Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(16):3639-3649. doi:10.7150/jca.33431 This issue Cite
Research Paper
Association between Polymorphisms of Vitamin D Receptor and Lung Cancer Susceptibility: Evidence from an Updated Meta-analysis
1. The Department of Respiratory and Critical Care Medicine of the First Affiliated Hospital of Xi'an Jiaotong University. 277 West Yanta Road, Xi'an, Shaanxi 710061, China.
2. The Department of Medical Oncology of the First Affiliated Hospital of Xi'an Jiaotong University. 277 West Yanta Road, Xi'an, Shaanxi 710061, China.
The first 2 authors contributed equally to this work.
Received 2019-1-23; Accepted 2019-5-3; Published 2019-6-9
Abstract
Purpose: The aim of this meta-analysis was to investigate polymorphism of Bsm1, Apal, Taq1 and Cdx-2 in vitamin D receptor (VDR) associations in relation to lung cancer (LC) susceptibility.
Methods: 9 literatures were recruited into this meta-analysis from PubMed, PMC, Embase, Web of Science, Cochrane library and CNKI. STATA version 15.1 was used for statistical tests. The heterogeneity was tested using I2 statistics. According to the value of I2, the random-effect model (REM) or fixed-effect model (FEM) was selected to combine data from studies, respectively. Potential publication bias was evaluated by Egger's test. Sensitivity analysis was also performed to evaluate the stability and reliability in results.
Results: Decreased susceptibility of LC was found in all genetic models contrast in Bsm1 gene of VDR (a vs. A: OR = 0.62, 95 % CI = 0.44-0.87; aa vs. AA: OR = 0.76, 95 % CI = 0.60-0.96; Aa vs. AA: OR = 0.59, 95 % CI = 0.39-0.88; aa vs. AA+Aa: OR = 0.80, 95 % CI = 0.64-0.99; Aa+aa vs. AA: OR = 0.57, 95 % CI = 0.37-0.86). The similar results were also found in partial genetic models of Taq1 (a vs. A: OR = 0.88, 95 % CI = 0.79-0.98; aa vs. AA+Aa: OR = 0.84, 95 % CI = 0.73-0.98) and Cdx-2 (Aa vs. AA: OR = 0.80, 95 % CI = 0.66-0.98; Aa+aa vs. AA: OR = 0.79, 95 % CI = 0.65-0.96). Likewise, significant correlation between Bsm1, Taq1 polymorphism and LC risk was detected among Asians. Cdx-2 polymorphism was considered as a protective factor in Caucasians, whereas no association of Apal polymorphism with LC risk was observed in Asians and Caucasians for all genetic models.
Conclusion: The results of this meta-analysis suggested that Bsm1, Taq1 and Cdx-2 polymorphism may contribute to lung cancer susceptibility, more studies need be conducted to confirm in the future.
Keywords: Vitamin D receptor, VDR, Lung cancer, Polymorphism, risk, Meta-analysis
Introduction
Cancer constitutes a significant disease burden worldwide, [1] there will be an estimated 18.1 million new cancer cases in 2018. [2] Lung cancer (LC) has been the most commonly diagnosed cancer and the leading cause of cancer-related death in the world. [2] However, there are distinct variations in LC incidence and mortality by region. [3] To date, the etiology of which remains unclear. Smoking is firmly‐established as a predominant factor in the incidence of LC. [4, 5] However, not all individuals exposed to smoking will develop LC and some patients with LC have never smoked. Other susceptibility factors such as viral infections, air pollution and exposure to occupational and environmental carcinogens could also increase the incidence of LC. [6, 7] It can be seen that excluding environmental factors, the difference in risk of LC among individuals may be related to genetic factors, which is also considered to be an influential factor that lead to the incidence of LC. [8, 9] Genome-wide association studies (GWAS) have been identified several susceptibility gene locus of cancer, some of which increase the susceptibility of LC. [10-12] The identification of gene mutations that are important to the susceptibility of LC will contribute to a better understanding of the pathogenesis of LC and may lead to new approaches of disease treatment or prevention.
Vitamin D, a seco-steroidal prohormone, plays a crucial role in regulating metabolism of calcium and phosphate, which is metabolized by enzymes into the active form 1,25(OH)2D3 (1,25-D3). [13, 14] Many previous studies have shown that vitamin D regulates the entire process of tumorigenesis, from initiation to metastasis, and the interaction of the cellular microenvironment. [15, 16]
Vitamin D receptor (VDR), located in chromosome 12q13.11 which spans ~100kb and have five promoters, eight coding exons, and six untranslated exons, is a nuclear biomacromolecule. [17] In target tissues, 1,25-D3 binds to VDR and induces both genomic and non-genomic regulation of downstream targets involving diverse biological functions such as anti-differentiation and anti-proliferation activities in cancer cell lines and modulating E-cadherin and EMT-related molecules gene expression. [18, 19] Studies have shown that 1,25-D3 and the VDR suppress c-MYC function via regulating the c-MYC/MXD1 network, providing a molecular basis for cancer preventive actions of vitamin D.[20] Variants of VDR was found associate with tuberculosis,[21] osteoporosis[22] and cancers including colorectal cancer and LC.[23, 24] Until now, certain VDR gene variants have been verified in relation to LC risk with different results including Fok1, Bsm1, Taq1, Apa1, Cdx2. In order to further provide theoretical support for the pathogenesis of LC, explore the association between Bsm1 (rs1544410 G>A), Apa1 (rs7975232 C>A), Taq1 (rs731236 T>C) and Cdx-2 (rs11568820 T>C) polymorphisms of VDR associations in relation to LC susceptibility was performed in this meta-analysis of 9 related studies.
Methods
Publication search strategy
We conducted a systematic literature search of the PubMed, PMC, Embase, Web of Science, Cochrane library and CNKI by using the following search terms, MeSH “VDR, Polymorphism, Genetic and Lung Neoplasms” and free words “vitamin D receptor, rs1544410, Bsm1, rs731236, Taql, rs7975232, Apa1, rs11568820, Cdx-2, Genetic Polymorphisms, Genetic Polymorphism, Polymorphism, Polymorphisms, Pulmonary Neoplasms, Lung Neoplasm, Pulmonary Neoplasm, Lung Cancer, Lung Cancers, Pulmonary Cancer and Pulmonary Cancers” with title or abstracts restrictions. Reference lists of the retrieved articles were also browsed for other potential correlation articles. We did not contact authors of the primary studies for complete information.
Study selection
All studies were included in this meta-analysis strictly to follow criteria includes/excludes in order to minimized the heterogeneity: (1) Case-control studies; (2) Analyzing the relationship between VDR genetic polymorphism and LC risk; (3) VDR original data of genotype distribution should be provide in detail; (4) Odds ratios (ORs) and 95% confidence interval (CI) were used to estimate genotype frequencies between cases and controls; (5) English and Chinese literatures. Articles that meet the following criteria will be excluded: (1) All review articles, editorials, conference summary, case reports and overlapping studies; (2) Insufficient information about outcomes and unrelated outcomes; (3) Not meeting language requirements; (4) Only the latest study with higher quality ratings and more detailed basic information will be selected into the analysis if more than one related paper is published from same research group.
Data extraction and quality assessment
All data were extracted and tabulated by two authors independently using a standardized data-collection form, any disagreements were settled down by discussion. If the two investigators failed to reach a consensus, an agreement was achieved after public discussion with the adjudicator. Information was recorded as follows: last name of the first author, year of publication, country, ethnicity of subjects, genotyping methods, source of control, quality score, number of cases and controls, frequencies of VDR Bsm1, Apa1, Taq1 and Cdx2 genotypes in all participants, ORs and 95% CI in all candidates gene locus and Hardy-Weinberg equilibrium (HWE). “A” and “a” were used to present wild-type allele and mutant alleles of candidate single nucleotide polymorphisms (SNPs), respectively (A>a). The quality of selected studies were evaluated by Newcastle Ottawa Quality Assessment Scale for Case-Control Studies (NOS, http://www.ohri.ca/programs/clinical_epidemiology/nosgen.pdf). [25] The quality score ranges from 0 to 10. 5 or more than 5 scores of NOS indicate studies with good quality.
Statistical analysis
The principal summary measurement of the strength of connection between the VDR gene polymorphisms and risk of LC was reported by pooled odds ratio (POR), 95% CI and P value. The 95% CI no overlap with 1 was deemed to be statistically significant. PORs were performed for allelic model (a vs. A), heterozygote model (Aa vs. AA), homozygote model (aa vs. AA), dominant model (Aa + aa vs. AA) and recessive model (aa vs. Aa + AA). The significance of POR was assessed by Z test with P<0.05. I2 statistics were used to evaluate the heterogeneity among studies. I2<50% with P>0.1 was considered have no heterogeneity. The FEM would be performed in the absence of heterogeneity, otherwise, REM would be performed. Publication bias was assessed by Egger's linear regression test. The P<0.1 and asymmetric plot was considered as the existence of publication bias. Sensitivity analysis was also used to evaluate the quality and stability in results by omitting each study in each turn. Subgroup analysis was further carried out by race. All statistical analyses were performed with Stata 15.1 software (StataCorp, College Station, TX, USA). Two-side P < 0.05 was considered significant.
Results
Literature search
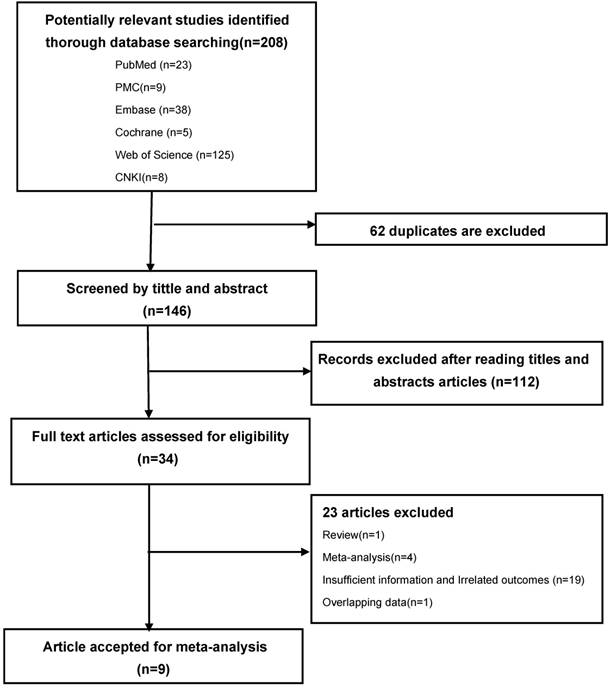
According to the search strategy, 208 potentially relevant published literatures were identified from electronic database (23 in PubMed, 9 in PMC, 38 in Embase, 5 in Cochrane library, 125 in Web of science, 8 in CNKI). Out of these, 62 were excluded due to duplicate records, 112 were excluded after reading titles and abstracts due to irrelated with LC and VDR polymorphism. The remaining 34 studies were full-text reviewed and 23 studies were excluded, of which, 1 was review, 1 was overlapping data, 4 were meta-analyses, 19 had insufficient information and unrelated outcomes. The remaining 9 literatures were included in this meta-analysis because accordance with the inclusion criteria. [24, 26-33] The flow chart of literature selection process is shown in Figure 1.
Baseline properties of studies
This study involved a total of 2324 cases and 2464 controls. Of these, 4 studies were conducted in Caucasians, 5 studies in Asians. All the articles included in this study were high-quality studies, NOS scores for all them were more than 5. The detailed baseline characteristics and quality score of all included articles are presented in Table 1, Table 2, respectively.
Association between the VDR polymorphism and LC risk
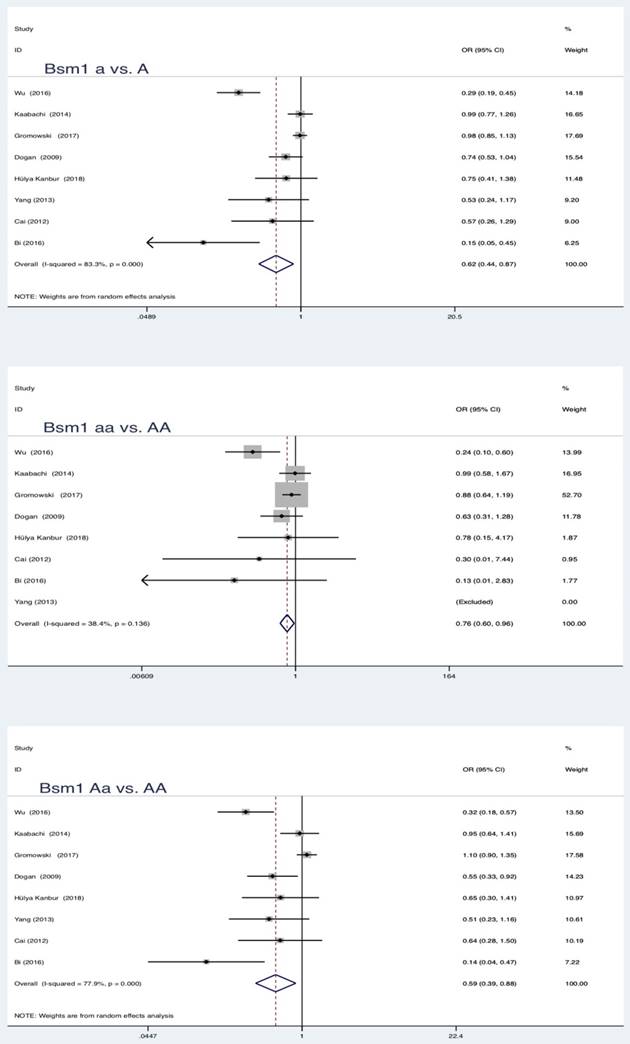
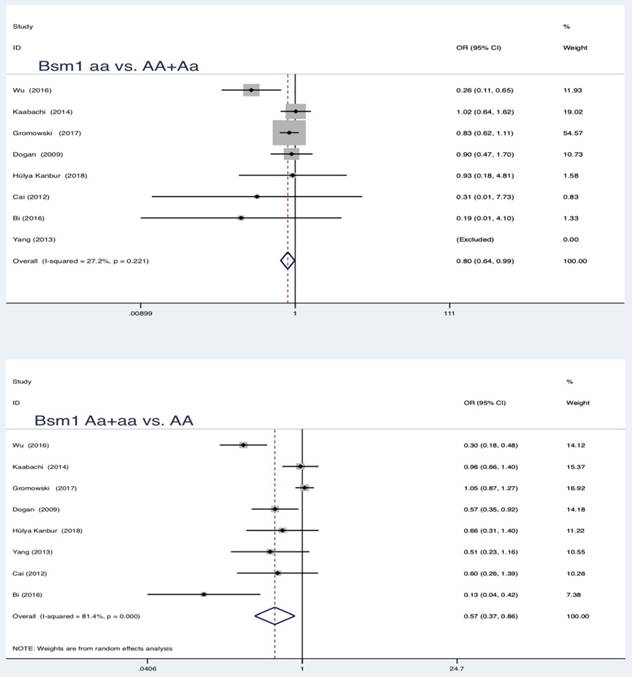
In the pooled analysis, statistically significant protection role of Bsm1 (rs1544410 G>A) polymorphism in LC was observed among allele model, homozygous and heterozygous models and all genetic models (a vs. A: OR = 0.62, 95 % CI = 0.44-0.87, P = 0.005; aa vs. AA: OR = 0.76, 95 % CI = 0.60-0.96, P = 0.019; Aa vs. AA: OR = 0.59, 95 % CI = 0.39-0.88, P = 0.010; aa vs. AA+Aa: OR = 0.80, 95 % CI = 0.64-0.99, P = 0.039; Aa+aa vs. AA: OR = 0.57, 95 % CI = 0.37-0.86, P = 0.007).
Characteristics of the studies evaluating the effects of VDR gene polymorphism on LC risk
| Control Source | Quality score | Case | Control | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNPs | Author | Year | Country | Ethnicity | Method | Case | Control | AA | Aa | aa | AA | Aa | aa | HWE | ||
| Bsm1 | Dogan [26] | 2009 | Turkey | Caucasian | PCR-RFLP | Healthy | 7 | 137 | 156 | 57 | 60 | 20 | 45 | 86 | 25 | p > 0.05 |
| Cai [27] | 2012 | China | Asian | PCR-RFLP | Healthy | 8 | 140 | 132 | 130 | 10 | 0 | 117 | 14 | 1 | p > 0.05 | |
| Yang [28] | 2013 | China | Asian | PCR-RFLP | Healthy | 7 | 144 | 142 | 134 | 10 | 0 | 124 | 18 | 0 | p > 0.05 | |
| Kaabachi [29] | 2014 | Tunisian | Caucasian | PCR-RFLP | Healthy | 7 | 240 | 280 | 74 | 126 | 40 | 84 | 150 | 46 | p > 0.05 | |
| Wu [30] | 2016 | China | Asian | PCR-RFLP | Healthy | 8 | 426 | 445 | 403 | 17 | 6 | 373 | 49 | 23 | p < 0.05 | |
| Bi [31] | 2016 | China | Asian | PCR-RFLP | Healthy | 8 | 50 | 50 | 46 | 4 | 0 | 30 | 18 | 2 | p > 0.05 | |
| Gromowski [24] | 2017 | Polish | Caucasian | PCR-TaqMan | Healthy | 8 | 840 | 920 | 330 | 388 | 92 | 384 | 410 | 122 | p > 0.05 | |
| Hülya Kanbur [33] | 2018 | Turkey | Caucasian | PCR-TaqMan | Healthy | 6 | 59 | 55 | 37 | 19 | 3 | 29 | 23 | 3 | p > 0.05 | |
| Apal | Dogan [26] | 2009 | Turkey | Caucasian | PCR-RFLP | Healthy | 7 | 137 | 156 | 44 | 64 | 29 | 58 | 76 | 22 | p > 0.05 |
| Yang [28] | 2013 | China | Asian | PCR-RFLP | Healthy | 7 | 144 | 142 | 76 | 63 | 5 | 74 | 60 | 8 | p > 0.05 | |
| Kaabachi [29] | 2014 | Tunisian | Caucasian | PCR-RFLP | Healthy | 7 | 240 | 280 | 101 | 118 | 21 | 100 | 134 | 46 | p > 0.05 | |
| Wu [30] | 2016 | China | Asian | PCR-RFLP | Healthy | 8 | 426 | 445 | 140 | 191 | 95 | 142 | 214 | 89 | p > 0.05 | |
| Bi [31] | 2016 | China | Asian | PCR-RFLP | Healthy | 8 | 50 | 50 | 27 | 22 | 1 | 12 | 36 | 2 | p < 0.05 | |
| Yang [32] | 2017 | China | Asian | PCR-RFLP | Healthy | 7 | 288 | 284 | 142 | 116 | 30 | 150 | 87 | 47 | p < 0.05 | |
| Gromowski [24] | 2017 | Polish | Caucasian | PCR-TaqMan | Healthy | 8 | 840 | 920 | 236 | 412 | 175 | 235 | 500 | 184 | p < 0.05 | |
| Taq1 | Dogan [26] | 2009 | Turkey | Caucasian | PCR-RFLP | Healthy | 7 | 137 | 156 | 64 | 59 | 14 | 49 | 83 | 24 | p > 0.05 |
| Yang [28] | 2013 | China | Asian | PCR-RFLP | Healthy | 7 | 144 | 142 | 135 | 9 | 0 | 129 | 12 | 1 | p > 0.05 | |
| Kaabachi [29] | 2014 | Tunisian | Caucasian | PCR-RFLP | Healthy | 7 | 240 | 280 | 90 | 118 | 32 | 98 | 146 | 36 | p > 0.05 | |
| Wu [30] | 2016 | China | Asian | PCR-RFLP | Healthy | 8 | 426 | 445 | 409 | 14 | 3 | 416 | 27 | 2 | p > 0.05 | |
| Yang [32] | 2017 | China | Asian | PCR-RFLP | Healthy | 7 | 288 | 284 | 258 | 27 | 3 | 240 | 38 | 6 | p < 0.05 | |
| Gromowski [24] | 2017 | Polish | Caucasian | PCR-TaqMan | Healthy | 8 | 840 | 920 | 340 | 390 | 95 | 375 | 423 | 122 | p > 0.05 | |
| Cdx-2 | Wu [30] | 2016 | China | Asian | PCR-RFLP | Healthy | 8 | 426 | 445 | 63 | 324 | 39 | 52 | 360 | 33 | p < 0.05 |
| Gromowski [24] | 2017 | Polish | Caucasian | PCR-TaqMan | Healthy | 8 | 840 | 920 | 649 | 170 | 3 | 653 | 207 | 11 | p > 0.05 | |
HWE, Hardy-Weinberg equilibrium; P value >0.05 showed that SNPs were in HWE
PRISMA flow-diagram. The screening process (inclusion/exclusion) of the studies dealing with VDR gene polymorphism and LC susceptibility
Quality assessment conducted according to the NOS for all selected studies
| First author and year | Quality indicators | |||
|---|---|---|---|---|
| Selection | Comparability | Exposure | Quality score | |
| Dogan, 2009 [26] | *** | * | ** | 7 |
| Cai, 2012 [27] | **** | * | ** | 8 |
| Yang, 2013 [28] | *** | * | ** | 7 |
| Kaabachi, 2014 [29] | *** | * | ** | 7 |
| Wu, 2016 [30] | **** | * | ** | 8 |
| Bi, 2016 [31] | **** | * | ** | 8 |
| Yang, 2017 [32] | *** | * | ** | 7 |
| Gromowski, 2017 [24] | **** | * | ** | 8 |
| Hülya Kanbur, 2018 [33] | ** | * | ** | 6 |
*indicates points of score
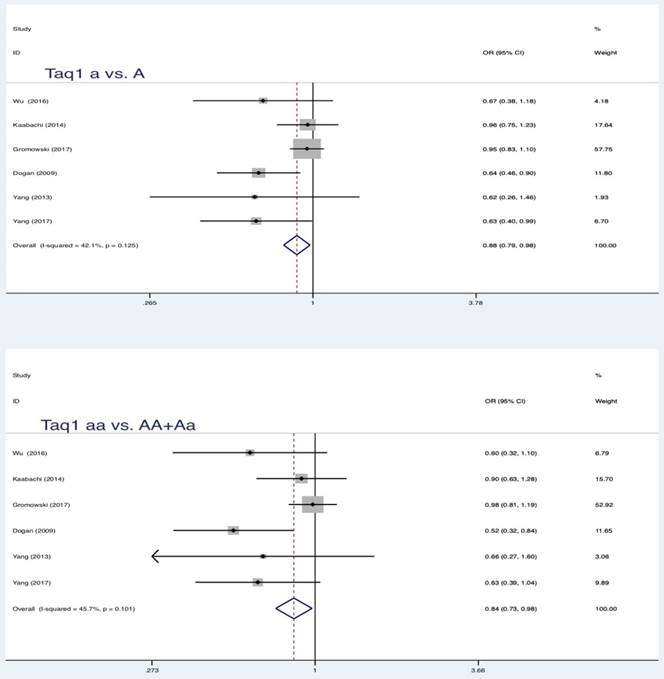
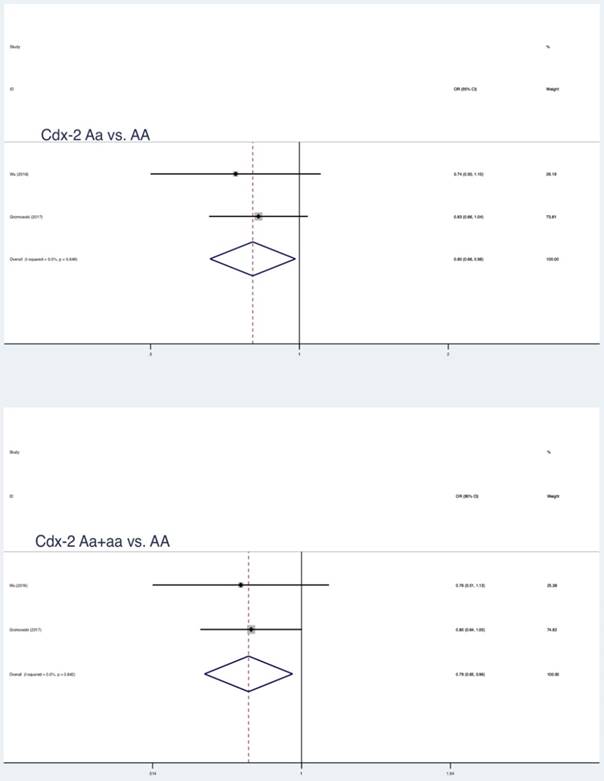
Similar results were also found in partial genetic models of Taq1 (rs731236 T>C, a vs. A: OR = 0.88, 95 % CI = 0.79-0.98, P = 0.017; aa vs. AA+Aa: OR = 0.84, 95 % CI = 0.73-0.98, P = 0.022) and Cdx-2 (rs11568820 T>C, Aa vs. AA: OR = 0.80, 95 % CI = 0.66-0.98, P = 0.032; Aa+aa vs. AA: OR = 0.79, 95 % CI = 0.65-0.96, P = 0.018) (Table 3, Figure 2, Figure 3, Figure 4). No statistically significant association were found between Apa1 (rs7975232 C>A) gene polymorphism and LC.
As shown in Table 4, significant correlation between Bsm1, Taq1 polymorphism and LC risk was detected among Asians when stratified by ethnicity. Such association was not observed for the Caucasians. Contrary to the above results, Cdx-2 polymorphism was considered as a protective factor in the Caucasians.
Heterogeneity test and Sensitivity analysis
The heterogeneity of all VDR gene polymorphisms allelic models, genotype, dominant models and recessive models was made to analyze in all selected studies. High-estimated heterogeneity was observed in Bsm1 (allele genetic model, heterozygous genotype and dominate model), Apa1 (all models), Cdx2 (allele genetic model, homozygous genotype and recessive model) indicating between-study heterogeneity. There was no heterogeneity in Taq1 (Table 3).
Summary estimates for the association between LC and all genetic models of Bsm1
Meta-analysis and publication bias between VDR gene polymorphisms and LC
| Test of association | Test of heterogeneity | Bias | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNPs | Comparison | Na | POR | 95%CI | Z | P-value | I2 | Ph | Model | Egger's test |
| Bsm1 | a vs A | 8 | 0.62 | 0.44-0.87 | 2.79 | 0.005* | 83.3 | 0.000 | Random | 0.036* |
| aa vs AA | 0.76 | 0.60-0.96 | 2.34 | 0.019* | 38.4 | 0.136 | Fixed | 0.009* | ||
| Aa vs AA | 0.59 | 0.39-0.88 | 2.57 | 0.010* | 77.9 | 0.000 | Random | 0.126 | ||
| aa vs AA+Aa | 0.80 | 0.64-0.99 | 2.07 | 0.039* | 27.2 | 0.221 | Fixed | 0.239 | ||
| Aa+aa vs AA | 0.57 | 0.37-0.86 | 2.68 | 0.007* | 81.4 | 0.000 | Random | 0.025* | ||
| Apal | a vs A | 7 | 0.93 | 0.81-1.07 | 1.00 | 0.318 | 52.8 | 0.048 | Random | 0.411 |
| aa vs AA | 0.85 | 0.62-1.16 | 1.03 | 0.302 | 53.4 | 0.045 | Random | 0.392 | ||
| Aa vs AA | 0.92 | 0.73-1.16 | 1.00 | 0.319 | 59.6 | 0.021 | Random | 0.690 | ||
| aa vs AA+Aa | 0.88 | 0.64-1.21 | 0.77 | 0.439 | 61.8 | 0.015 | Random | 0.337 | ||
| Aa+aa vs AA | 0.90 | 0.74-1.11 | 0.98 | 0.327 | 53.5 | 0.044 | Random | 0.508 | ||
| Taq1 | a vs A | 6 | 0.88 | 0.79-0.98 | 2.38 | 0.017* | 42.1 | 0.125 | Fixed | 0.043* |
| aa vs AA | 0.81 | 0.63-1.03 | 1.73 | 0.084 | 0.00 | 0.504 | Fixed | 0.450 | ||
| Aa vs AA | 0.86 | 0.74-1.00 | 1.93 | 0.054 | 45.3 | 0.104 | Fixed | 0.029* | ||
| aa vs AA+Aa | 0.84 | 0.73-0.98 | 2.29 | 0.022* | 45.7 | 0.101 | Fixed | 0.560 | ||
| Aa+aa vs AA | 0.85 | 0.67-1.07 | 1.42 | 0.156 | 0.00 | 0.740 | Fixed | 0.038* | ||
| Cdx-2 | a vs A | 2 | 0.88 | 0.72-1.08 | 1.22 | 0.224 | 51.3 | 0.152 | Random | - |
| aa vs AA | 0.59 | 0.17-2.00 | 0.85 | 0.397 | 68.1 | 0.077 | Random | - | ||
| Aa vs AA | 0.80 | 0.66-0.98 | 2.14 | 0.032* | 0.0 | 0.649 | Fixed | - | ||
| aa vs AA+Aa | 0.68 | 0.16-2.85 | 0.53 | 0.597 | 77.9 | 0.033 | Random | - | ||
| Aa+aa vs AA | 0.79 | 0.65-0.96 | 2.36 | 0.018* | 0.0 | 0.842 | Fixed | - | ||
Summary estimates for the association between LC and partial genetic models of Taq1
Meta-analysis between VDR gene polymorphisms and LC based on stratification analysis
| Ethnicity | N | a vs A | aa vs AA | Aa vs AA | aa vs AA+Aa | Aa+aa vs AA | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| POR | 95%CI | P* | POR | 95%CI | P* | POR | 95%CI | P* | POR | 95%CI | P* | POR | 95%CI | P* | ||
| Bsm1 | 8 | |||||||||||||||
| Asian | 4 | 0.33 | 0.24-0.46 | 0.000* | 0.23 | 0.10-0.54 | 0.001* | 0.37 | 0.25-0.54 | 0.000* | 0.23 | 0.10-0.54 | 0.001* | 0.34 | 0.24-0.48 | 0.000* |
| Caucasian | 4 | 0.95 | 0.84-1.06 | 0.319 | 0.86 | 0.68-1.10 | 0.240 | 0.97 | 0.83-1.15 | 0.749 | 0.86 | 0.68-1.10 | 0.240 | 0.95 | 0.81-1.11 | 0.513 |
| Apa1 | 7 | |||||||||||||||
| Asian | 4 | 0.95 | 0.83-1.09 | 0.452 | 0.84 | 0.58-1.21 | 0.347 | 0.88 | 0.56-1.39 | 0.579 | 0.80 | 0.49-1.29 | 0.357 | 0.95 | 0.79-1.15 | 0.628 |
| Caucasian | 3 | 0.95 | 0.85-1.06 | 0.325 | 0.89 | 0.48-1.66 | 0.721 | 0.86 | 0.72-1.03 | 0.108 | 0.95 | 0.54-1.69 | 0.867 | 0.87 | 0.74-1.04 | 0.118 |
| Taq1 | 6 | |||||||||||||||
| Asian | 3 | 0.64 | 0.46-0.89 | 0.008* | 0.66 | 0.24-1.80 | 0.415* | 0.62 | 0.43- 0.90 | 0.013* | 0.69 | 0.25-1.86 | 0.459 | 0.63 | 0.44-0.89 | 0.009* |
| Caucasian | 3 | 0.91 | 0.81-1.02 | 0.115 | 0.82 | 0.64-1.05 | 0.116 | 0.92 | 0.78- 1.09 | 0.333 | 0.86 | 0.68-1.09 | 0.200 | 0.90 | 0.77-1.05 | 0.186 |
| Cdx-2 | 2 | |||||||||||||||
| Asian | 1 | 0.97 | 0.81-1.17 | 0.776 | 0.98 | 0.54-1.76 | 0.934 | 0.74 | 0.50-0.66 | 0.142 | 1.26 | 0.78-2.04 | 0.352 | 0.76 | 0.51-1.13 | 0.177 |
| Caucasian | 1 | 0.79 | 0.64-0.98 | 0.029* | 0.27 | 0.07- 0.99 | 0.048* | 0.83 | 0.66- 1.04 | 0.104 | 0.29 | 0.08-1.03 | 0.056 | 0.80 | 0.64-1.00 | 0.052 |
* indicates P <0.05
Sensitivity analysis was conducted to assess the influence of each individual study on the POR by deleting one single study each time. The results showed that the corresponding PORs were not materially altered by removing any individual study in Bsm1, Apa1, Taq1 and Cdx2 genes. Therefore, the sensitivity analysis confirmed that the results of this meta-analysis were statistically reliable and stable (Table 5).
Sensitive analyses for candidate genes
| Author, Year | Bsm1 | Apa1 | Taq1 | Cdx-2 |
|---|---|---|---|---|
| OR(95%CI) | OR(95%CI) | OR(95%CI) | OR(95%CI) | |
| Dogan, 2009 [26] | 0.74(0.53-1.04) | 1.28(0.92-1.79) | 0.64(0.46-0.90) | - |
| Cai, 2012 [27] | 0.57(0.26-1.29) | 0.47(0.26-0.87) | - | - |
| Yang, 2013 [28] | 0.53(0.24-1.17) | - | 0.62(0.27-1.46) | - |
| Kaabachi, 2014 [29] | 0.99(0.77-1.26) | 0.74(0.57-0.95) | 0.96(0.75-1.23) | - |
| Wu ,2016 [30] | 0.30(0.19-0.45) | 1.03(0.85-1.24) | 0.67(0.38-1.18) | 0.97(0.81-1.17) |
| Bi, 2016 [31] | 0.15(0.05-0.45) | - | - | - |
| Yang, 2017 [32] | - | 0.94(0.73-1.21) | 0.63(0.40-0.99) | - |
| Gromowsk, 2017 [24] | 0.98(0.86-1.13) | 0.96(0.84-1.10) | 0.95(0.83-1.10) | 0.79(0.64-0.98) |
| Hülya Kanbur, 2018 [33] | 0.75(0.41-1.39) | 0.93(0.64-1.35) | - | - |
| Pooled data | 0.62(0.44-0.87) | 0.93(0.81-1.07) | 0.88(0.79-0.98) | 0.88(0.72-1.08) |
Summary estimates for the association between LC and partial genetic models
Publication bias
As shown in Table 3, significant results of Egger's test were reveled which showed publication bias was present in partial genetic models of Bsm1 and Taq1. No Egger's test was performed for the association between Cdx-2 and LC susceptibility owing to the limited number of included studies.
Discussion
As the most common type of cancer all over the world, the role of genetic factors in etiology of LC has aroused strong attention. SNP refers to the DNA sequence polymorphism caused by the variation of single nucleotide at the level of genome. [34] It affects the process of transcription, translation, expression and function of protein, which determine the difference in genetic susceptibility of different individuals. As we know, Vitamin D, a steroid hormone, plays a crucial role in bone metabolism and calcium homeostasis, the deficiency of which is widely regarded as a primary cause of rickets. [35] The most active metabolite form of vitamin D is 1,25-D3, which not only participates in maintaining calcium homeostasis, but also has some non-endocrine effects such as influencing cardiovascular disease, diabetes and cancers. [36, 37] Previous studies have proved that high circulating levels of 1,25-D3 have the ability to prevent the development of cancer. [38] Expression and nuclear activation of the VDR are necessary for the function of vitamin D. Thus, genetic alternation of the VDR gene could lead to important defects in gene activation, which is bound to affect the biological effects of vitamin D. At present, a lot of studies have investigated the association between VDR polymorphism and LC susceptibility, but the specific correlation of them still unclear. Therefore, this meta-analysis was performed to provide more accurate statistical evidence of association between VDR polymorphism and LC risk.
In this meta-analysis, we comprehensively assessed 4 candidate SNPs (Bsm1, Apa1, Taq1, Cdx-2) of VDR gene for association with LC susceptibility from 9 selected studies, and we found many differences between the results of this study and previous studies. [39-41] Previous studies have shown that Apa1 and Bsm1 were associated with LC risk in overall populations, Caucasians and Asians in some genetic models. However, according to our study, all genetic models of Apa1 failed to find correlation with LC incidence both in overall and stratified analysis, no association was found between Bsm1 and susceptibility to LC in Caucasians although it could reduce the risk of LC in overall populations and Asians under all 5 genetic models. Taq1 decreased LC risk in overall populations and Caucasians but not in Asians in previous meta-analyses, whereas Taq1 variations in decreased risk of LC were verified in overall and Asians in our study. What's more, this is the first meta-analysis to investigate the relationship between Cdx-2 polymorphism and LC susceptibility. Heterozygote model and dominate model of Cdx-2 were considered as a protective factor to LC in Caucasiana. The above observations suggested that the polymorphism of Bsm1, Taq1 and Cdx-2 leads to increase resistance to LC susceptibility. The functional polymorphisms, which are located near 3′ UTR of the gene polymorphisms, might affect the function of VDR by regulating the stability of mRNA and the translation efficiency of protein to influence the effect of vitamin D on tumor inhibition.[42]
The results of stratification analysis suggested that polymorphisms of the same locus might play different roles in affecting LC susceptibility in different ethnic groups. Racial and regional differences of LC incidence and mortality in global statistics further illustrate this point. [3] Therefore, this study may attribute the current results to racial differences. However, it is noteworthy that LC is a very heterogeneous disease with the interaction of multiple genes, factors and multiple stages. [6] There may be potential confounding factors between different races to weaken or exaggerate the statistical power such as differences in geographical location, living condition and customs. What's more, the susceptibility to LC of people is polygenic and multiple candidate genes may jointly participate in the risk of LC. Due to multifactorial nature of LC incidence and complexity of the genetic factors, VDR genetic polymorphism cannot be responsible for the susceptibility of LC alone. Hence, more related genes need to be included in follow-up studies to investigate the etiology of LC.
To the best of our knowledge, just 3 previous researches have investigated the effect of VDR genetic polymorphism in relation to LC risk. [39-41] In this present study, we identified 3 locus in VDR genes have significant association with LC. Currently, compared with the previous, there are various advantages in our study, more gene loci were included in analysis, especially to Cdx-2, which is the first gene loci included in this meta-analysis, more databases were retrieved (Pubmed, PMC, Embase, Cochrane library, Web of Science and CNKI), more studies with better quality were selected for analysis, the relationship between polymorphism of candidate gene locus and lung cancer susceptibility was analyzed under 5 genetic models, stratified analysis was further conducted by ethnicity and larger sample size increased the statistical capacity. Accordingly, our study may be the most powerful investigation in illuminating the effect of VDR polymorphism in LC risk. However, although the results have strong statistical significance, there are still several potential limitations of the present research. First, language bias and selection bias could not be ruled out, as only partial databases and studies published in Chinese and English were included and browsed, it is possible that some relevant studies published in other languages and indexed in other electronic databases may have been omitted. Second, we did not test gene-environment interactions and not stratified by other factors such as smoking and Vitamin D concentrations in vivo due to the deficiency of original data. Third, significant heterogeneity between studies was detected, which would impair the validity of conclusion.
Conclusion
In summary, the present meta-analysis revealed that polymorphisms of Bsm1, Taq1and Cdx-2 in VDR are associated with susceptibility of LC. Bsm1 and Taq1 variations reduced LC risk among Asian, and the similar association was found in Cdx-2 polymorphism among Caucasian. However, Apa1 gene failed to find correlation with LC incidence both in two ethnicities.
Abbreviations
VDR: vitamin D receptor; LC: lung cancer; REM: random-effect mode; FEM: fixed-effect model; GWAS: Genome-wide association studies; OR: odds ratio; 95%CI: 95% confidence interval; HWE: Hardy-Weinberg equilibrium; SNPs: single nucleotide polymorphisms; NOS: Newcastle Ottawa Quality Assessment Scale; POR: pooled odds ratio; 1,25D3: 1,25(OH)2D3.
Acknowledgements
Financial support was provided by Shaanxi International Cooperation Project (2018KW-039).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Global Burden of Disease Cancer C, Fitzmaurice C, Akinyemiju TF, Al Lami FH, Alam T, Alizadeh-Navaei R. et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2016: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2018;4:1553-68
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
3. Wong MCS, Lao XQ, Ho KF, Goggins WB, Tse SLA. Incidence and mortality of lung cancer: global trends and association with socioeconomic status. Sci Rep. 2017;7:14300
4. Doll R, Hill AB. Smoking and carcinoma of the lung. Preliminary report. 1950. Bull World Health Organ. 1999;77:84-93
5. Bach PB. Smoking as a factor in causing lung cancer. Jama. 2009;301:539-41
6. Akhtar N, Bansal JG. Risk factors of Lung Cancer in nonsmoker. Curr Probl Cancer. 2017;41:328-39
7. Couraud S, Zalcman G, Milleron B, Morin F, Souquet PJ. Lung cancer in never smokers-a review. Eur J Cancer. 2012;48:1299-311
8. Brennan P, Hainaut P, Boffetta P. Genetics of lung-cancer susceptibility. Lancet Oncol. 2011;12:399-408
9. Lin H, Huang YS, Yan HH, Yang XN, Zhong WZ, Ye HW. et al. A family history of cancer and lung cancer risk in never-smokers: A clinic-based case-control study. Lung Cancer. 2015;89:94-8
10. Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D. et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633-7
11. Wang Y, McKay JD, Rafnar T, Wang Z, Timofeeva MN, Broderick P. et al. Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nat Genet. 2014;46:736-41
12. Broderick P, Wang Y, Vijayakrishnan J, Matakidou A, Spitz MR, Eisen T. et al. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer research. 2009;69:6633-41
13. Zhang R, Naughton DP. Vitamin D in health and disease: current perspectives. Nutr J. 2010;9:65
14. DeLuca HF. Overview of general physiologic features and functions of vitamin D. Am J Clin Nutr. 2004;80:1689S-96S
15. Giammanco M, Di Majo D, La Guardia M, Aiello S, Crescimannno M, Flandina C. et al. Vitamin D in cancer chemoprevention. Pharmaceutical biology. 2015;53:1399-434
16. Feldman D, Krishnan AV, Swami S, Giovannucci E, Feldman BJ. The role of vitamin D in reducing cancer risk and progression. Nature reviews Cancer. 2014;14:342-57
17. Miyamoto K, Kesterson RA, Yamamoto H, Taketani Y, Nishiwaki E, Tatsumi S. et al. Structural organization of the human vitamin D receptor chromosomal gene and its promoter. Molecular endocrinology (Baltimore, Md). 1997;11:1165-79
18. Upadhyay SK, Verone A, Shoemaker S, Qin M, Liu S, Campbell M. et al. 1,25-Dihydroxyvitamin D3 (1,25(OH)2D3) Signaling Capacity and the Epithelial-Mesenchymal Transition in Non-Small Cell Lung Cancer (NSCLC): Implications for Use of 1,25(OH)2D3 in NSCLC Treatment. Cancers. 2013;5:1504-21
19. Trump DL, Hershberger PA, Bernardi RJ, Ahmed S, Muindi J, Fakih M. et al. Anti-tumor activity of calcitriol: pre-clinical and clinical studies. The Journal of steroid biochemistry and molecular biology. 2004;89-90:519-26
20. Salehi-Tabar R, Nguyen-Yamamoto L, Tavera-Mendoza LE, Quail T, Dimitrov V, An BS. et al. Vitamin D receptor as a master regulator of the c-MYC/MXD1 network. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:18827-32
21. Zhang HQ, Deng A, Guo CF, Wang YX, Chen LQ, Wang YF. et al. Association between FokI polymorphism in vitamin D receptor gene and susceptibility to spinal tuberculosis in Chinese Han population. Archives of medical research. 2010;41:46-9
22. Zhao B, Zhang W, Du S, Zhou Z. Vitamin D receptor BsmI polymorphism and osteoporosis risk in post-menopausal women. Archives of medical science: AMS. 2016;12:25-30
23. Cho YA, Lee J, Oh JH, Chang HJ, Sohn DK, Shin A. et al. Vitamin D receptor FokI polymorphism and the risks of colorectal cancer, inflammatory bowel disease, and colorectal adenoma. Scientific reports. 2018;8:12899
24. Gromowski T, Gapska P, Scott RJ, Kaklewski K, Marciniak W, Durda K. et al. Serum 25(OH)D concentration, common variants of the VDR gene and lung cancer occurrence. Int J Cancer. 2017;141:336-41
25. Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. European journal of epidemiology. 2010;25:603-5
26. Dogan I, Onen HI, Yurdakul AS, Konac E, Ozturk C, Varol A. et al. Polymorphisms in the vitamin D receptor gene and risk of lung cancer. Medical science monitor: international medical journal of experimental and clinical research. 2009;15:Br232-42
27. Cai L, Yang Y, Qi X, Fu Z, Tang L, Yu Z. THE RELATIONSHIP BETWEEN VITAMIN D RECEPTOR GENE BSM1 SITES POLYMORPHISMS AND HEREDITARY SUSCEPTIBILITY TO NON-SMALL CELL LUNG CANCER. Chinese Journal of Coal Industry Medicine. 2012:15
28. Yang Y, Zhou W, Tang L, Qi X, Hou Y, Cai L. et al. Interactions of Vitamin D receptor gene polymorphism with smoking factor on non-small lung cancer risk. Chin J Gerontol. 2013;33:3844-6
29. Kaabachi W, Kaabachi S, Rafrafi A, Amor AB, Tizaoui K, Haj Sassi F. et al. Association of vitamin D receptor FokI and ApaI polymorphisms with lung cancer risk in Tunisian population. Mol Biol Rep. 2014;41:6545-53
30. Wu X, Cheng J, Yang K. Vitamin D-Related Gene Polymorphisms, Plasma 25-Hydroxy-Vitamin D, Cigarette Smoke and Non-Small Cell Lung Cancer (NSCLC) Risk. Int J Mol Sci. 2016;17:1597 -
31. Bi X, Jia Y, Liu W, Liu L, Zang A. Vitamin D receptor gene polymorphism and genetic susceptibility of nonsmall cell lung cancer. J Trop Med. 2016;16:905-8 +18
32. Yang Y, Wu J, Zhou W, Hou Y, Yin R, Wang Z. Association of vitamin D receptor gene Apa1 and Taq1 polymorphisms with susceptibility to lung squamous cell carcinoma. JOURNAL OF HEBEI MEDICAL UNIVERSITY. 2017;38:20-3 +71
33. Çiçek H, Güleken N, Öztuzcu S, Sevinç A. Vitamin D receptor polymorphisms and related biochemical parameters in various cancer species. Turkish Journal of Biochemistry. 2017;42:161-8
34. Shaw G. Polymorphism and single nucleotide polymorphisms (SNPs). BJU international. 2013;112:664-5
35. Holick MF, Chen TC. Vitamin D deficiency: a worldwide problem with health consequences. Am J Clin Nutr. 2008;87:1080S-6S
36. Vojdeman FJ, Madsen CM, Frederiksen K, Durup D, Olsen A, Hansen L. et al. Vitamin D levels and Cancer Incidence in 217.244 individuals from Primary Health Care in Denmark. Int J Cancer. 2019 [Epub ahead of print]
37. Sanghera DK, Sapkota BR, Aston CE, Blackett PR. Vitamin D Status, Gender Differences, and Cardiometabolic Health Disparities. Ann Nutr Metab. 2017;70:79-87
38. Deeb KK, Trump DL, Johnson CS. Vitamin D signalling pathways in cancer: potential for anticancer therapeutics. Nature reviews Cancer. 2007;7:684-700
39. Zhong H, Zhou R, Feng Y, Zheng GX, Liang Y, Zhang JY. et al. Association of vitamin D receptor gene polymorphism with the risk of lung cancer: a meta-analysis. Journal of receptor and signal transduction research. 2014;34:500-5
40. Fu Y, Li J, Zhang Y. Polymorphisms in the vitamin D receptor gene and the lung cancer risk. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:1323-30
41. Yu ZH, Chen M, Zhang QQ, Hu X. The Association of Vitamin D Receptor Gene Polymorphism with Lung Cancer Risk: An Update Meta-analysis. Combinatorial chemistry & high throughput screening. 2018;21:704-10
42. Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108:545-56
Author contact
![]() Corresponding author: Mingwei Chen, Ph.D., The Department of Respiratory and Critical Care Medicine of the First Affiliated Hospital of Xi'an Jiaotong University. 277 West Yanta Road, Xi'an, Shaanxi 710061, People's Republic of China; Tel: +86-29-85323850; Fax: +86-29-85323850; Email: chenmw36com.
Corresponding author: Mingwei Chen, Ph.D., The Department of Respiratory and Critical Care Medicine of the First Affiliated Hospital of Xi'an Jiaotong University. 277 West Yanta Road, Xi'an, Shaanxi 710061, People's Republic of China; Tel: +86-29-85323850; Fax: +86-29-85323850; Email: chenmw36com.

 Global reach, higher impact
Global reach, higher impact