdu 610041, PR China
3. Department of Dermatology and State Key Laboratory of Biotherapy, West China Hospital, West China Medical School, Sichuan University, Chengdu 610041, PR China
* These authors contributed equally.
Received 2018-10-1; Accepted 2019-5-7; Published 2019-6-9
Abstract
Cutaneous melanoma is one of the most common malignant skin tumors and advanced melanoma is usually associated with a poor prognosis. In the current study, we demonstrated the tumor suppressing role of epithelial membrane protein-2 (EMP2) by inducing apoptosis in a A375 human melanoma cell line. Mechanistically, the low expression of EMP2 in melanoma is partially due to autophagic protein degradation mediated by the mTOR pathway. These results suggest there is regulation of autophagy as well as EMP2 levels might be an interesting novel targeted therapeutic strategy for melanoma. Although the further investigation is needed to deeply understand the regulatory mechanisms of EMP2 in melanoma progression and metastasis, our results clarify the functions and mechanisms of autophagy in melanoma, and shed new light on novel targeted therapeutics for melanoma.
Keywords: EMP2, mTOR, Apoptosis, Autophagy, Melanoma.
Introduction
Melanoma is one of the most common types of skin malignant neoplasms. Many targeted therapeutic strategies for melanoma are still under evaluation in clinical trials.[1] Although various surgical and immunotherapeutic approaches have been developed to improve the prognosis of cutaneous melanoma patients, such as checkpoint inhibitors, chimeric antigen receptor T-cell (CAR-T) immunotherapy, oncolytic viruses and dendritic cell (DC)/neoantigen vaccines, their clinical usage is seriously limited by their high cost and complicated individualized manufacture.[2-7] Hence, the discovery of novel therapeutic targets for cutaneous melanoma are urgently needed.
Epithelial membrane protein 2 (EMP2) is a tetraspan protein belonging to the PMP22 (peripheral myelin protein 22) family.[8] EMP2 is typically located in the cytoplasm or cellular membrane, and plays diverse roles in tumor carcinogenesis, proliferation and metastasis. It is notable that the expression level of EMP2 is remarkably dynamic among different cancer types but is always associated with an altered structure and function of extracellular membrane (ECM).[9-14] Within EMP2, the amino acid domains between each transmembrane alpha-helix domain are generally located in extracellular zones and interact with the ECM as well as other tumor membrane proteins.[15-17] To the best of our knowledge, the relationship between aberrant expression of EMP2 and melanoma have not been previously reported. There are only a few studies that suggest that an EMP homologue PMP22 is related to the prognosis of cutaneous melanoma. EMP2 also regulates the cell cycle in addition to programmed cell death in various tumors.[18, 19] For example, its function includes G2/M cell cycle arrest and apoptosis, suppression of anchorage-independent cell growth, and response to estrogenic hormone signals, etc.
Autophagy is an evolutionarily conserved and universal process for the degradation of misfolded proteins, protein aggregations and damaged cytoplasmic organelles. This process facilitates devouring of these adherent molecule in an autophagy vacuole which then fuse with lysosomes.[20-22] There are three main autophagy subtypes that have been described: macrophagy, microphagy and chaperone-mediated autophagy.[23] There are many reports that autophagy has a protective role in tumor cells in competitive environments such as starvation, cytotoxic agents and hypoxia, among others.[24-30] After the formation of the autophagosome, its contents are irreversibly degraded by merging with a lysosome to form an autolysosome.
In mammalian cells, there is another protein degradation pathway besides autophagy, called the ubiquitin-proteasome pathway. The ubiquitin-proteasome pathway is also involved in the inflammatory response, carcinogenesis and stress signals. Although there are increasing reports about protein homeostasis in carcinogenesis and tumor progression regulated by autophagy and/or ubiquitin-proteasome pathways,[31-33] the interactions, downstream signaling pathways of these two systems in solid tumors are still unclear.
In the current study, we found that EMP2 is downregulated in cutaneous melanoma tissues at both the mRNA and protein levels, as well as in melanoma cell lines. Further, studies have indicated that the knockdown of EMP2 does not significantly interfere A375 melanoma cell proliferation. However, the overexpression of EMP2 remarkably inhibits proliferation of A375 cells. Results of gene set enrichment analysis (GSEA) suggest that mTOR pathway is associated with differentially expressed genes between melanoma and normal skin tissues related to EMP2 gene. It is interesting that the phosphorylation level of mTOR, rather than total mTOR level, is significantly changed in melanoma. We further identified mTOR-mediated autophagy as a trigger for the down-regulation of EMP2, but changes in EMP2 levels did not change autophagy levels in A375 melanoma cells. In addition, the overexpression of EMP2 suppressed melanoma cell proliferation via induction of mitochondrial autophagy. In summary, we identified EMP2 as a novel melanoma suppressor, which is regulated by mTOR-mediated autophagy. These results amended the function and mechanisms of autophagy in melanoma, and shed new light on novel targeted therapeutics of melanoma.
Materials and Methods
Cell Lines and reagents
HaCaT, HSF-1 and four melanoma cell lines were originally obtained from the ATCC (American type culture collection) and were cultured as previously described. Cells were maintained in Dulbecco's Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS, Gibco), 100 U/mL penicillin, and 100 µg/mL streptomycin at 37°C in a humidified atmosphere of 5% CO2. The reagents applied in the experiments including DAPI stain, 3- (4,5- dimethyl- 2- thiazolyl)- 2, 5- diphenyl- 2- H- tetrazolium bromide (MTT), dimethyl sulfoxide (DMSO), rapamysin, MHY1485 and Bafilomycin A1 and other antibodies, were almost obtained from Sigma (St. Louis, MO, USA), except where noted. TRIzol and Lipofectamine 2000 was purchased from Invitrogen. Two individual EMP2-specific small interfering RNAs (siRNAs), universal scrambled negative control siRNA and EMP2-overexpression vector were constructed and purchased from Ruibo (Guangzhou, China).
Cell proliferation assay
Cell proliferation assays were estimated by the MTT. Growing cells were seeded in 96-well plates at a cell density of 10,000 cells /well. Cells were transfected with two individual EMP2-specific siRNAs and negative control siRNA by using Lipofectamine 2000 according to manufacturer's recommendation, and incubated for 20, 40, 60 and 80 h at 37°C and 5% CO2 incubator, respectively. MTT (5 mg/ml) was added to each well and continued the incubation at 37°C for 3 h, the purple formazan crystal was dissolved in 100μL dimethyl sulfoxide (DMSO), and the optical density (OD) was measured at a wavelength 570 nm using a spectrophotometer.
GFP-LC3 transfection and Immunofluorescence staining
Approximately 1×105 A375 cells were plated on a coverslip, incubated overnight, and then transfected with GFP-LC3 at a concentration of 1,000 virus particles/cell in DMEM. After incubation for 24 h, cells were treated with rapamycin or EMP2-overexpression vector. For visualization of EMP2 and nucleus, coverslips were mounted with antibody against EMP2 with RED or DAPI before being analyzed by Confocal Microscope.
Western blotting analysis
Western blot was performed according to the standard method as previously described. Total cell extracts were prepared by disrupting cells in RIPA buffer with PMSF. Equal amounts of protein were resolved by SDS-PAGE and transferred onto PVDF membranes. After blocking with 5% skimmed milk, membranes were hybridized with the specific primary antibodies at 4°C overnight, and visualized after incubating secondary antibodies. The antibodies used in this study included those against EMP2 (1:1000, ABCAM), p-mTOR, mTOR, LC3, SQSTM1, FADD, Fas, FasL, Mcl-1, Bax (1:1000; Cell Signaling Technology), Cyto.C, PARP, Bcl-2 (1:1000; Cell Signaling Technology), Caspase3 (1:1000; Cell Signaling Technology), Caspase8, Caspase9. GADPH was detected as a loading control. Secondary antibodies were horseradish peroxidase conjugated goat anti-mouse (1:5000; Abcam, Cambridge, UK) and goat anti-rabbit (1:5000; Abcam).
Apoptosis analysis
A375 cells were seeded in 6-well plates, and transfected with negative control siRNA, EMP2-specific siRNA, negative control vector and EMP2-overexpression vector respectively, then harvested with trypsin for further analysis. To analyze apoptosis, the cells were washed in PBS and stained with FITC annexin V apoptosis detection kit in conjunction with PI, and assessed by flow cytometer to determine the apoptosis of A375 cells with proper machine settings. For immunofluorescence assay, A375 cells were stained with DAPI and the images were captured by a fluorescent microscope.
RT-PCR analysis
Total RNA was extracted from cultured cells using TRIzol reagent following the manufacturer's instructions, and then quantified with the Nanodrop 1000, followed by cDNA synthesis and RT-PCR. RT-PCR were performed using ExTaq polymerase (Takara) and gene expression was analyzed by CFX-manager software (CFX96 Real-Time System; Bio-Rad). Expression data were normalized against the housekeeping gene GAPDH, which is stable across all samples, to control the variability in expression levels. EMP2: forward: 5'- ATTCTCTGCTGCATCGCCTT-3'; reverse: 5'- CGTAGCTGCCTTCTCTGGTC-3'. GADPH forward: 5'-ACCACAGTCCATGCCATCAC-3'; reverse: 5'- TCCACCACCCTGTTGCTGTA -3'.
Immunohistochemical of tumor tissue sections
Melanoma and adjacent normal dermatic tissue microarrays were purchased from Shanghai Outdo Biotechnology (Shanghai, China). Immunohistochemical (IHC) analyses were performed according to standard method and microarray instruction. The sections were incubated overnight at 4°C with primary antibodies against EMP2, mTOR or p-mTOR(S2448). To analyze the immunohistochemistry, the staining of EMP2, mTOR or p-mTOR(S2448) were scored relative to staining intensity. The antigen repair of TMA sections was incubated in 0.1 M citrate at pH 6.0 at 100°C for 30 min. The EMP2, mTOR or p-mTOR(S2448) was detected using the corresponding primary antibody at 1:150-200 dilution followed by visualization using DAB according to the manufacturer's instructions. All sections were evaluated by two experience investigators.
Results
EMP2 is down-regulated in melanoma tissues
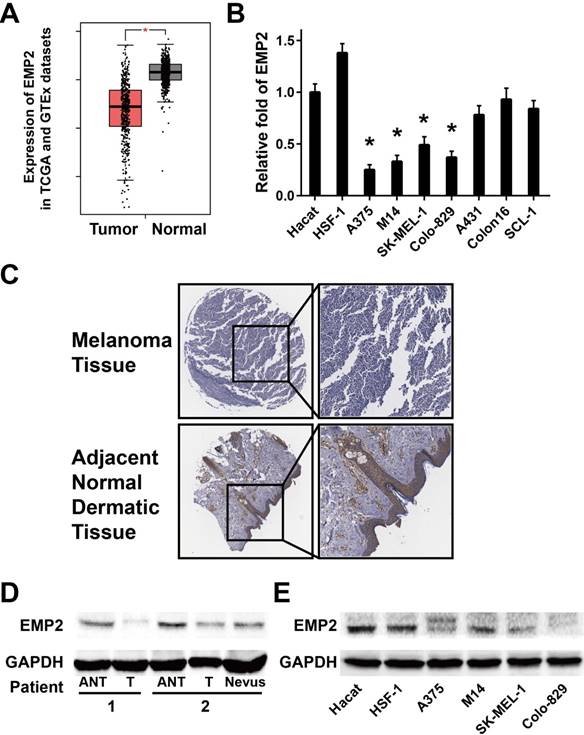
Analysis of EMP2 expression in published genetic profiles from melanoma patients in the TCGA and GTEx datasets showed that EMP2 levels were down-regulated in melanoma samples compared with normal tissue samples (P < 0.001, Figure 1A and S1).[34, 35] Real-time PCR analysis verified that EMP2 mRNA expression was indeed down-regulated (by at least 2-fold) in cultured melanoma cells, and none significantly changes in human squamous cell carcinoma cells in comparison with human keratinocytes (Hacat) and human skin fibroblasts (HSF-1) (Figure 1B). Furthermore, EMP2 protein expression was significantly down-regulated in all four melanoma cell lines when compared with normal lines via western blotting (Figure 1E). When EMP2 expression was evaluated by IHC staining, the positive intensity was obviously higher in adjacent normal skin tissue (ANT) compared with the melanoma tissue (Figure 1C). Western blotting analysis also revealed that EMP2 protein was markedly suppressed in melanoma tissues compared with ANT and nervous tissue (Figure 1D). Taken together, these results strongly indicated that EMP2 was down-regulated in melanoma.
EMP2 modulates proliferation of melanoma cells
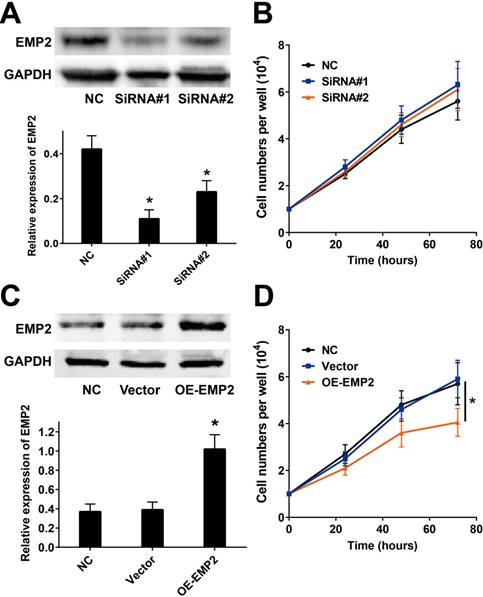
We measured the impacts of EMP2 in cutaneous melanoma by employing the commonly used A375 melanoma cell line. We then employed two EMP2-specific siRNAs to suppress EMP2 mRNA (Figure 2A). Cell number counting assays demonstrated that there were no significant changes in cellular proliferation after RNAi (Figure 2B). On the other hand, overexpression of EMP2 significantly decreased cell populations compared with vector-transfected cells (Figure 2C and D).
EMP2 expression was down-regulated in melanoma tissues compared with normal human skin tissue. A. Expression of EMP2 was obviously down-regulated in melanoma tissues (Tumor) compared with normal human skin tissue samples (Normal) in the TCGA and GTEx datasets; B. Real-time PCR analysis of EMP2 expression in Hacat, HSF-1 and four melanoma cell lines; C. IHC assays showed lower EMP2 expression in melanoma tissues correlates with adjacent normal dermatic tissues; D. EMP2 protein expression level in two paired melanoma tissues (T) and their adjacent normal tissues (ANT) or nevus. E. Western blotting of EMP2 expression in Hacat- ,HSF-1 and four melanoma cell lines.
EMP2 overexpression and RNAi effects in A375 cell lines. A & B. EMP2 mRNA was suppressed by specific siRNA. As shown, the protein level of EMP2 in A375 cells was obviously down-regulated, but the cellular proliferation did not show distinctly change before and after RNA interference; C & D. EMP2 overexpression was performed by lentivirus transfection and significantly inhibited A375 proliferation
EMP2 modulates proliferation of melanoma cells
We measured the impacts of EMP2 in cutaneous melanoma by employing the commonly used A375 melanoma cell line. We then employed two EMP2-specific siRNAs to suppress EMP2 mRNA (Figure 2A). Cell number counting assays demonstrated that there were no significant changes in cellular proliferation after RNAi (Figure 2B). On the other hand, overexpression of EMP2 significantly decreased cell populations compared with vector-transfected cells (Figure 2C and D).
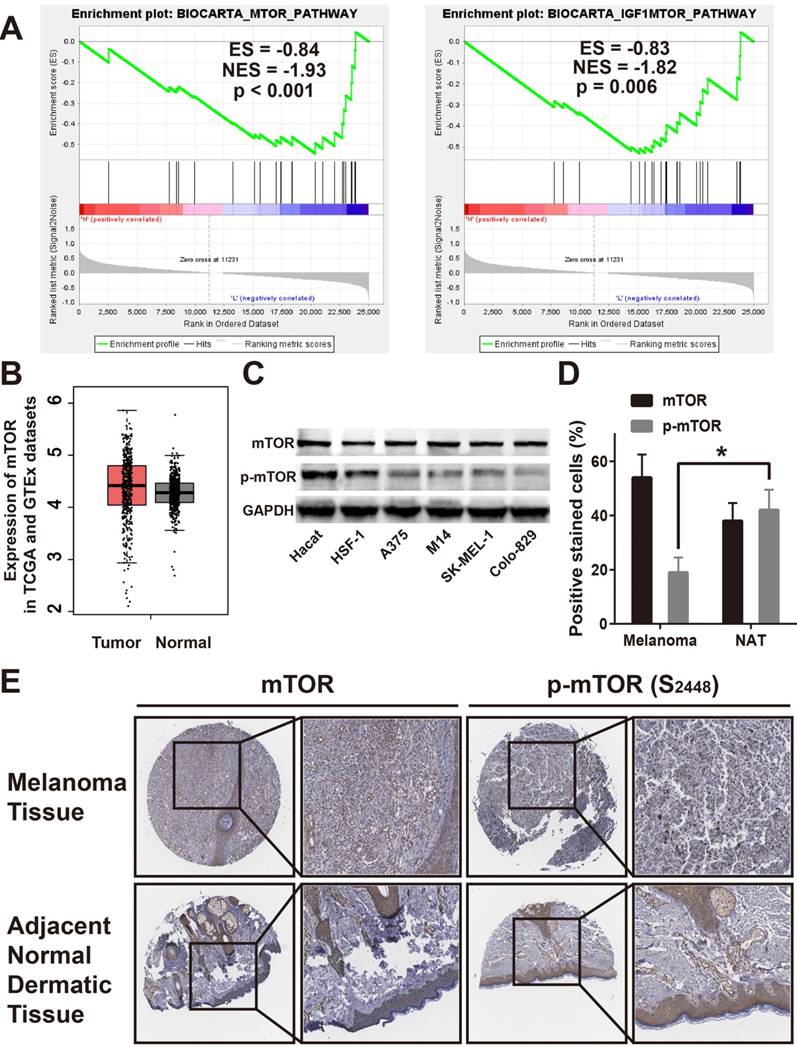
EMP2 expression positively correlated by mTOR activation. A. GSEA suggested that EMP2 expression positively correlated with mTOR and IGF1-mTOR pathway; B. Expression of mTOR was no significant differences in melanoma tissues (Tumor) compared with normal human dermaic tissue samples (Normal) in the TCGA and GTEx datasets; C. Expression of mTOR and p-mTOR were dected in Hacat, HSF-1, A375, M14, SK-MEL-1 and Colo-829 cells by Western Blotting, and GADPH was used as a loading control; D. IHC assays showed lower p-mTOR expression in melanoma tissues correlates with adjacent normal skin tissues and no mTOR expression difference within two samples; E. Statistic analysis for the number of the positive stain cells in IHC.
EMP2 expression positively correlated with mTOR activation
In an attempt to determine which pathway may be involved in EMP2-mediated melanoma progression, gene set enrichment analysis (GSEA), and the Cancer Genome Atlas (TCGA) profiles were performed.[36] As shown in Figure 3A, we found that EMP2 levels were positively correlated with the mTOR-activated gene signature, suggesting that the mTOR pathway and IGF1/mTOR pathway may be involved in the function of EMP2 (Figure 3A). By determining mTOR expression in TCGA and GTEx datasets, we found that mTOR levels had no differences between tumor and normal samples (Figure 3B). We tested mTOR protein expression in all four melanoma cell lines compared with normal lines by western blotting, which verified the results of this analysis. In addition, the phosphorylation of mTOR, p-mTOR was clearly down-regulated in melanoma cell lines, suggesting a change in mTOR activity (Figure 3C). As shown in Figure 3D, mTOR were similarly stained in melanoma and ANT, while p-mTOR were weakly stained in melanoma tissue. These results were the same when counting positive stain cells as well (Figure 3E).
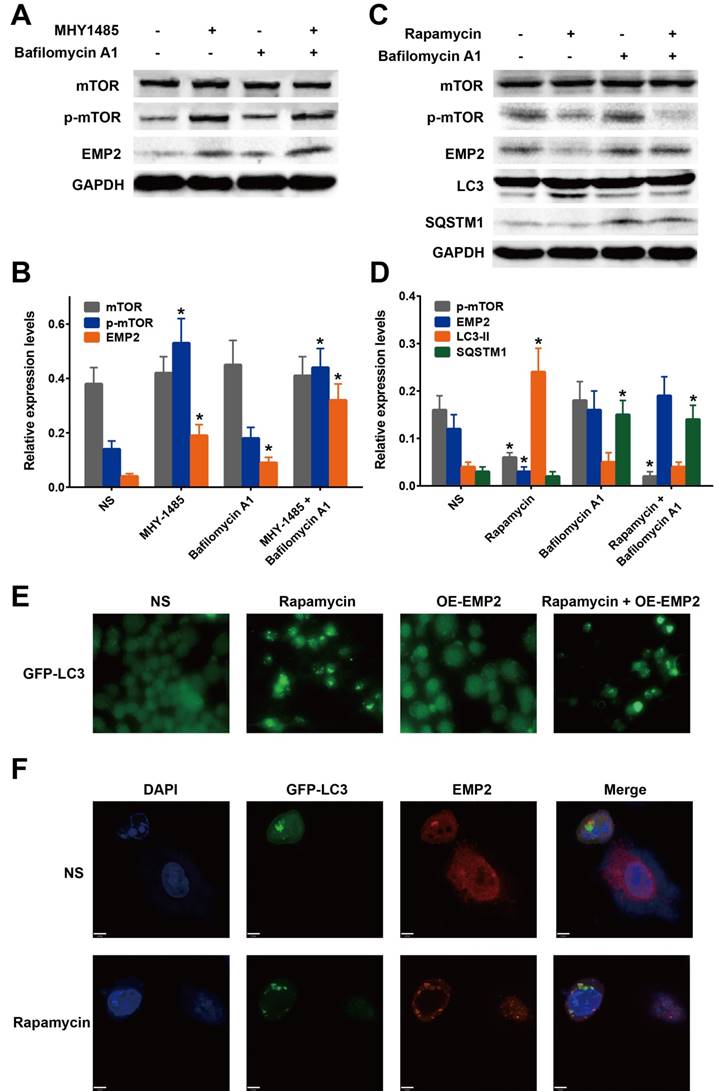
mTOR activation and inhibition regulate EMP2 levels via autophagic protein degradation. A. A375 cells were treated with MHY1485 (mTOR activator) and Bafilomycin A1(autophagy inhibitor). Western blot analysis was performed to detect the change of mTOR, p-mTOR and EMP2; B. Statistical analysis of Figure 4A; C. A375 cells were treated with Rapamycin (mTOR inhibitor) and Bafilomycin A1(autophagy inhibitor). Western blot analysis was performed to detect the change of mTOR, p-mTOR, EMP2, LC-3 and SQSTM1; D. Statistical analysis of Figure 4C; E. GFP-LC3 marked autophagosomes in A375 treated with Rapamycin, EMP2-specific overexpression vector(OE-EMP2) or OE-EMP2+Rapamycin. GFP fluorescence was analyzed using confocal microscopy; F. To confirm the cellular localization of autophagosomes and EMP2, A375 cells were counterstained with DAPI to identify nucleus, and EMP2 was stained as red. The fluorescence were analyzed using confocal microscopy. A375 cells were treated with Rapamycin as experimental group.
EMP2 expression negatively correlated with autophagy activation
mTOR was shown to play a major role in the autophagy pathway by inhibiting autophagy through phosphorylation. Rapamycin is an inhibitor of the mTOR pathway, whereas MHY1485 is an activator. To confirm the effect of p-mTOR on EMP2, we confirmed the level of EMP2 protein after A375 cells were treated with MHY1485 or rapamycin. As a result, EMP2 expression was demonstrated to be positively correlated with the expression of p-mTOR (Figure 4A-D). Taken together, we were able to determine that the autophagic process decreased EMP2 expression.
Bafilomycin A1 is known as an inhibitor of regulating the latter stages of autophagy. Specifically this molecule has been shown to inhibit fusion between autophagosomes and lysosomes, which leads to the inhibition of LC3 cleavage and up-regulation of SQSTM1 (also called p62). Western blot analysis was employed to assess whether BafA1 influenced the expression of EMP2. Our results showed that, exposure of A375 cells to bafilomycin A1 increased the level of EMP2 (Figure 4A-D). In addition, the expression of EMP2 was much higher with MHY1485 co-treatment, indicating that the suppression of autophagic process was indeed upregulated EMP2 in A375 cells. On the other hand, compared with A375 cells treated with rapamycin only, bafilomycin A1 co-treatment group showed a higher level of EMP2.
Taken together, we concluded the degradation of EMP2 in A375 is regulated by the autophagic process, especially the autolysosome. To test our hypothesis, we evaluated the colocalization of the autolysosome and EMP2 to affirm the level of this process. A375 cells were transfected with GFP-LC3 and stained with DAPI. Fluorescence-labeled EMP2 antibody was used to visualize the nucleus, autolysosome and EMP2 respectively. LC3 and EMP2 signaling overlapped in cells under rapamycin treatment (Figure 4F), illustrating that autophagy-activation indeed included the degradation of EMP2 protein.
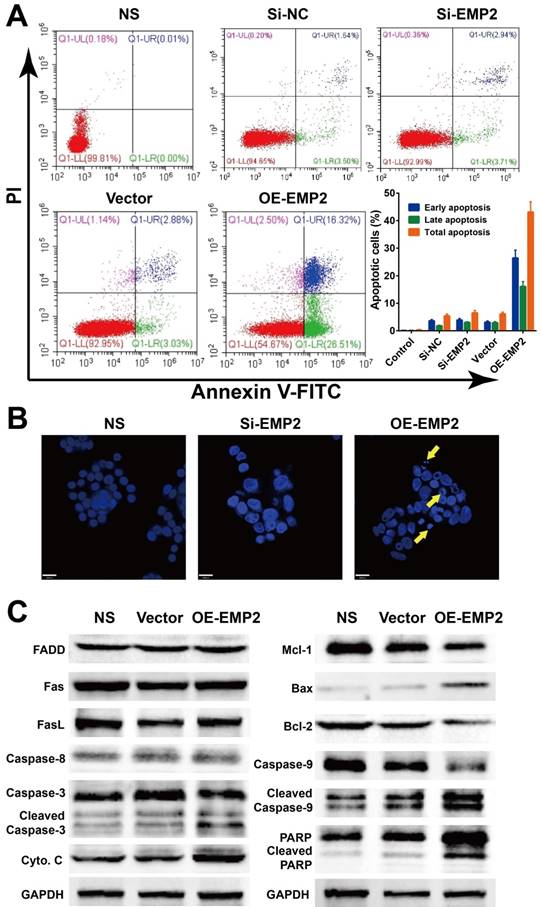
EMP2 RNAi and overexpression effect cell apoptosis in A375 cells. A375 cells were transfected with negative control siRNA, EMP2-specific siRNA, negative control vector or EMP2-specific overexpression vector. A. Flow cytometry assays were performed to examine cell apoptosis by annexin V-FITC/propidium iodide (PI) double staining. B. A375 cells were stained with DAPI (blue). C. Total protein was isolated and western blot performed.
EMP2 did not directly regulated autophagy
EMP2 expression was examined as a regulated autophagic process. We transfected A375 cells with GFP-LC3 to assess this process. In comparison with the control group, the cells treated with OE-EMP2 did not show any differences. Furthermore, in contrast to cells treated by rapamycin, the activation of green fluorescence was not changed in OE-EMP2 co-treated cells. We thus inferred that EMP2 did not participate in the regulation of autophagy.
Overexpressed EMP2 induced apoptosis in A375 cells
We further tested whether the level of EMP2 was correlated with apoptosis in A375 cells. The Annexin V/PI apoptosis flow cytometry assay revealed an higher apoptosis ratio in A375 cells with OE-EMP2 than with si-EMP2 (Figure 5A). Additionally, DAPI staining of nuclei with deep blue fluorescence signal was considered to be evidence of damaged DNA as a result of apoptosis in OE-EMP2 treated cells; thus confirming the induction of apoptosis via the up-regulation of EMP2 (Figure 2B). Western blotting analysis (Figure 5C) demonstrated that the levels of cytochrome C and BAX were increased, while the level of Bcl-2 was decreased in OE-EMP2 treated cells. Cleavage of caspase 3, 9 and PARP were also observed in this group. This finding implied the intrinsic cell death pathway was activated and resulted in apoptosis.
Discussion
Our results revealed a potential targeted therapeutic strategy for cutaneous melanoma: suppression of the autophagic protein degradation of tumor suppressor EMP2 could result in mitochondrial apoptosis in A375 melanoma cells. Our main findings were numerous. For example, we found EMP2 to be significantly downregulated in melanoma cells and tissues while autophagy was activated in melanoma cells. Additionally, EMP2 protein degradation was dependent on autophagy and autophagic degradation of EMP2 was mainly mediated by mTOR pathway. In summary, our results provided novel insights into the therapeutic potential of EMP2 in melanoma, which is regulated by mTOR-mediate autophagy.
EMP2 is a cytoplasmic and cellular surface protein that has been recognized as an object of broad interest due to its potential as a tumor biomarker and therapeutic target. In this study, we first reported on the downregulated expression of EMP2 in melanoma tissues and cell lines. The overexpression of EMP2 significantly induced inhibition of proliferation and mitochondrial apoptosis in A375 melanoma cells. The expression of EMP2 protein was correlated with autophagy activation levels. In addition, the autophagic degradation of EMP2 was mainly mediated by the mTOR pathway. Our results not only agreed with previous reports that EMP2 is aberrantly expressed in tumor tissues, but also provides the first evidence that EMP2 suppresses melanoma cell proliferation.[37] The mechanisms detailed here include suppression via inducing mitochondrial apoptosis and the EMP2 defects in melanoma which is mediated by mTOR-depended autophagic protein degradation. Tumor suppressor capacity of EMP2 in melanoma may be utilized as a novel therapeutic strategy to further identify and develop novel targeted therapies. In addition, the functions and potential mechanism of EMP2 are varied in different type of cancers. Recently, Chen et al. reported that EMP2 is highly expressed in circulating breast cancer cells.[38] And the elevated EMP2 expression is associated with poor prognosis of glioma patients.[39] EMP2 also promotes ALDH expression and activity in uterine endometrial cancer stem cells.[15]
Autophagy is a functionally and evolutionarily preserved biological process that can assist cells in responding to external stimuli. Here, the mTOR-complex is a master sensor regulating autophagy negatively via diverse signaling pathways. Although there are limited reports about the direct relationship between EMP2 and autophagy, recent studies have already shown that autophagy mediates tumor suppressor proteins degradation in various types of cancers. In our current study, we observed RNA interference or overexpression of EMP2 as having no evident influence on basal autophagy in melanoma cells. On the other hand, the mTOR-dependent autophagy activation executed EMP2 protein degradation. Considering that autophagy plays a versatile role in determining the cellular fate, further research about the protective effects and mechanisms of increased autophagy levels, and the antitumor effects of EMP2 overexpression in melanoma cells needed among animal models which will be the next step in our research.[40-43]
In conclusion, our results identified EMP2 as a novel melanoma suppressor, which is regulated by mTOR-mediate autophagy. These results provide novel insights into the functions and mechanisms of autophagy in melanoma, as well as providing evidence for novel targeted therapeutics of melanoma.
Supplementary Material
Supplementary figure.
Acknowledgements
We are grateful for financial support from the National Natural Science Foundation of China (81872535 and 81571009), the Sichuan Youth Science and Technology Foundation (2016JQ0060) and Sichuan Applied Basic Research Project (2017JY0299).
Competing Interests
All authors have given approval to the final version of the manuscript. All authors have declared that there were not conflicts of interests.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA: a cancer journal for clinicians. 2018;68:7-30
2. Cavalcante L, Chowdhary A, Sosman JA, Chandra S. Combining Tumor Vaccination and Oncolytic Viral Approaches with Checkpoint Inhibitors: Rationale, Pre-Clinical Experience, and Current Clinical Trials in Malignant Melanoma. American journal of clinical dermatology. 2018;19:657-70
3. Chen Y, Yuan F, Jiang X, Lv Q, Luo N, Gong C. et al. Discovery of a self-assembling and self-adjuvant lipopeptide as a saccharide-free peptide vaccine targeting EGFRvIII positive cutaneous melanoma. Biomaterials science. 2018;6:1120-8
4. Fotaki G, Jin C, Kerzeli IK, Ramachandran M, Martikainen MM, Karlsson-Parra A. et al. Cancer vaccine based on a combination of an infection-enhanced adenoviral vector and pro-inflammatory allogeneic DCs leads to sustained antigen-specific immune responses in three melanoma models. Oncoimmunology. 2018;7:e1397250
5. Sainz V, Moura LIF, Peres C, Matos AI, Viana AS, Wagner AM. et al. alpha-Galactosylceramide and peptide-based nano-vaccine synergistically induced a strong tumor suppressive effect in melanoma. Acta biomaterialia. 2018;76:193-207
6. Simon B, Harrer DC, Schuler-Thurner B, Schaft N, Schuler G, Dorrie J. et al. The siRNA-mediated downregulation of PD-1 alone or simultaneously with CTLA-4 shows enhanced in vitro CAR-T-cell functionality for further clinical development towards the potential use in immunotherapy of melanoma. Experimental dermatology. 2018;27:769-78
7. Wang Y, Zhang L, Xu Z, Miao L, Huang L. mRNA Vaccine with Antigen-Specific Checkpoint Blockade Induces an Enhanced Immune Response against Established Melanoma. Molecular therapy: the journal of the American Society of Gene Therapy. 2018;26:420-34
8. Liehr T, Kuhlenbaumer G, Wulf P, Taylor V, Suter U, Van Broeckhoven C. et al. Regional localization of the human epithelial membrane protein genes 1, 2, and 3 (EMP1, EMP2, EMP3) to 12p12.3, 16p13.2, and 19q13.3. Genomics. 1999;58:106-8
9. Forbes A, Wadehra M, Mareninov S, Morales S, Shimazaki K, Gordon LK. et al. The tetraspan protein EMP2 regulates expression of caveolin-1. The Journal of biological chemistry. 2007;282:26542-51
10. Fu M, Maresh EL, Helguera GF, Kiyohara M, Qin Y, Ashki N. et al. Rationale and preclinical efficacy of a novel anti-EMP2 antibody for the treatment of invasive breast cancer. Molecular cancer therapeutics. 2014;13:902-15
11. Gee HY, Ashraf S, Wan X, Vega-Warner V, Esteve-Rudd J, Lovric S. et al. Mutations in EMP2 cause childhood-onset nephrotic syndrome. American journal of human genetics. 2014;94:884-90
12. Gordon LK, Kiyohara M, Fu M, Braun J, Dhawan P, Chan A. et al. EMP2 regulates angiogenesis in endometrial cancer cells through induction of VEGF. Oncogene. 2013;32:5369-76
13. Morales SA, Mareninov S, Wadehra M, Zhang L, Goodglick L, Braun J. et al. FAK activation and the role of epithelial membrane protein 2 (EMP2) in collagen gel contraction. Investigative ophthalmology & visual science. 2009;50:462-9
14. Wadehra M, Goodglick L, Braun J. The tetraspan protein EMP2 modulates the surface expression of caveolins and glycosylphosphatidyl inositol-linked proteins. Molecular biology of the cell. 2004;15:2073-83
15. Kiyohara MH, Dillard C, Tsui J, Kim SR, Lu J, Sachdev D. et al. EMP2 is a novel therapeutic target for endometrial cancer stem cells. Oncogene. 2017;36:5793-807
16. Qin Y, Fu M, Takahashi M, Iwanami A, Kuga D, Rao RG. et al. Epithelial membrane protein-2 (EMP2) activates Src protein and is a novel therapeutic target for glioblastoma. The Journal of biological chemistry. 2014;289:13974-85
17. Qin Y, Takahashi M, Sheets K, Soto H, Tsui J, Pelargos P. et al. Epithelial membrane protein-2 (EMP2) promotes angiogenesis in glioblastoma multiforme. Journal of neuro-oncology. 2017;134:29-40
18. Williams CJ, Chu A, Jefferson WN, Casero D, Sudhakar D, Khurana N. et al. Epithelial membrane protein 2 (EMP2) deficiency alters placental angiogenesis, mimicking features of human placental insufficiency. The Journal of pathology. 2017;242:246-59
19. Zhang Q, Fan X, Xu B, Pang Q, Teng L. miR-133b acts as a tumor suppressor and negatively regulates EMP2 in glioma. Neoplasma. 2018;65:494-504
20. Xie FJ, Zheng QQ, Qin J, Zhang LL, Han N, Mao WM. Autophagy Inhibition Stimulates Apoptosis in Oesophageal Squamous Cell Carcinoma Treated with Fasudil. Journal of Cancer. 2018;9:1050-6
21. Ye M, Wang S, Wan T, Jiang R, Qiu Y, Pei L. et al. Combined Inhibitions of Glycolysis and AKT/autophagy Can Overcome Resistance to EGFR-targeted Therapy of Lung Cancer. Journal of Cancer. 2017;8:3774-84
22. Zhao R, Chen M, Jiang Z, Zhao F, Xi B, Zhang X. et al. Platycodin-D Induced Autophagy in Non-Small Cell Lung Cancer Cells via PI3K/Akt/mTOR and MAPK Signaling Pathways. Journal of Cancer. 2015;6:623-31
23. Ke B, Tian M, Li J, Liu B, He G. Targeting Programmed Cell Death Using Small-Molecule Compounds to Improve Potential Cancer Therapy. Medicinal research reviews. 2016;36:983-1035
24. Ouyang L, Chen Y, Wang XY, Lu RF, Zhang SY, Tian M. et al. Polygonatum odoratum lectin induces apoptosis and autophagy via targeting EGFR-mediated Ras-Raf-MEK-ERK pathway in human MCF-7 breast cancer cells. Phytomedicine: international journal of phytotherapy and phytopharmacology. 2014;21:1658-65
25. Ouyang L, Zhang L, Liu J, Fu L, Yao D, Zhao Y. et al. Discovery of a Small-Molecule Bromodomain-Containing Protein 4 (BRD4) Inhibitor That Induces AMP-Activated Protein Kinase-Modulated Autophagy-Associated Cell Death in Breast Cancer. Journal of medicinal chemistry. 2017;60:9990-10012
26. Pan Z, Chen Y, Liu J, Jiang Q, Yang S, Guo L. et al. Design, synthesis, and biological evaluation of polo-like kinase 1/eukaryotic elongation factor 2 kinase (PLK1/EEF2K) dual inhibitors for regulating breast cancer cells apoptosis and autophagy. European journal of medicinal chemistry. 2018;144:517-28
27. Guo W, Wang H, Yang Y, Guo S, Zhang W, Liu Y. et al. Down-regulated miR-23a Contributes to the Metastasis of Cutaneous Melanoma by Promoting Autophagy. Theranostics. 2017;7:2231-49
28. Martin S, Dudek-Peric AM, Garg AD, Roose H, Demirsoy S, Van Eygen S. et al. An autophagy-driven pathway of ATP secretion supports the aggressive phenotype of BRAF(V600E) inhibitor-resistant metastatic melanoma cells. Autophagy. 2017;13:1512-27
29. Ndoye A, Budina-Kolomets A, Kugel CH 3rd, Webster MR, Kaur A, Behera R. et al. ATG5 Mediates a Positive Feedback Loop between Wnt Signaling and Autophagy in Melanoma. Cancer research. 2017;77:5873-85
30. Wang L, Guo W, Ma J, Dai W, Liu L, Guo S. et al. Aberrant SIRT6 expression contributes to melanoma growth: Role of the autophagy paradox and IGF-AKT signaling. Autophagy. 2018;14:518-33
31. Knecht E, Aguado C, Carcel J, Esteban I, Esteve JM, Ghislat G. et al. Intracellular protein degradation in mammalian cells: recent developments. Cellular and molecular life sciences: CMLS. 2009;66:2427-43
32. Spencer AL, Bagai I, Becker DF, Zuiderweg ER, Ragsdale SW. Protein/protein interactions in the mammalian heme degradation pathway: heme oxygenase-2, cytochrome P450 reductase, and biliverdin reductase. The Journal of biological chemistry. 2014;289:29836-58
33. Sun T, Wang X, Lu Q, Ren H, Zhang H. CUP-5, the C. elegans ortholog of the mammalian lysosomal channel protein MLN1/TRPML1, is required for proteolytic degradation in autolysosomes. Autophagy. 2011;7:1308-15
34. Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nature genetics. 2013;45:580-5
35. Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD. et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell. 2018;173:400-16 e11
36. Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23:3251-3
37. Wang Y, Wang C, Chen C, Wu F, Shen P, Zhang P. et al. Long non-coding RNA NEAT1 regulates epithelial membrane protein 2 expression to repress nasopharyngeal carcinoma migration and irradiation-resistance through miR-101-3p as a competing endogenous RNA mechanism. Oncotarget. 2017;8:70156-71
38. Chen Q, Yao L, Burner D, Minev B, Lu L, Wang M. et al. Epithelial membrane protein 2: a novel biomarker for circulating tumor cell recovery in breast cancer. Clinical & translational oncology: official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2018
39. Chung LK, Pelargos PE, Chan AM, Demos JV, Lagman C, Sheppard JP. et al. Tissue microarray analysis for epithelial membrane protein-2 as a novel biomarker for gliomas. Brain tumor pathology. 2018;35:1-9
40. Leng HJ, Peng F, Zingales S, Huang W, Wang B, Zhao Q. et al. Core-Scaffold-Inspired Asymmetric Synthesis of Polysubstituted Chiral Hexahydropyridazines that Potently Inhibit Breast Cancer Cell Proliferation by Inducing Apoptosis. Chemistry. 2015;21:18100-8
41. Chen Y, Zheng Y, Jiang Q, Qin F, Zhang Y, Fu L. et al. Integrated bioinformatics, computational and experimental methods to discover novel Raf/extracellular-signal regulated kinase (ERK) dual inhibitors against breast cancer cells. European journal of medicinal chemistry. 2017;127:997-1011
42. Zhang N, Huang Y, Wu F, Zhao Y, Li X, Shen P. et al. Codelivery of a miR-124 Mimic and Obatoclax by Cholesterol-Penetratin Micelles Simultaneously Induces Apoptosis and Inhibits Autophagic Flux in Breast Cancer in vitro and in vivo. Molecular pharmaceutics. 2016;13:2466-83
43. Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A. et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12:1-222
Author contact
![]() Corresponding authors: Dr. Peng Zhang: izhangpengedu.cn; Prof. Dr. Ding Bai: baidingedu.cn; Prof. Dr. Xian Jiang: jennyxianjcom
Corresponding authors: Dr. Peng Zhang: izhangpengedu.cn; Prof. Dr. Ding Bai: baidingedu.cn; Prof. Dr. Xian Jiang: jennyxianjcom

 Global reach, higher impact
Global reach, higher impact