Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(19):4679-4688. doi:10.7150/jca.29398 This issue Cite
Research Paper
Glutathione S-transferase genes variants and glioma risk: A case-control and meta-analysis study
1. Department of Neurology, Xiangya Hospital, Central South University, Changsha 410008, People's Republic of China
2. Department of Neurosurgery, Xiangya Hospital, Central South University, Changsha 410008, People's Republic of China
3. Department of Pharmacy, the Second Xiangya Hospital, Central South University; Institute of Clinical Pharmacy, Central South University, Changsha 410011, People's Republic of China.
4. Department of Pharmacy, Xiangya Hospital, Central South University, Changsha 410078, People's Republic of China
Received 2018-8-21; Accepted 2019-6-23; Published 2019-8-19
Abstract
Background: The glutathione S-transferase (GST) genes encode enzymes that metabolize carcinogenic compounds, and their variants, GSTP1 (Ile105Val and Ala114Val), GSTT1 (null/present), and GSTM1 (null/present), reduce enzyme activity that may affect the risk of developing cerebral glioma. This study undertook a case-control study and a meta-analysis to evaluate associations between these GST gene variants and the risk of glioma.
Methods: The study enrolled 384 glioma patients (194 men and 190 women; mean age, 48.3 ± 9.2 years) and 340 healthy controls (174 men and 166 women; mean age, 46.5 ± 9.8 years). The amplification refractory mutation system assay was performed to identify GST gene variants of all 724 subjects. A meta-analysis enrolled 15 studies (including our case-control results) was performed.
Results: Our case-control study found that the frequency of GSTP1 Ile105Val Val/Val genotype was significantly higher in the glioma group than that in the healthy controls (11.7% vs. 6.4%) (OR=1.50; 95% CI=1.05-2.04; P=0.01); the frequency of the Val/Ile + Ile/Ile genotypes was different from glioma patients and controls (88.3% vs. 93.6%) (OR=1.47(1.04-2.10); P=0.015); there were no associations between GSTP1 Ala114Val, GSTT1 (null/present) and GSTM1 (null/present) variants and glioma risk. Our meta-analysis confirmed that the GSTP1 Ile105Val variant was associated with an overall increased glioma risk. Moreover, our meta-analysis also confirmed the GSTP1 Ala114Val and GSTT1 null/present variants were associated with an increased glioma risk in the Caucasian population, rather than the Asian population.
Conclusions: This study showed that GST gene variants were associated with an increased risk of glioma with ethnic differences. Future large-scale, multi center, controlled, prospective studies are required to support these findings and to determine how these GST gene variants may affect the pathogenesis of glioma.
Keywords: glutathione S-transferase genes, glioma, gene polymorphism, risk, meta-analysis
Introduction
Glioma accounts for approximately 80% of all brain tumors and are associated with poor patient survival [1]. To the best of our knowledge, no environmental carcinogens, apart from ionizing radiation, have been implicated in the etiology of gliomas [2-4]. However, studies in rats demonstrated that brain tumors maybe induced by various carcinogenic substances, such as acrylonitrile, ethylene oxide and acrylamide [5-7]. However, studies in humans reported no definitive association with occupational or environmental exposure and the risk of primary brain tumor [8-10]. Several individuals may be exposed to toxic agents over time. Therefore, some researchers have focused on the polymorphisms in the glutathione-S-transferase (GST) A (GSTM1), u (GSTT1) and k (GSTP1) variants, which have been reported to be associated with the etiology of cerebral tumors [11-23]. These enzymes, as part of the phase II detoxification process, are involved in the metabolism of several electrophilic compounds, including carcinogens, mutagens, cytotoxic drugs and metabolites, and the detoxification of the products of reactive oxidation [24-26].
Previously published studies have shown that variants of the GSTP1 gene (Ile105Val and Ala114Val), variants of the GSTT1 gene (null/present), and variants of the GSTM1 gene (null/present) might result in significant changes in the function of the GST enzymes [1, 25, 27-33]. The associations with these GST gene variants and cancer risk has been investigated in previous studies [34-36], particularly in association with the risk of developing glioma [11-13]. However, the cumulative results remain inconclusive due to differences in ethnicity, the number of research subjects, patient age, and subtypes of glioma. There have been only two studies, but with inconsistent results, that have addressed the association with these GST gene variants and the risk of glioma in the Han Chinese population [11, 37]. Therefore, the aim of this case-control study was to investigate the relationship between GSTP1 (Ile105Val and Ala114Val), GSTT1 (null/present) and GSTM1 (null/present) variants and the risk of glioma in the Han Chinese population compared with the Caucasian population.
Due to the inconsistent findings in previously published studies regarding the association with GST gene variants and the risk of glioma, it was deemed necessary to perform a meta-analysis. At this time, there have been several previously published and formally analyzed systematic reviews and meta-analysis of published studies on the GSTP1, GSTM1 and GSTT1 variants and the risk of primary brain tumors, including glioma [1, 2, 38-40]. However, the results of these previous meta-analyses are inconclusive, or have not been updated, or do not include racial subgroup analyses. Therefore, a meta-analysis was performed to specifically investigate the association with GSTP1 (Ile105Val and Ala114Val), GSTT1 (null/present) and GSTM1 (null/present) variants and the risk of glioma that included data from this case-control study of 724 patients combined with other recently published and relevant studies.
Materials and methods
Case-control study subjects
The study subjects, including glioma patients and healthy controls, were enrolled from the Xiangya Hospital of Central South University (Changsha, China) between 2006 and 2013. All cases included in this study were histologically confirmed as glioma. The healthy controls were Han Chinese people, with no tumor and normal on physical examination and who were randomly selected from the Medical Examination Center of Xiangya Hospital.
The Chinese Clinical Trial Register approved the clinical study admission (Registration No. ChiCTR-COC-15006186), and the Ethics Committees of the Xiangya School of Medicine and the Institute of Clinical Pharmacology of Central South University approved the study protocol. A standardized questionnaire was used to collect demographic details and clinical data, including patient age, sex, smoking status, and glioma types. All the patients provided written informed consent in compliance with the Code of Ethics of the World Medical Association (Declaration of Helsinki) prior to participating in the study.
Genotyping
DNA was isolated from 3ml of whole blood samples, using the phenol-chloroform extraction method, and then stored at 4°C. An amplification refractory mutation system assay, described by Hemmingsen et al, was performed to identify four alleles of GSTP1 using two different sets of primers amplifying exon 5 and exon 6 [41].
The primers included a forward primer upstream of the codon 105 substitution (5′-ACCCCAGGGCTCTATGGGAA-3′) and two reverse primers, primer 'A' (5′-TCACATAGTCATCCTTGCCGG-3′; Ala114-specific) and primer 'B' (5′-TCACATAGTCATCCTTGCCGA-3′; Val114-specific).
The polymerase chain reaction products were digested with the BsmA1 restriction enzyme (New England BioLabs, Inc., Ipswich, MA, USA) and resolved using a 3.5% agarose gel to identify the Ile105Val variant. GSTT1 and GSTM1 null/present variants were detected by the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method, as previously described [13].
Meta-analysis literature and search strategy
Two investigators (MQZ and YJW) searched PubMed, PMC, EMBASE, Web of Science, and WanFang databases using the following search terms: 'glutathione S-transferase P1', 'glutathione S-transferase M1', 'glutathione S-transferase T1', 'GSTP1', 'GSTM1', 'GSTT1' combined with 'glioma' or 'brain tumor'. Studies that met the following inclusion criteria were included in the analysis: studies on GSTP1, GSTM1, GSTT1 variants and risk of brain cancer, including glioma; and available genotypic frequencies. The exclusion criteria included: the lack of a control group; duplicate publications; and no available data on genotypic frequencies. Literature search updated on January 23, 2019. The information collected from each study included author name, year of publication, patient ethnicity, and frequency of variants in both groups. The Newcastle-Ottawa scale was used for quality assessment of literature reviewed [55].
Statistical analysis
The SPSS software for Windows, version 13.0 (SPSS Inc., Chicago, IL, USA) was used for statistical analysis. The Hardy-Weinberg equilibrium was analyzed with the χ2 test. Data on age, sex and other characteristics were compared to glioma patients and healthy controls with the Student's t-test or χ2 analysis. The different distributions of the GSTP1 variants of the two groups were examined by the χ2 test. P<0.05 was considered to indicate statistically significant differences. The meta-analysis was performed using STATA version 12 (Stata Corp, College Station, TX, USA).The significances of the pooled odds ratio (OR) were tested by the Z-test, and P<0.05 was considered to be statistically significant. Heterogeneity was calculated with the χ2 analysis based on the Q-test and I2. If P<0.05 and I2>50% were present, the Mantel-Haenszel random effect model was used, otherwise a fixed effect model was used [42, 43]. Data was combined using fixed-effects models if the heterogeneity was significant. The potential publication bias was assessed using Egger's test and Begg's test.
Results
Patient characteristics
Of the 384 patients with glioma (194 men and 190 women) (mean age, 48.3 ± 9.2 years), and the 340 healthy controls (174 men and 166 women) (mean age, 46.5 ± 9.8 years) were enrolled in the case-controlled study. The demographic and clinical characteristics of the patients and controls are summarized in Table 1. The 384 patients included 128 (33.3%) cases of glioblastoma; 126 (32.8%) cases of aplastic astrocytoma, diffuse astrocytoma or other types of astrocytoma; and 130 (33.9%) cases of other gliomas. There was no difference in gender and age distribution between patient and controls.
Demographic and clinical characteristics of the subjects.
| Parameters | Patients, N (%) (N=384) | Healthy controls, N (%) (N=340) | P- value |
|---|---|---|---|
| Male gender | 194 (50.5) | 174 (51.2) | - |
| Female gender | 190 (48.5) | 166 (48.8) | 0.84 |
| Age (years), mean ± SD | 48.3 ± 9.2 | 46.5 ± 9.8 | 0.73 |
| Smoking (yes/no) | 223/161 | 182/158 | 0.45 |
| Glioma type | |||
| Glioblastoma | 128 (33.3) | - | - |
| Astrocytomas (except for glioblastoma) | 126 (32.8) | - | - |
| Other | 130 (33.9) | - | - |
SD, standard deviation.
GSTP1 (Ile105Val and Ala114Val), GSTM1 (null/present) and GSTT1 (null/present) variants and glioma risk
GSTP1 (Ile105Val and Ala114Val), GSTM1 (null/present) and GSTT1 (null/present) variants were genotyped in 384 glioma patients and 340 healthy controls. The investigated SNPs were all in Hardy-Weinberg equilibrium. As shown in Table 2, the frequency of the Ile105Val GG genotype was significantly greater in the glioma group compared with that in the healthy controls (11.7% vs. 6.4%) (OR=1.50(1.05-2.04); P=0.01). The frequencies of the GA + AA genotypes were significantly different from the two groups (88.3 vs. 93.6%) (OR=1.47(1.04-2.10); P=0.015). There was no difference in the allele or genotype frequencies of Ala114Val (rs1138272) between glioma patients and healthy controls. As shown in Table 2, there were no differences in the frequencies of GSTM1 (null/present) and GSTT1 (null/present) variants between Han Chinese glioma patients and healthy controls: GSTM1 (43% vs. 48%; OR=1.07(0.93-1.21); P=0.31); GSTT1 (41.1% vs. 34.1%; OR=0.89(0.81-1.06); P=0.052).
Meta-analysis study review and selection
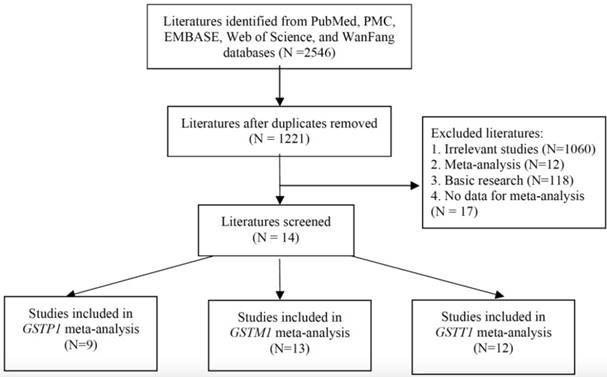
Figure 1 shows the literature review selection procedure. A total of 2546 publications were selected from the PubMed, PMC, EMBASE, WanFang, and Web of Science databases. Following removal of duplicate publications and 1221 abstracts were retrieved. A total of 1060 irrelevant studies, 12 meta-analyses, 118 basic research studies, and e17 studies without detailed information were excluded, according to the publication selection criteria. Finally, 15 studies (including our case-control data) were deemed eligible for review (Table 3). There were 9 publications that included GSTP1 variants, 13 publications that included GSTM1 variants and 12 publications that included GSTT1 variants. The characteristics of the included publications are summarized in Table 3.
Meta-analysis about GSTP1 Ile105Val and Ala114Val variants and glioma risk
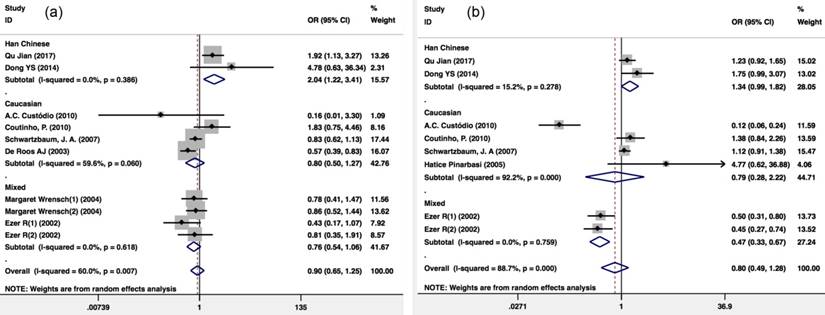
A total of 9 publications cover GSTP1 variants. According to the GSTP1 Ile105Val recessive model (Val/Val vs. Ile carriers), the pooling ORs of eight publications including 4,128 glioma patients and 2,134 healthy controls showed a significant association. Because there was heterogeneity (I2=60%; P=0.007), a Mantel-Haenszel random-pooling model was used to analyze the published data. The results showed that there was significant difference risk of glioma between the GSTP1 Ile105Val Val/Val genotype and Ile carriers among the Han Chinese population (OR=2.035(1.216-3.406); P=0.007) compared with that expected in the overall population (OR=0.902(0.650-1.251); P=0.535), the Caucasian population (OR=0.799(0.504-1.268); P=0.342) and a mixed population (OR=0.756(0.541-1.057); P=0.102) (Figure 2a).
There were 7 publications that included 3,265 glioma patients and 1,273 healthy controls that included the GSTP1 Ile105Val variant dominant model (Val carriers vs. Ile/Ile). Because there was heterogeneity (I2=88.7%; P<0.001), a Mantel-Haenszel random-pooling model was used to analyze the published data. The results showed that the distribution of GSTP1 Ile105Val Val variant carriers in glioma and healthy controls showed no difference (OR=0.797(0.495-1.285); P=0.352) (Figure 2b).
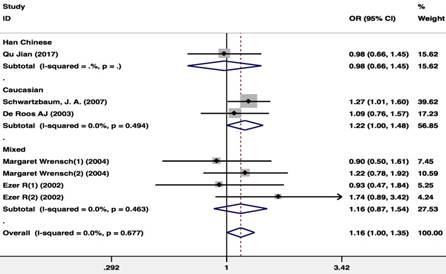
There were 5 publications including 2,342 glioma patients and 3,203 healthy controls included in the publications that included the GSTP1 Ala114Val polymorphism dominant model (Val carriers vs. Ala/Ala). Because there was no study heterogeneity (I2=0%; P=0.677), a Mantel-Haenszel fixed pooling model was used for analysis. The GSTP1 Ala114Val carriers were associated with an increased risk of glioma in the overall population (OR=1.163(1.002- 1.350); P=0.047) and in the Caucasian population (OR=1.215(1.001-1.476); P=0.049) (Figure 3).
Allele and genotype frequencies of the two SNPs in glioma patients (n=384) and healthy controls (n=340).
| SNP | Genotype | Case | Control | P-value | Odds ratio |
|---|---|---|---|---|---|
| (384) | (340) | (95%CI) | |||
| GSTP1 Ile105Val rs1695 | A | 542(70.6%) | 515(75.7%) | Reference | |
| G | 226(29.4%) | 191(24.3%) | 0.31 | 1.06(0.94-1.20) | |
| AA | 203(52.9%) | 197(58.0%) | Reference | ||
| AG | 136(35.4%) | 121(35.6%) | 0.59 | 1.05(0.89-1.23) | |
| GG | 45(11.7%) | 22(6.4%) | 0.01 | 1.50(1.05-2.04) | |
| AG+GG vs. AA | 181(47.1%) | 143(42%) | 0.17 | 1.12(0.95-1.31) | |
| AA+AG vs. GG | 339(88.3%) | 318(93.6%) | 0.015 | 1.47(1.04-2.10) | |
| GSTP1 Ala114Val rs1138272 | C | 687(89.5%) | 613(90.1%) | Reference | |
| T | 81(10.5%) | 67(9.9%) | 0.66 | 1.04(0.87-1.26) | |
| CC | 322(83.9%) | 284(83.5%) | Reference | ||
| CT | 43(11.2%) | 45(13.2%) | 0.45 | 0.92(0.74-1.14) | |
| TT | 19(4.9%) | 11(3.3%) | 0.27 | 1.28(0.79-2.06) | |
| CT+TT vs. CC | 62(16.1%) | 56(16.5%) | 0.91 | 0.99(0.80-1.22) | |
| CC+CT vs.TT | 365(95.1%) | 329(96.7%) | 0.25 | 1.29(0.80-2.08) | |
| GSTM1 | Null | 165(43%) | 159(46.8%) | 0.31 | 1.07(0.93-1.21) |
| Present | 219(57.0%) | 181(53.2%) | Reference | ||
| GSTT1 | Null | 158(41.1%) | 116(34.1%) | 0.052 | 0.89(0.81-1.06) |
| Present | 226(58.9%) | 224(65.9%) | Reference |
Bold print indicates statistical significance; SNP, single-nucleotide polymorphism; CI, confidence interval.
Selection of studies for inclusion in the meta-analysis.
Characteristics of studies included in the meta-analysis.
| Authors | Year | Ethnicity | NOS Score | GSTP1 rs1695 Case | GSTP1 rs1695 Control | GSTP1 rs1138272 Case | GSTP1 rs1138272 Control | GSTM1 Control | GSTM1 Case | GSTT1 Control | GSTT1 Case | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | AA | AG | GG | CC | CT | TT | CC | CT | TT | (-) | (+) | (-) | (+) | (-) | (+) | (-) | (+) | ||||
| Qu Jian | 2017 | Han Chinese | 8 | 203 | 136 | 45 | 197 | 121 | 22 | 322 | 43 | 19 | 284 | 45 | 11 | 159 | 181 | 165 | 219 | 116 | 224 | 158 | 226 |
| Dong YS[11] | 2014 | Han Chinese | 7 | 180 | 102 | 19 | 52 | 19 | 1 | ||||||||||||||
| A.C. Custódio[12] | 2010 | Caucasian | 7 | 87 | 13 | 0 | 35 | 43 | 2 | ||||||||||||||
| Coutinho, P.[13] | 2010 | Caucasian | 6 | 129 | 172 | 46 | 35 | 37 | 6 | 153 | 194 | 37 | 41 | 76 | 271 | 11 | 67 | ||||||
| Schwartzbaum, J. A.[14] | 2007 | Caucasian | 9 | 682 | 716 | 193 | 216 | 189 | 67 | 580 | 138 | 1344 | 252 | 237 | 193 | 119 | 111 | 68 | 362 | 46 | 185 | ||
| Hatice Pinarbasi[15] | 2005 | Caucasian | 7 | 132 | 21 | 30 | 1 | 37 | 116 | 15 | 16 | 31 | 122 | 10 | 21 | ||||||||
| Margaret Wrensch(1)[16] | 2004 | Mixed | 7 | 159 | 21 | 136 | 23 | 153 | 28 | 128 | 26 | 84 | 82 | 93 | 91 | 35 | 132 | 36 | 147 | ||||
| Margaret Wrensch(2)[16] | 2004 | Mixed | 7 | 241 | 28 | 297 | 40 | 228 | 43 | 292 | 45 | 177 | 160 | 140 | 124 | 74 | 263 | 54 | 210 | ||||
| Ezer R(1)[17] | 2002 | Mixed | 7 | 139 | 71 | 10 | 46 | 44 | 10 | 192 | 28 | 1 | 86 | 13 | 1 | 49 | 51 | 120 | 101 | 18 | 82 | 29 | 191 |
| Ezer R(2)[17] | 2002 | Mixed | 7 | 111 | 44 | 14 | 46 | 44 | 10 | 127 | 27 | 9 | 86 | 13 | 1 | 49 | 51 | 60 | 112 | 18 | 82 | 25 | 145 |
| De Roos AJ[18] | 2003 | Caucasian | 6 | 508 | 59 | 329 | 67 | 345 | 59 | 498 | 78 | 321 | 254 | 212 | 191 | 100 | 445 | 77 | 309 | ||||
| Salnikova LE[19] | 2013 | Caucasian | 7 | 202 | 261 | 99 | 32 | 96 | 367 | 37 | 124 | ||||||||||||
| Elexpuru-Camiruaga J[20] | 1995 | Caucasian | 6 | 351 | 262 | 130 | 88 | 91 | 403 | 70 | 148 | ||||||||||||
| Trizna[21] | 1998 | Caucasian | 7 | 39 | 51 | 47 | 43 | 27 | 63 | 25 | 65 | ||||||||||||
| Hand PA[22] | 1996 | Caucasian | 6 | 121 | 90 | 51 | 38 | 56 | 228 | 30 | 59 | ||||||||||||
| Wiencke[23] | 1997 | Caucasian | 6 | 305 | 157 | 334 | 158 | ||||||||||||||||
| Butler[62] | 2003 | Caucasian | 8 | 294 | 285 | 164 | 161 | 85 | 494 | 56 | 269 | ||||||||||||
Meta-analysis of the GSTP1 Ile105Val polymorphism and glioma risk. (a) Val/Val vs. Ile carriers; (b) Val carriers vs. Ile/Ile. CI, confidence interval.
Meta-analysis of the GSTP1 Ala114Val polymorphism and glioma risk (Val carriers vs. Ala/Ala).CI, confidence interval.
Meta-analysis of GSTT1 and GSTM1 null/present variants and glioma risk. (a) GSTM1 null/present; (b) GSTT1 null/present. CI, confidence interval.
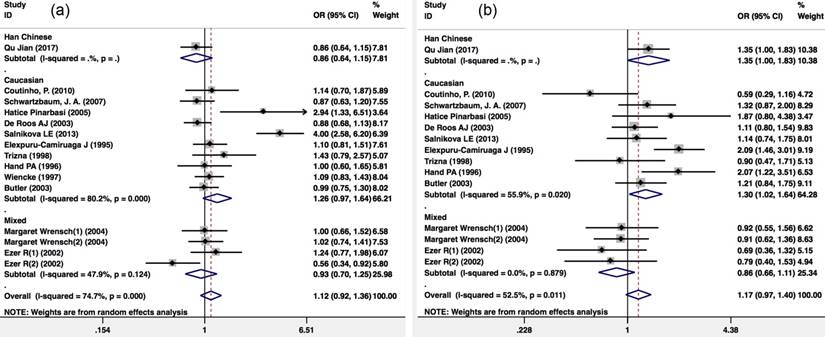
Meta-analysis about GSTM1 (null/present) and GSTT1 (null/present) variants and the risk of glioma
There were 13 publications that included 2,312 glioma patients and 4,966 healthy controls included in the publication analysis of the GSTM1 null/present variant and glioma risk. Because there was heterogeneity (I2=74.7%; P<0.001), a Mantel-Haenszel random-pooling model was used in the analysis. No significant association was found between the GSTM1 null/present variant and glioma risk of the overall population (OR=1.121(0.924-1.360); P=0.247), the Caucasian population (OR=1.260(0.966-1.643); P=0.088) and the overall mixed population (OR=0.935(0.698-1.252); P=0.651) (Figure 4a).
There were 12 publications that included 2,830 glioma patients and 4,429 healthy controls included in the publication analysis that included the GSTT1 (null/present) variant and glioma risk. Because there was heterogeneity (I2=52.5%; P=0.011), a Mantel-Haenszel random-pooling model was used in the analysis. There was no significant association with GSTT1 (null/present) variant and glioma risk in the Caucasian population (OR=1.296(1.023-1.641); P=0.031) (Figure 4b).
Publication bias and sensitivity analysis
Publication bias was examined by Egger's test and Begg's test. For all pooled OR analysis, Egger's test and Begg's test showed no publication bias (Figure S1). For the pooling ORs of GSTP1 Ile105Val Val/Val vs. Ile carriers in the Han Chinese population and GSTP1 Ala114Val Val carriers vs. Ala/Ala in the overall population and Caucasian population, sensitivity analysis showed that changing the analysis model (fixed or random model) had no significant effects on the pooled ORs and the final strength of the association (Table 4). For the pooling ORs of GSTT1 null vs. present in Caucasians, sensitivity analysis found that changing the analysis model (fixed or random model) had no significant effect on the pooled OR and the final strength of the association.
Discussion
The GST gene variants, GSTP1, GSTT1, and GSTM1, encode enzymes that metabolize carcinogenic compounds and reduced enzyme activity that may affect the risk of developing cerebral glioma. This case-control study investigated the associations with GSTP1 (Ile105Val and Ala114Val), GSTT1 (null/present) and GSTM1 (null/present) variants and the risk of cerebral glioma. The study was supported by meta-analysis on GST gene variants and cerebral glioma in the Han Chinese population, which showed that GSTP1 Ile105Val and GSTT1 null/present variants were associated with the risk of glioma.
Susceptibility genes for the development of glioma have been identified with this study, and the findings are supported by previous studies. It has been previously reported that the rs25489 (Arg280His) and Arg399Gln (rs25487) polymorphisms of the XRCC1 gene may affect the risk of developing glioma in the Chinese population [44]. However, the development of cerebral glioma can only be partly explained by these gene findings, because glioma is a complex tumor, and its development may be associated with susceptibility to multiple genes as well as environmental factors.GST is known to catalyze the detoxification of reactive electrophilic compounds, such as cytotoxic drugs, and GSTs are a large family of cytosolic phase II xenobiotic metabolizing enzymes [45, 46]. GST enzymes are associated with the accumulation of increased amounts of reactive oxygen species, which is a phenomenon involved in carcinogenesis [47]. The individual variability in the GST enzyme activity may lead to different effects of detoxification of potential carcinogens and result in susceptibility to cancer [45, 46]. The enzymes, GSTP1, GSTT1 and GSTM1 are the predominant enzymes in the GST enzyme family, and they have been proposed to play a critical role in protecting the brain from toxic compounds [48, 49]. GSTP1 I105V and A114V and deletions in the GSTM1 and GSTT1 genes could substantially reduce the activity of these protective GST enzymes [1, 32, 33, 35].
It has been previously reported that GSTP1 is associated with susceptibility to other types of malignancy, including prostate cancer[50] and breast cancer [31]. Also, previous studies have shown that polymorphisms of the GSTP1gene may affect the response to chemotherapy and treatment outcome in breast cancer [51], and may also affect epirubicin treatment outcome as well as epirubicin-related toxicity [29]. These findings indicate that the GSTP1 gene is an important gene associated with the development and treatment of cancer.
Previous case-control studies assessed the association with GSTP1 polymorphisms and the risk of developing glioma, but the reported findings have been inconsistent. Gao et al. analyzed the association with GSTP1 polymorphisms and the risk of developing glioma in 301 patients with glioma and 302 healthy controls and reported a positive result for GSTP1 Ile105Val (rs1695) [37]. Casartelliet al. also found the GSTP1 Ile105Val polymorphism to be involved in the susceptibility to astrocytoma and the development of glioblastoma multiforme (GBM) [12]. However, there have also been studies reporting negative results [1, 2, 13]. This inconsistency may be due to the differences in patient age, glioma subtype, and ethnicity. Some studies combined subtypes of glioma to analyze the distribution of GSTP1 gene polymorphisms. Therefore, more patients are required to classify the subtypes of glioma and analyze the polymorphisms by subtype.
The results of this study were consistent with those of Qin et al. and Gao et al. both involving the Han Chinese population [11, 37]. In the Qin et al. study, the distribution of rs1695 in GSTP1 allelic frequencies was significantly different from 72 cases of GBM and 302 control cases in the Han Chinese population. Furthermore, rs1695 in GSTP1 decreased the risk of GBM in the log-additive model (OR=0.56; 95% CI, 0.34-0.94; P=0.022) [11]. In the present study, GBM comprised 33.3% of total glioma cases. Gao et al. investigated 301 glioma patients and 302 controls in a case-controlled study and found a positive association between GSTP1 rs1695 and glioma risk. In the present study, Ile105Val GG genotype patients had a 1.5 times increase in risk compared with wild-type patients, and GG genotype patients had a 1.47 times increase in risk compared with AA+AG patients. Furthermore, both the literature review and the analysis undertaken in this study confirmed the positive associations.
Meta-analysis of the association between GSTP1, GSTM1 and GSTT1 variants and glioma risk.
| Genetic comparisons | No. of studies | Study groups | Test of association | Test of heterogeneity | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR/HR(95% CI) | Z | P-value | Model | χ2 | P-value | I2(%) | Tau-squared | |||
| GSTP1 | ||||||||||
| Val/Val vs. Ile carriers | 8 | Overall | 0.902(0.650-1.251) | 0.62 | 0.535 | R | 22.5 | 0.007 | 60% | 0.137 |
| 2 | Han Chinese | 2.035(1.216-3.406) | 2.7 | 0.007 | R | 0.75 | 0.386 | 0.00% | 0 | |
| 2.115(1.269-3.524) | 2.87 | 0.004 | F | |||||||
| 4 | Caucasian | 0.799(0.504-1.268) | 0.95 | 0.342 | R | 7.42 | 0.06 | 59.60% | 0.1085 | |
| 2 | Mixed | 0.756(0.541-1.057) | 1.63 | 0.102 | R | 1.79 | 0.618 | 0.00% | 0 | |
| Val carriers vs. Ile/Ile | 7 | Overall | 0.797(0.495-1.285) | 0.93 | 0.352 | R | 61.84 | < 0.001 | 88.70% | 0.3722 |
| 2 | Han Chinese | 1.345(0.994-1.819) | 1.92 | 0.055 | R | 1.18 | 0.278 | 15.20% | 0.0095 | |
| 4 | Caucasian | 0.794(0.284-2.218) | 0.44 | 0.66 | R | 38.38 | < 0.001 | 92.20% | 0.888 | |
| Val carriers vs. Ala/Ala | 5 | Overall | 1.163(1.002-1.350) | 1.98 | 0.047 | F | 4 | 0.677 | 0.00% | - |
| 1.162(1.001-1.349) | 1.98 | 0.048 | R | |||||||
| 2 | Caucasian | 1.215(1.001-1.476) | 1.97 | 0.049 | F | 0.47 | 0.494 | 0 | - | |
| 1.216(1.002- 1.477) | 1.98 | 0.048 | R | |||||||
| 2 | Mixed | 1.160(0.871-1.543) | 1.02 | 0.31 | F | 2.57 | 0.463 | 0 | - | |
| GSTM1 null vs. present | 13 | Overall | 1.121(0.924-1.360) | 1.16 | 0.245 | R | 55.3 | < 0.001 | 74.70% | 0.102 |
| 10 | Caucasian | 1.260(0.966-1.643) | 1.71 | 0.088 | R | 45.52 | < 0.001 | 80.20% | 0.1372 | |
| 2 | Mixed | 0.935(0.698-1.252) | 0.45 | 0.651 | R | 5.76 | 0.124 | 47.90% | 0.0424 | |
| GSTT1null vs. present | 12 | Overall | 1.168(0.974-1.400) | 1.67 | 0.094 | R | 27.39 | 0.011 | 52.50% | 0.0587 |
| 9 | Caucasian | 1.296(1.023-1.641) | 2.15 | 0.031 | R | 18.14 | 0.02 | 55.90% | 0.0686 | |
| 1.301(1.122-1.509) | 3.48 | 0.001 | F | |||||||
| 2 | Mixed | 0.857(0.660-1.112) | 1.16 | 0.245 | F | 0.67 | 0.879 | 0.00% | - | |
OR, odds ratio; CI, confidence interval; vs., versus; F, fixed effect model; R, random effect model.
Among the published genome-wide association studies (GWAS) relevant to the literature review and the analysis undertaken in this study, there was no direct information or data that could be applied to this analysis [52-57]. A previous study reported that the GSTP1 gene Ala114Val (rs1138272) polymorphism might be used as a predictive factor for cisplatin-based chemotherapy in osteosarcoma patients [30]. Another study reported that children carrying GSTP1 rs1138272 minor alleles might represent a susceptible population with increased risk of asthma associated with air pollution [58]. These findings suggest that this variant may be involved in oxidative stress, which is a common phenomenon in the development of glioma. However, no association with this polymorphism with the risk of glioma was observed in the present study. However, meta-analysis undertaken in this study showed that GSTP1 Ala114Val (rs1138272) was associated with the risk of glioma in the overall population and Caucasian population studied.
Ethnicity is an important factor associated with GST gene variants and glioma risk. In the meta-analysis in this study, a significant association was found between GSTP1 Ile105Val (Val/Val vs. Ile carriers) and glioma risk in the Han Chinese population, but not in the Caucasian or mixed population. The GSTT1 null/present variant was only significantly associated with increased risk of glioma in the Caucasian population. These results implied that the effects of GST gene variants affecting glioma was associated with ethnic differences. Further studies are required in future that includes populations of different ethnicities.
In the literature review and analysis undertaken in this study, there were several publications on that showed an association with GSTP1 variants and the risk of glioma [1, 2, 38-40]. The results for GSTP1 Ile105Val (rs1695) were positive [37], but negative results were also reported [1, 2, 39, 40]. The literature review and analysis undertaken in this study showed that the GSTP1 Ala114Val variant was significantly associated with the risk of glioma [38, 39]. The present study showed a positive result of GSTP1 Ile105Val (rs1695). To draw more precise conclusions, meta-analysis was done to investigate GST gene variants and the risk of glioma.
Meta-analysis undertaken in this study included nine publications that showed no significant association with the GSTP1 Ile105Val and Ala114Val variants. The funnel plots of the results of the three comparisons demonstrated that there was publication bias in the GSTP1 Ile105Val analysis; this may have been due to the limited research samples and unpublished negative results. There were no robust results from the exact effects of the GSTP1 variants. A widely expanded large-sample case-control study or GWAS may be necessary to identify the key pathogenic gene or genes associated with glioma.
Heterogeneity is an important element in the meta-analysis, and in this study, publication heterogeneity was found when the ORs were pooled for GSTP1 Ile105Val (Val/Val vs. Ile carriers). To find the source of heterogeneity, an ethnicity subgroup analysis was performed and found significance only in the Han Chinese group of heterogeneity, which implied that ethnicity was part of the source of heterogeneity. Also, a random model and a fixed model were used to confirm the positive results. The Egger's test and Begg's test also showed no publication bias for all pooled OR analysis (Figure S1).
This study had several limitations. The study was performed in a single center and had a limited patient sample size. Because of the limited number of patients, the cases of glioma were combined, without distinguishing between the histological type of glioma, grade, stage, or isocitrate dehydrogenase (IDH) gene status. While there were some studies that found the different genetic susceptibility to glioma subgroups, such as GBM and non-GBM tumors, IDH-mutant and IDH wild-type tumors [59-61].
In conclusion, a case-controlled study, including a total of 724 subjects was performed, and showed that the GSTP1 gene Ile105Val (rs1695) polymorphism was associated with an increased risk of glioma in the Han Chinese population, and the meta-analysis results confirmed this association and also showed that the GSTP1 Ala114Val variant was associated with overall increased risk of glioma and an increased risk in the Caucasian population, and the GSTT1 null/present variant was associated with an increased risk of glioma in the Caucasian population. Future large-scale, multi center, controlled, prospective studies are required to support these findings and to determine how these GST gene variants may affect the pathogenesis of cerebral glioma.
Supplementary Material
Supplementary figures.
Acknowledgements
WPL, QQ and JQ were involved in the conception and design of the study. HYL, MQZ and YJW collected the samples and DNA extraction. MQZ and YJW did the genotyping, literature searches, data extraction, and analyses. HYL, MQZ assessed the data quality. QQ and JQ contributed to data analysis and interpretation of the results. QL and JQ modified manuscript. All authors reviewed and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Xie P, Liang Y, Liang G. et al. Association between GSTP1 Ile105Val polymorphism and glioma risk: a systematic review and meta-analysis. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:493-9
2. Lai R, Crevier L, Thabane L. Genetic polymorphisms of glutathione S-transferases and the risk of adult brain tumors: a meta-analysis. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2005;14:1784-90
3. Xu D, Chen X, Chen K. et al. Tetra-sulfonate phthalocyanine zinc-bovine serum albumin conjugate-mediated photodynamic therapy of human glioma. J Biomater Appl. 2014;29:378-85
4. Yan Y, Peng Y, Ou Y. et al. MicroRNA-610 is downregulated in glioma cells, and inhibits proliferation and motility by directly targeting MDM2. Mol Med Rep. 2016;14:2657-64
5. Rice JM, Wilbourn JD. Tumors of the nervous system in carcinogenic hazard identification. Toxicologic pathology. 2000;28:202-14
6. Liu W, Lv G, Li Y. et al. Downregulation of CDKN2A and suppression of cyclin D1 gene expressions in malignant gliomas. J Exp Clin Cancer Res. 2011;30:76
7. Cui Y, Zhao J, Yi L. et al. microRNA-153 Targets mTORC2 Component Rictor to Inhibit Glioma Cells. PloS one. 2016;11:e0156915
8. Wrensch M, Minn Y, Chew T. et al. Epidemiology of primary brain tumors: current concepts and review of the literature. Neuro-oncology. 2002;4:278-99
9. Downs SH, Black N. The feasibility of creating a checklist for the assessment of the methodological quality both of randomised and non-randomised studies of health care interventions. Journal of epidemiology and community health. 1998;52:377-84
10. Yan Y, Jiang Y. RACK1 affects glioma cell growth and differentiation through the CNTN2-mediated RTK/Ras/MAPK pathway. Int J Mol Med. 2016;37:251-7
11. Dong YS, Hou WG, Li XL. et al. Genetic association of CHEK2, GSTP1, and ERCC1 with glioblastoma in the Han Chinese population. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:4937-41
12. Custodio AC, Almeida LO, Pinto GR. et al. GSTP1 Ile105Val polymorphism in astrocytomas and glioblastomas. Genetics and molecular research: GMR. 2010;9:2328-34
13. Coutinho P, Sandim V, Oliveira JA. et al. Lack of association between glutathione S-transferase polymorphisms and primary glioma in a case-control study in Rio de Janeiro. Genetics and molecular research: GMR. 2010;9:539-44
14. Schwartzbaum JA, Ahlbom A, Lonn S. et al. An international case-control study of glutathione transferase and functionally related polymorphisms and risk of primary adult brain tumors. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2007;16:559-65
15. Pinarbasi H, Silig Y, Gurelik M. Genetic polymorphisms of GSTs and their association with primary brain tumor incidence. Cancer Genet Cytogenet. 2005;156:144-9
16. Wrensch M, Kelsey KT, Liu M. et al. Glutathione-S-transferase variants and adult glioma. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2004;13:461-7
17. Ezer R, Alonso M, Pereira E. et al. Identification of glutathione S-transferase (GST) polymorphisms in brain tumors and association with susceptibility to pediatric astrocytomas. J Neurooncol. 2002;59:123-34
18. De Roos AJ, Rothman N, Inskip PD. et al. Genetic polymorphisms in GSTM1, -P1, -T1, and CYP2E1 and the risk of adult brain tumors. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2003;12:14-22
19. Salnikova LE, Belopolskaya OB, Zelinskaya NI. et al. The potential effect of gender in CYP1A1 and GSTM1 genotype-specific associations with pediatric brain tumor. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2013;34:2709-19
20. Elexpuru-Camiruaga J, Buxton N, Kandula V. et al. Susceptibility to astrocytoma and meningioma: influence of allelism at glutathione S-transferase (GSTT1 and GSTM1) and cytochrome P-450 (CYP2D6) loci. Cancer research. 1995;55:4237-9
21. Trizna Z, de Andrade M, Kyritsis AP. et al. Genetic polymorphisms in glutathione S-transferase mu and theta, N-acetyltransferase, and CYP1A1 and risk of gliomas. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 1998;7:553-5
22. Hand PA, Inskip A, Gilford J. et al. Allelism at the glutathione S-transferase GSTM3 locus: interactions with GSTM1 and GSTT1 as risk factors for astrocytoma. Carcinogenesis. 1996;17:1919-22
23. Wiencke JK, Wrensch MR, Miike R. et al. Population-based study of glutathione S-transferase mu gene deletion in adult glioma cases and controls. Carcinogenesis. 1997;18:1431-3
24. Vasieva O. The many faces of glutathione transferase pi. Current molecular medicine. 2011;11:129-39
25. Strange RC, Spiteri MA, Ramachandran S. et al. Glutathione-S-transferase family of enzymes. Mutation research. 2001;482:21-6
26. Townsend D, Tew K. Cancer drugs, genetic variation and the glutathione-S-transferase gene family. American journal of pharmacogenomics: genomics-related research in drug development and clinical practice. 2003;3:157-72
27. Ishimoto TM, Ali-Osman F. Allelic variants of the human glutathione S-transferase P1 gene confer differential cytoprotection against anticancer agents in Escherichia coli. Pharmacogenetics. 2002;12:543-53
28. Ginsberg G, Smolenski S, Hattis D. et al. Genetic Polymorphism in Glutathione Transferases (GST): Population distribution of GSTM1, T1, and P1 conjugating activity. Journal of toxicology and environmental health Part B, Critical reviews. 2009;12:389-439
29. Deng X, Yang X, Cheng Y. et al. GSTP1 and GSTO1 single nucleotide polymorphisms and the response of bladder cancer patients to intravesical chemotherapy. Scientific reports. 2015;5:14000
30. Goricar K, Kovac V, Jazbec J. et al. Genetic variability of DNA repair mechanisms and glutathione-S-transferase genes influences treatment outcome in osteosarcoma. Cancer epidemiology. 2015;39:182-8
31. Kimi L, Ghatak S, Yadav RP. et al. Relevance of GSTM1, GSTT1 and GSTP1 Gene Polymorphism to Breast Cancer Susceptibility in Mizoram Population, Northeast India. Biochemical genetics. 2015
32. Hu X, Herzog C, Zimniak P. et al. Differential protection against benzo[a]pyrene-7,8-dihydrodiol-9,10-epoxide-induced DNA damage in HepG2 cells stably transfected with allelic variants of pi class human glutathione S-transferase. Cancer research. 1999;59:2358-62
33. Ali-Osman F, Akande O, Antoun G. et al. Molecular cloning, characterization, and expression in Escherichia coli of full-length cDNAs of three human glutathione S-transferase Pi gene variants. Evidence for differential catalytic activity of the encoded proteins. The Journal of biological chemistry. 1997;272:10004-12
34. Rednam S, Scheurer ME, Adesina A. et al. Glutathione S-transferase P1 single nucleotide polymorphism predicts permanent ototoxicity in children with medulloblastoma. Pediatric blood & cancer. 2013;60:593-8
35. Watson MA, Stewart RK, Smith GB. et al. Human glutathione S-transferase P1 polymorphisms: relationship to lung tissue enzyme activity and population frequency distribution. Carcinogenesis. 1998;19:275-80
36. Kilburn L, Okcu MF, Wang T. et al. Glutathione S-transferase polymorphisms are associated with survival in anaplastic glioma patients. Cancer. 2010;116:2242-9
37. Li G, Jin TB, Wei XB. et al. Selected polymorphisms of GSTP1 and TERT were associated with glioma risk in Han Chinese. Cancer epidemiology. 2012;36:525-7
38. Geng P, Li J, Wang N. et al. Genetic Contribution of Polymorphisms in Glutathione S-Transferases to Brain Tumor Risk. Molecular neurobiology. 2016;53:1730-40
39. Fan Z, Wu Y, Shen J. et al. Glutathione S-transferase M1, T1, and P1 polymorphisms and risk of glioma: a meta-analysis. Molecular biology reports. 2013;40:1641-50
40. Yao L, Ji G, Gu A. et al. An updated pooled analysis of glutathione S-transferase genotype polymorphisms and risk of adult gliomas. Asian Pacific journal of cancer prevention: APJCP. 2012;13:157-63
41. Hemmingsen A, Fryer AA, Hepple M. et al. Simultaneous identification of GSTP1 Ile105->Val105 and Ala114->Val114 substitutions using an amplification refractory mutation system polymerase chain reaction assay: studies in patients with asthma. Respiratory research. 2001;2:255-60
42. Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719-48
43. Coory MD. Comment on: Heterogeneity in meta-analysis should be expected and appropriately quantified. Int J Epidemiol. 2010;39:932 author reply 3
44. Li J, Qu Q, Qu J. et al. Association between XRCC1 polymorphisms and glioma risk among Chinese population. Medical oncology. 2014;31:186
45. Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annual review of pharmacology and toxicology. 2005;45:51-88
46. Hayes JD, Strange RC. Glutathione S-transferase polymorphisms and their biological consequences. Pharmacology. 2000;61:154-66
47. Xu Z, Zhu H, Luk JM. et al. Clinical significance of SOD2 and GSTP1 gene polymorphisms in Chinese patients with gastric cancer. Cancer. 2012;118:5489-96
48. Corrigall AV, Kirsch RE. Glutathione S-transferase distribution and concentration in human organs. Biochemistry international. 1988;16:443-8
49. Hayes JD, Pulford DJ. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Critical reviews in biochemistry and molecular biology. 1995;30:445-600
50. Mian OY, Khattab MH, Hedayati M. et al. GSTP1 Loss results in accumulation of oxidative DNA base damage and promotes prostate cancer cell survival following exposure to protracted oxidative stress. The Prostate. 2015
51. Yao S, Qian K, Wang R. et al. [Comparison of the Efficacy and Safety of Icotinib with Standard Second-line Chemotherapy in Previously Treated Advanced Non-small Cell Lung Cancer]. Zhongguo fei ai za zhi = Chinese journal of lung cancer. 2015;18:369-73
52. Kinnersley B, Labussiere M, Holroyd A. et al. Genome-wide association study identifies multiple susceptibility loci for glioma. Nature communications. 2015;6:8559
53. Kinnersley B, Kamatani Y, Labussiere M. et al. Search for new loci and low-frequency variants influencing glioma risk by exome-array analysis. European journal of human genetics: EJHG. 2016;24:717-24
54. Shete S, Hosking FJ, Robertson LB. et al. Genome-wide association study identifies five susceptibility loci for glioma. Nature genetics. 2009;41:899-904
55. Wrensch M, Jenkins RB, Chang JS. et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nature genetics. 2009;41:905-8
56. Stacey SN, Sulem P, Jonasdottir A. et al. A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nature genetics. 2011;43:1098-103
57. Rajaraman P, Melin BS, Wang Z. et al. Genome-wide association study of glioma and meta-analysis. Human genetics. 2012;131:1877-88
58. MacIntyre EA, Brauer M, Melen E. et al. GSTP1 and TNF Gene variants and associations between air pollution and incident childhood asthma: the traffic, asthma and genetics (TAG) study. Environmental health perspectives. 2014;122:418-24
59. Melin BS, Barnholtz-Sloan JS, Wrensch MR. et al. Genome-wide association study of glioma subtypes identifies specific differences in genetic susceptibility to glioblastoma and non-glioblastoma tumors. Nat Genet. 2017;49:789-94
60. Kinnersley B, Mitchell JS, Gousias K. et al. Quantifying the heritability of glioma using genome-wide complex trait analysis. Sci Rep. 2015;5:17267
61. Leu S, von Felten S, Frank S. et al. IDH mutation is associated with higher risk of malignant transformation in low-grade glioma. J Neurooncol. 2016;127:363-72
62. Butler MA, Ruder AM, Daly AK. et al. Polymorphisms in GSTM1, GSTT1, GSTP1 and NAT2 and susceptibility to primary intracranial brain gliomas. Proc Am Assoc Cancer Res. 2003;44:128
Author contact
![]() Corresponding authors: Dr. Weiping Liu, Department of Neurology, Xiangya Hospital, Central South University. No.87 Xiangya Road, Changsha, China, 410008; Tel.: +86-13507484320, Fax number: +86-0731-84327216, E-mail: Aliu1220com OR Dr. Jian Qu, Department of Pharmacy, the Second Xiangya Hospital, Central South University; Institute of Clinical Pharmacy, Central South University. No.139 Middle Renmin Road, Changsha, China, 410011; Tel.: +86-15973190614, Fax number: +86-0731-85292128, E-mail: qujianstanleyedu.cn OR Dr. Qiang Qu, Department of Pharmacy, Xiangya Hospital, Central South University. No.87 Xiangya Road, Changsha, China, 410008; Tel.: +86-1511466106, Fax number: +86-0731-85295864, Email: quqiangedu.cn
Corresponding authors: Dr. Weiping Liu, Department of Neurology, Xiangya Hospital, Central South University. No.87 Xiangya Road, Changsha, China, 410008; Tel.: +86-13507484320, Fax number: +86-0731-84327216, E-mail: Aliu1220com OR Dr. Jian Qu, Department of Pharmacy, the Second Xiangya Hospital, Central South University; Institute of Clinical Pharmacy, Central South University. No.139 Middle Renmin Road, Changsha, China, 410011; Tel.: +86-15973190614, Fax number: +86-0731-85292128, E-mail: qujianstanleyedu.cn OR Dr. Qiang Qu, Department of Pharmacy, Xiangya Hospital, Central South University. No.87 Xiangya Road, Changsha, China, 410008; Tel.: +86-1511466106, Fax number: +86-0731-85295864, Email: quqiangedu.cn

 Global reach, higher impact
Global reach, higher impact