Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(21):5130-5138. doi:10.7150/jca.34741 This issue Cite
Research Paper
Comprehensive biomarker analyses identifies HER2, EGFR, MET RNA expression and thymidylate synthase 5'UTR SNP as predictors of benefit from S-1 adjuvant chemotherapy in Japanese patients with stage II/III gastric cancer
1. Department of Gastric Surgery, National Cancer Center Hospital, Tokyo, Japan
2. Department of Gastrointestinal Surgery, Kanagawa Cancer Center, Yokohama, Japan
3. Department of Biostatistics and Epidemiology, Yokohama City University, Yokohama, Japan
4. Cancer Therapeutics and Stratified Oncology, Genome Institute of Singapore, Singapore
5. Cancer and Stem Cell Biology Program, Duke-NUS Graduate Medical School, Singapore
6. Cancer Science Institute of Singapore, National University of Singapore, Singapore, Singapore
7. Department of Pathology, National University of Singapore, Singapore, Singapore
8. Cancer Therapeutics and Stratified Oncology, Genome Institute of Singapore, Singapore
9. Division of Pathology and Data Analytics, Leeds Institute of Medical Research at St James's, University of Leeds, Leeds, UK
10. Department of Pathology, GROW School for Oncology and Developmental Biology, Maastricht University Medical Center, Maastricht, NL
11. Department of Pathology, Tokyo Metropolitan Geriatric Hospital and Institute of Gerontology, Tokyo, Japan
12. Department of Pathology, Kanagawa Cancer Center, Yokohama, Japan
13. Molecular Pathology and Genetics Division, Kanagawa Cancer Center Research Institute, Yokohama, Japan
14. Department of Biomedical Statistics and Bioinformatics, Graduate School of Medicine, Kyoto University, Kyoto, Japan
#Takaki Yoshikawa and Toru Aoyama equally contributed this article.
* Joint senior authors
Received 2019-3-9; Accepted 2019-6-25; Published 2019-8-28
Abstract
Purpose: A comprehensive molecular analysis was conducted to identify prognostic and predictive markers for adjuvant S-1 chemotherapy in stage II/III Japanese gastric cancer (GC) patients and to evaluate their potential suitability for alternative cytotoxic or targeted drugs.
Experimental Design: We investigated genetic polymorphisms of enzymes potentially involved in 5-fluoruracil (5-FU) metabolism as well as platinum resistance, previously identified genomic subtypes potentially predicting 5-FU benefit, and mRNA expression levels of receptor tyrosine kinases and KRAS as potential treatment targets in a single institution cohort of 252 stage II/III GC patients treated with or without S-1 after D2 gastrectomy.
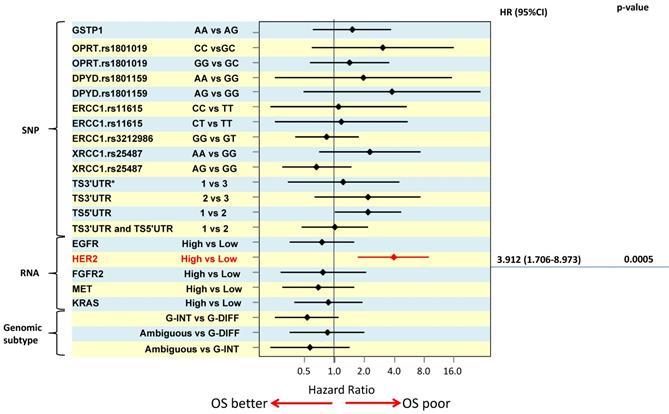
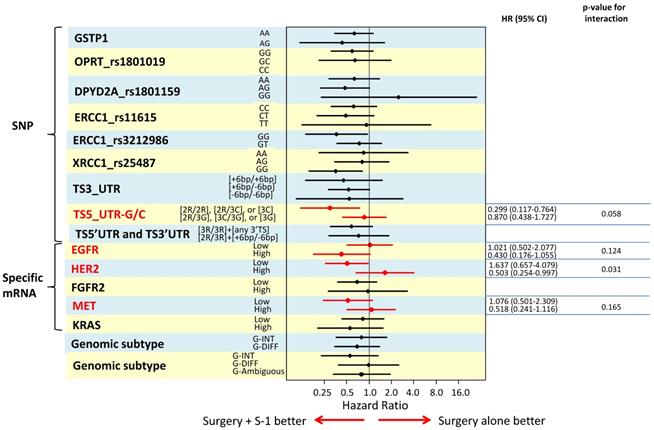
Results: 88% and 62% GC had a potentially 5-FU sensitive phenotype by SNP analyses of TS 3'UTR, and TS 5'UTR, respectively. 24%, 46%, 40%, 5%, and 44% GC had a potentially platinum sensitive phenotype by SNP analyses of GSTP1, ERCC1 rs11615, ERCC1 rs3212986, ERCC2, and XRCC1, respectively. High HER2, EGFR, FGFR2, or MET mRNA expression was observed in 49%, 66%, 72%, and 54% GC, respectively. High HER2 expression was the only significant prognosticator (HR=3.912, 95%CI: 1.706-8.973, p=0.0005). High HER2 (p=0.031), low EGFR (p=0.124), high MET (p=0.165) RNA expression, and TS 5'UTR subtype 2R/2R, 2R/3C, or 3C (p=0.058) were significant independent predictors for S-1 resistance.
Conclusions: The present study suggests that platinum-based or RTK targeted agents could be alternative treatment options for a substantial subgroup of Japanese GC patients currently treated with S-1. HER2, EGFR, MET, and TS 5'UTR SNP appear to be promising predictive markers for S-1 resistance warranting validation in an independent GC series.
Introduction
Gastric cancer (GC) is the fourth most prevalent cancer and the third leading cause of cancer-related death worldwide (1). D2 gastrectomy is the mainstay of treatment for resectable GC in Japan (2), Europe and the US (3, 4). Two recent phase III trials in Eastern Asia demonstrated in patients with pathological stage II/III GC that adjuvant chemotherapy after D2 gastrectomy improved overall survival compared to surgery alone changing clinical practice in Japan (5, 6).
However, adjuvant S-1 chemotherapy does not seem to benefit all patients with stage II/III GC and severe or serious S-1 toxicity including anorexia, nausea, and diarrhea has been reported (5). Furthermore, 29% of ACTS-GC trial patients in the surgery plus adjuvant S-1 arm were not cured, whereas 61% of patients were cured by surgery alone (7) indicating that there is an urgent clinical need to (a) identify biomarkers that can better stratify patients for available treatment options than the currently used TNM stage after surgery and (b) to assess alternative treatment options such as other cytotoxic drugs (e.g. platinum) and/or targeted therapy.
Several previous GC studies have attempted to identify predictive or prognostic markers for S-1 chemotherapy. These studies included the investigation of protein expression of metabolic enzymes of 5-FU or folate such as thymidylate synthase (TS) (8-14), dihydropyrimidine dehydrogenase (DPYD) (8-10), orotate phosphoribosyl transferase (OPRT) (10-12, 14-16), thymidine phosphorylase (11, 13), or methylenetetrahydrofolate reductase (17, 18), the investigation of genetic polymorphisms of excision repair cross-complementing gene 1 (ERCC1) (17, 19, 20), or the investigation of expression of EGFR (11, 17, 21) or VEGF (22). However, all previous studies investigated usually only a small number of markers in each series and were often limited by the absence of a control group, small sample size, or use of a mixture of pre- and post-operative chemotherapy or resectable and unresectable disease. Furthermore, only two studies have investigated the relationship between biomarker and S-1 monotherapy (8, 21), while all other studies have used S-1 in combination with other cytotoxic drugs which may have influenced the results.
The aim of the current study was (a) to conduct a comprehensive molecular analysis of potential prognostic and predictive factors for adjuvant S-1 chemotherapy and (b) to evaluate the potential applicability of alternative cytotoxic or targeted drugs in a series of Japanese stage II/III gastric cancer patients treated with or without adjuvant S-1. Our analyses covered a wide range of biomarkers previously suggested to be related to benefit from S-1, 5-FU or platinum based chemotherapy, such as gene polymorphisms of TS, OPRT, DPD, ERCC1, GSTP-1, and to potential benefit from receptor tyrosine kinases (RTKs) targeting therapy.
Patients and Methods
Patients
This study was approved by the institutional review board of the Kanagawa Cancer Center, Yokohama, Japan. Gastric cancer patients were selected from a prospective database of the Kanagawa Cancer Center, Department of Gastrointestinal Surgery, Yokohama, Japan, according to the following criteria: (i) histologically proven gastric adenocarcinoma, (ii) curative gastrectomy as primary treatment between June 2002 and March 2010 following Japanese gastric cancer treatment guidelines published in 2010 (2), (iii) pathologically stage II or III gastric cancer according to the 7th edition of the TNM classification (23), and (iv) availability of at least 5 years follow up data from all patients.
A total of 252 patients were selected for this study. A flow diagram of patients involved in each analysis step is presented in Figure 1. Associations with clinicopathologic factors were examined in all patients (cohort 1, n=252), while 22 patients who received UFT instead of S-1 as adjuvant chemotherapy and 15 patients who received palliative S-1 chemotherapy due to peritoneal cytology positive were excluded from survival analyses (cohort 2, n=215). Survival data were obtained from hospital records or from the Governmental registry system. The median follow-up period was 48.6 months (range 3.3-136.7 months).
A flow diagram of patients involved in each analysis step
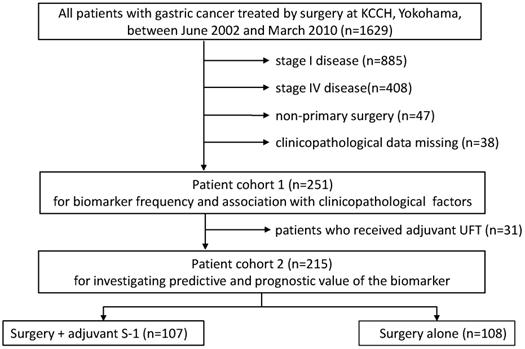
Preparation of the specimen and pathological diagnosis
Gastric cancer resection specimens were worked up according to the Japanese guidelines for gastric cancer (24). In addition, histological subtype according to the Lauren classification was determined. All slides from all resection specimens were reviewed by four senior gastrointestinal pathologists (HG, YM, TA, YK) to select and mark a representative block with the highest tumor cell density and a block with normal tissue for RNA and DNA extraction, respectively.
DNA and RNA extraction
DNA for polymorphism analyses was extracted from normal tissue (non-metastatic lymph nodes or normal gastric wall) using the QIAamp DNA Micro kit (Qiagen, Hilden, Germany) and quality controlled as described previously (25). RNA was extracted from tumor tissue block sections after microdissection using the High Pure RNA Paraffin Kit (Roche, Mannheim, Germany) according to the instructions of the manufacturer. The extracted RNA was quantified by a Nanodrop UV-Vis Spectrophotometer (Thermo Fisher Scientific, Waltham, MA) and 100 ng was used for the Nanostring assay.
Analyses of polymorphisms
DPYD, OPRT, ERCC1, ERCC2 and XRCC1 genotyping by MassARRAY
Genotype analysis was performed using the MassARRAY iPLEX system (Sequenom, San Diego, CA) according to manufacturer's instructions. A 7-plex assay interrogating DPYD (A1627G7&IVS14+1G>A), OPRT (G638C), ERCC1 (C118T&C8092A), ERCC2 (K751Q) and XRCC1 (A399G) was designed using MassARRAY Online Design Tools (Sequenom) as described previously (26). Sample genotypes were determined by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) analysis using the MassARRAY Compact system (Sequenom). The mass spectra analysis and genotype calls were generated using the Sequenom TYPER 4.0 software.
GSTP1 and TS 3'UTR genotyping by Sanger sequencing
GSTP1 (I105V) and TS 3'UTR (agttcat variant) genotyping was performed by Sanger sequencing. Purified pellets were dissolved in Hi-Di Formamide (Life Technologies) and analyzed using an ABI PRISM 3730 Genetic Analyzer (Life Technologies). Chromatograms were analyzed by SeqScape V2.5 (Life Technologies) and manual review.
TS 5'UTR genotyping by gel electrophoresis
TS 5'UTR were investigated as described previously (27). For TS 5'UTR, the predominant alleles expected at TS 5'UTR are the 2R and 3R alleles. PCR products from 2R and 3R alleles differ by a single 28 bp repeat, which can be resolved using gel electrophoresis. To determine the TS 5' UTR 2R/3R genotypes, 10 µl of PCR product was purified by 4 µl of Exonuclease I (New England Biolabs) and Thermosensitive Shrimp Alkaline Phosphatase (Promega) mixture treatment. After Exo-TSAP purification, PCR products were digested with 1 µl HaeIII (10,000 units/ml, New England Biolabs) followed by electrophoreses in 4% agarose/1xTBE gel.
Nanostring Assay
A set of 203 genes selected based upon the molecular gastric cancer signature published by Tan et al (28) as well as other genes implicated in gastric carcinogenesis such EGFR, HER2, FGFR2, cMET and KRAS were selected for a customized Nanostring assay (Nanostring Technologies, Seattle, WA). The assay was performed according to the instructions of the manufacturer and the Nanostring workflow was employed for digital detection of expression counts for each gene. The 'NanoStringNorm' R package (version 2.15) was used to normalize the raw expression data adjusting for technical assay variations, background and RNA content. All samples flagged by the software were removed and the normalized expression data was used for all downstream analyses.
Statistical analyses
Tumors were classified based on their expression signature as G-INT/G-DIF using the nearest template prediction algorithm as described previously (28). For the current study, we employed two different classification schemes. First, tumors were classified into two subtypes e.g. G-INT or G-DIF. Secondly, tumors were classified into 3 subtypes e.g. G-INT or G-DIF or G-ambiguous.
Associations between each biomarker and clinicopathological factors such as age, gender, location, tumor depth (pT), lymph node status (pN), distant metastasis (M), lymphatic channel invasion, blood vessel invasion, tumor size, macroscopic tumor type were examined in all patients. The analyses of the association between biomarker and histological subtype was restricted to those cases which were classified as either intestinal type or diffuse type GC. GC which were classified as mixed/unclassifiable according to Lauren (n=13) were excluded. Continuous factors (age, tumor size) were analyzed by student t-test, and categorical factors were analyzed by chi-square test. For analyses using distinct SNP subtypes, we excluded minor subtypes where the number of patients associated with a particular genotype (GG of GSTP1 in 5 patients and TT of ERCC1 rs3212986-C8092A in 10 patients) was felt to be too small for reliable statistical analysis. ERCC2 SNP was not analyzed for statistical analysis because most patients showed uniformal SNP type of TT (94.9%).
For survival analyses, the gene expression level cut off (high expression versus low expression) was determined by the maximum chi-square method (29). The validity of the selected cut off point was confirmed by a two-fold cross-validation approach for multivariate analysis (30). Overall survival probability curves were calculated using the Kaplan-Meier method and compared by the log-rank test. For the analyses of prognostic factors, a Cox's proportional hazard model was used to perform univariate and multivariate survival analyses in the surgery alone arm. A p-value of <0.05 was defined to be statistically significant for the analyses of the relationship of the biomarker with the clinicopathological factors including survival.
For the test of predictive factors, the interaction of each biomarker with S-1 treatment in the Cox's proportional hazard model was analyzed in the surgery alone and the surgery+S-1 arm. As this was a post hoc analysis that may not have enough power to detect an interaction effect, a raised significance level of p<0.2 was used to classify an interaction as being significant (31).
Results
Table 1 shows the clinicopathological characteristics by treatment and cohort. Cohort 1 (all patients) was used for the analysis of the biomarker frequency and association with clinicopathological analyses, cohort 2 (excluding patients who received adjuvant chemotherapy other than S-1) was used for the prognostic and predictive analyses. As expected, patients in the surgery+S-1 group had cancers with higher pT category, higher TNM stage, larger tumor size, and were more frequent of scirrhous macroscopic tumor type (all p-values < 0.05). Although there was no statistically significant difference in overall survival (OS) between the surgery alone and surgery+S-1 group by log-rank test (p=0.177) in our series, the hazard ratio for surgery+S-1 chemotherapy was 0.694 (p=0.196) in multivariate analysis which was similar to that observed in the ACTS-GC trial (5).
Frequency of biomarkers and association with clinicopathological factors
SNP genotypes suggested to predict 5-FU resistance e.g. TS 3'UTR (+6bp/-6bp and -6bp/-6bp) and TS 5'UTR (2R/3G, 3C/3G, and 3G/3G) were found in 88% and 62% GC, respectively. SNP phenotypes suggested to predict 5-FU toxicity e.g. OPRT (GC+CC) and DPYD (AG+GG) were present in 36% and 48% GC, respectively, while those suggested to predict platinum sensitivity e.g. GSTP1 (AG+GG), ERCC1 rs11615 (CT+TT), ERCC1 rs3212986 (GT+TT), ERCC2 variant (GT), and XRCC1 rs25487 (AG+GG) were present in 24%, 46%, 40%, 5%, and 44% GC, respectively. High HER2, EGFR, FGFR2, or MET RNA expression identifying GC patients potentially benefitting from targeted therapy, was observed in 49%, 66%, 72%, and 54% GC, respectively.
DPYD2A was associated with the presence of distant metastasis (p=0.040). GT type of ERCC1 and AA type of XRCC1 were associated with distal location of the cancer (p=0.018) and female gender (p=0.043), respectively. The [+6bp/-6bp] type of TS 3'UTR and [2R/3C] or [3C/3C] type of TS 5'UTR were associated with younger age (p=0.022 and p=0.008, respectively). No other associations between individual SNP biomarker status and clinicopathological factors were seen.
Clinicopathological characteristics of patient cohort 1 (n=252) and patient cohort 2 (n=215; excluding 22 patients treated with UFT adjuvant chemotherapy and 15 patients who had positive peritoneal cytology and received palliative S-1 chemotherapy (*))
| Surgery alone | Surgery + S-1 | ||||
|---|---|---|---|---|---|
| n | % | n / n* | %/ %* | ||
| Age | Median (range) years | 66 (35-85) | 64 (35-83) 64 (35-80)* | ||
| Gender | Male | 74 | 68.5 | 102 / 73 | 70.8 / 68.2 |
| Female | 34 | 31.5 | 42 / 34 | 29.2 / 31.8 | |
| Tumor location | Upper third | 29 | 26.9 | 47 / 35 | 32.6 / 32.7 |
| Middle third | 41 | 38 | 62 / 48 | 43.1 / 44.9 | |
| Lower third | 38 | 35.2 | 35 / 24 | 24.3 / 22.4 | |
| Tumor size | Median (range) mm | 50 (15-180) | 60 (25-212) 60 (28-212)* | ||
| Macroscopic tumour type | 0 | 25 | 23.1 | 14 / 10 | 9.7 / 9.3 |
| 1 | 6 | 5.6 | 7 / 6 | 4.9 / 5.6 | |
| 2 | 26 | 23.1 | 36 / 26 | 25.0 / 24.3 | |
| 3 | 28 | 26.9 | 37 / 24 | 25.6 / 22.4 | |
| 4 | 2 | 1.9 | 18 / 16 | 12.5 / 15 | |
| 5 | 21 | 19.4 | 32 / 25 | 22.3 / 23.4 | |
| Depth of invasion (pT) | T1/T2 | 31 | 28.7 | 18 / 16 | 12.5 / 15 |
| T3/T4 | 77 | 71.3 | 126 / 91 | 87.5 / 85 | |
| Lymph node status (pN) | N0 | 20 | 18.5 | 22 / 19 | 15.3 / 17.8 |
| N1/N2/N3 | 88 | 81.5 | 122 / 88 | 84.7 / 82.2 | |
| TNM stage | 2A | 18 | 16.7 | 8 / 5 | 5.6 / 4.7 |
| 2B | 36 | 33.3 | 36 / 31 | 25.0 / 29 | |
| 3A | 25 | 23.1 | 18 / 11 | 12.5 / 11.2 | |
| 3B | 14 | 13 | 29 / 25 | 20.1 / 22.4 | |
| 3C | 15 | 13.9 | 53 / 35 | 36.8 / 32.7 | |
| Lymphatic invasion | negative | 42 | 38.9 | 45 / 39 | 31.3 / 36.4 |
| positive | 66 | 61.1 | 99/ 68 | 68.7 / 63.6 | |
| Venous invasion | negative | 29 | 26.9 | 34 / 32 | 23.6 / 29.9 |
| positive | 79 | 73.1 | 110 / 75 | 76.4 / 70.1 | |
| Histological tumor type | Intestinal | 40 | 37.4 | 42 / 34 | 29.1 / 31.8 |
| Diffuse | 62 | 57.4 | 95 / 67 | 66.0 / 62.6 | |
| unclassifiable | 6 | 5.5 | 7 / 6 | 4.9 / 5.6 | |
Second, we examined associations between clinicopathological factors and gene expression, either at the single gene or gene signature level. High EGFR and high FGFR2 RNA expression were associated with younger age (p=0.02 and p=0.01, respectively). High EGFR, low HER2 and high FGFR2 RNA expression were associated with diffuse type histology (p=0.001, p=0.009 and p=0.016, respectively). High FGFR2 was associated with increased frequency of vascular invasion (p=0.047), high MET with distal location of the cancer (p=0.012) and low KRAS with presence of distant metastasis (p=0.022).
125 patients (51%) were classified as G-INT and 120 patients (49%) as G-DIFF when the two-category classification was employed. When the three-category classification was used, 88 (35.9%) patients were classified as G-INT, 71 (29%) patients as G-DIFF, and 86 (35.1%) patients as G-ambiguous. G-DIFF was associated with higher pT category (e.g. deeper invasion of the primary tumor) compared to G-INT or G-ambiguous type.
Association of biomarker status and patient prognosis
In order to distinguish between predictive and prognostic value of a biomarker, associations with survival were analysed separately by treatment e.g. the prognostic value of a marker was determined using data from the surgery alone treated patient group. For multivariate overall survival analyses, TNM stage, tumor size and macroscopic tumor type were included in the model together with the biomarker of interest. The results of the survival analyses are illustrated using a Forest plot of hazard ratios (Figure 2). High HER2 RNA expression was associated with poor survival and was the only significant independent prognosticator (HR=3.912, p=0.0005). Patients with TS 5'UTR 2R/3G or 3C/3G or 3G/3G tended to have poorer survival than those with 2R/2R, 2R/3C, or 3C/3C (HR =2.185, p=0.058) but the difference did not reach statistical significance.
Association of biomarker status and patient benefit from adjuvant S-1 chemotherapy
To identify biomarkers that might predict which GC patient subgroup will benefit from adjuvant S-1 therapy, we determined the interaction of each biomarker with S-1 treatment using a Cox's proportional model by including S-1 treatment, TNM stage, tumor size, macroscopical tumor type, and biomarker in the model. The results are illustrated in a Forest plot of hazard ratios (Figure 3). We found that TS 5'UTR subtype 2R/2R, 2R/3C, or 3C/3C (p=0.058), high RNA expression of HER2 (p=0.031), low EGFR expression (p=0.124) and high MET expression (p=0.165), were significant independent predictors for S-1 resistance.
Discussion
The present study reports our comprehensive analysis of prognostic and predictive biomarkers in a cohort of Japanese gastric cancer treated by surgery with and without adjuvant S-1 chemotherapy. S-1 is a drug which consists of tegafur, a prodrug of 5-FU, gimeracil, a competitive inhibitor of DPYD, and oteracil potassium, an inhibitor of 5-FU phosphorylation. We explored a number of different biomarkers including (i) genetic polymorphisms of enzymes involved in 5-FU metabolism such as OPRT (15), DPYD (32), and TS (33), (ii) genetic polymorphisms of proposed predictors for platinum resistance (34, 35) such as GSTP-1, ERCC1, and XRCC1, (iii) previously identified GC genomic subtypes (G-INT and G-DIFF (28)) as potential prognosticators as well as predictors for 5-FU sensitivity and (iv) RNA expression of EGFR, HER2, FGFR2, MET, and KRAS to evaluate the potential for therapies targeting the RTK/KRAS signaling pathway in Japanese gastric cancer currently treated with surgery plus S-1 chemotherapy.
Our study showed no relationship between OPRT and DPYD SNPs and S-1 treatment benefit, only the TS 5'UTR SNP predicted benefit from S-1 adjuvant chemotherapy. This is in contrast to previous studies which suggested that protein, mRNA, or genetic polymorphism of TS (8-14), DPYD (8-10), and OPRT (10-12, 14-16) were predictors of response to 5-FU based chemotherapy. However, most previous studies did not have a non-chemotherapy control arm and thus were not able to distinguish between prognostic and predictive value of a biomarker or suffered from relatively small sample size. Our study is the first to clarify the predictive and prognostic impact of these enzyme SNPs on S-1.
The results of the survival analyses are illustrated using a Forest plot of hazard ratios
The results are illustrated in a Forest plot of hazard ratios
TS is an enzyme which catalyzes the methylation of fluoro-dUMP to dTMP, an essential precursor for DNA synthesis (36). 5-FU inhibits TS activity and blocks DNA synthesis (18). It is thus biologically plausible that S-1 (a 5-FU prodrug) efficacy might also be influenced by TS activity. The promoter enhancer region of TS gene is polymorphic, containing a double (2R) or triple (3R) tandem repeat known to be involved in the translational auto-regulation mechanism of TS expression (37). Previous investigators reported that 2R/2R, 2R/3C, and 3C/3C type of TS polymorphisms were related with better survival in colorectal and gastric cancer patients receiving fluoropyrimidine (38). However, their study did not include a surgery alone group and thus cannot distinguish between prognostic and predictive value of the SNP. There is currently no study published investigating a relationship between TS polymorphism and S-1 efficacy. The present study was able to clarify that TS 5'UTR polymorphisms have no prognostic value in Japanese gastric cancer patients (e.g. do not influence the survival in the surgery alone group) but does predict benefit from adjuvant S-1 treatment depending on the SNP subtype of TS 5'UTR. This finding, if replicated in a second independent series, could indicate that determination of the TS 5'UTR SNP status which would be possible in any normal tissue including a routine blood sample might be a useful marker to stratify patients for adjuvant S-1 therapy and/or other therapy options.
It has been shown recently that classifying gastric cancer based on their RNA expression signature into G-INT and G-DIFF identifies groups of patients with different prognosis (28). Moreover, the genomic subtype seemed to predict benefit from adjuvant 5-FU based chemotherapy (28). However, when using the same classifier in the present study, the genomic classifier did neither predict patient prognosis nor benefit from adjuvant S-1. This discrepancy could be related to the stage selection bias in the current study which included only stage II/III gastric cancer patients whereas the previous study included all disease stages. Secondly, differences in drug efficacy between S-1 and 'conventional 5-FU' have been described for diffuse type gastric cancer (39) and might explain the different results.
In addition to the investigations into prognostic and predictive marker for S-1 benefit, we also used this gastric cancer cohort to explore whether there is any indication that patients might benefit from potential other cytotoxic drugs such as platinum based chemotherapy. Polymorphisms for GSTP-1, ERCC1 and XRCC1 have been previously identified as potential predictive biomarker for platinum based chemotherapy in colon cancer patients (34, 35, 40, 41). ERCC1 SNPs have shown to have prognostic and/or predictive value in the advanced (metastatic) gastric cancer setting (17, 19, 35, 42-44). In the current series, SNPs of GSTP-1, ERCC1, and XRCC1 were neither a prognosticator nor a predictor for S-1 adjuvant chemotherapy suggesting that adding platinum drugs to S-1 might improve patient's prognosis.
Similarly, we explored whether patients in our series would potentially be eligible for RTK targeting drug therapy such as HER2 targeting trastuzumab, pertuzumab or T-DM1; EGFR targeting nimotuzumab, FGFR2 targeting AZD4547 or MET targeting rilotumumab or onartuzumab or MEK inhibitors which may be attractive therapies for KRAS-driven tumors. The present study demonstrated that high HER2 RNA expression was associated with poor prognosis and low EGFR, high HER2 and high MET RNA expression were important predictor for S-1 resistance.
Our data seem to indicate that whereas therapy targeting HER2 or MET should not be combined with S-1 chemotherapy, a combination of EGFR targeting drugs with S-1 chemotherapy might be of added value. However, a recent study in a subset of patients from the ACTS-GC trial (5) reported that HER2 'positivity' by immunohistochemistry or copy number analyses was not a significant prognosticator and that neither EGFR nor HER2 status predicted benefit from S-1. The results from this study are not directly comparable with the current study as different methodology (RNA expression level in the current study versus protein expression and DNA copy number in the previous study) was used to establish the HER2/EGFR status of the individual case. Ma et al reported a low correlation between HER2 protein expression and RNA expression and suggested that HER2 protein and RNA may have a different clinical role in gastric cancer patients (45). On the other hand, there is currently no information available about the relationship between RNA and protein expression levels in EGFR in gastric cancer.
The present study has some limitations. This is a retrospective, single center case-control study with no treatment randomization. Thus, in contrast to the results from the ACTS-GC trial, the univariate overall survival curves were similar between the S-1 group and surgery alone group in the current study. However, the hazard ratio was almost identical to that observed in S-1 arm of ACTS phase III study when the survival was adjusted by Cox's multivariate analysis. In addition, the present study was only analyzed the Eastern cohort. Therefore, the further study should focus on the Western cohort. Furthermore, our study results require validation in a second independent cohort of gastric cancer patients treated with adjuvant S-1 which was not achievable within the current study cohort due to its sample size.
In conclusion, this is the first comprehensive biomarker analysis comparing biomarker status in stage II/III gastric cancer patients treated with surgery alone to those treated with surgery plus adjuvant S-1 chemotherapy. We identified several candidate markers such as HER2, EGFR, MET RNA expression and TS 5'UTR SNP which are able to identify patients with a high risk of recurrence and/or no potential benefit from adjuvant S-1 therapy. The findings of this study may potentially inform future trial design to test platinum drugs and targeting agents after biomarker stratification in order to improve survival of Japanese stage II/III gastric cancer. The promising results from this comprehensive but exploratory biomarker analyses require validation in a second series before clinical application.
Acknowledgements
This work was supported by following grants to PT: a core grant from GIS, National Medical Research Council grant NMRC/BnB/0005b/2013 and NMRC/TCR/009-NUHS/2013.
Competing Interests
The authors have declared that no competing interest exists.
References
1. GLOBOCAN2012: Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012. 2012. http://globocan.iarc.fr/Pages/fact_sheets_population.aspx
2. Association TJGC. Japanese gastric cancer treatment guidelines 2010 (ver. 3). Gastric Cancer. 2011;14(2):113-23
3. Okines A, Verheij M, Allum W, Cunningham D, Cervantes A. Gastric cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2010;21(Suppl 5):v50-4
4. NCCN. NCCN Clinical Practice Guidelines in Oncology. Gastric Cancer Version 2.2011 [online] 2011. Available from: http://www.nccn.org/professionals/physician_gls/pdf/gastric.pdf
5. Sakuramoto S, Sasako M, Yamaguchi T, Kinoshita T, Fujii M, Nashimoto A. et al. Adjuvant chemotherapy for gastric cancer with S-1, an oral fluoropyrimidine. N Engl J Med. 2007;357(18):1810-20
6. Bang YJ, Kim YW, Yang HK, Chung HC, Park YK, Lee KH. et al. Adjuvant capecitabine and oxaliplatin for gastric cancer after D2 gastrectomy (CLASSIC): a phase 3 open-label, randomised controlled trial. Lancet. 2012;379(9813):315-21
7. Sasako M, Sakuramoto S, Katai H, Kinoshita T, Furukawa H, Yamaguchi T. et al. Five-year outcomes of a randomized phase III trial comparing adjuvant chemotherapy with S-1 versus surgery alone in stage II or III gastric cancer. Journal of clinical oncology. 2011;29(33):4387-93
8. Sasako M, Terashima M, Ichikawa W, Ochiai A, Kitada K, Kurahashi I. et al. Impact of the expression of thymidylate synthase and dihydropyrimidine dehydrogenase genes on survival in stage II/III gastric cancer. Gastric Cancer. 2014;18(3):538-48
9. Miyamoto S, Boku N, Ohtsu A, Yoshida S, Ochiai A, Okabe H. et al. Clinical implications of immunoreactivity of thymidylate synthase and dihydropyrimidine dehydrogenase in gastric cancer treated with oral fluoropyrimidine (S-1). Study Group of S-1 for Gastric Cancer. Int J Oncol. 2000;17(4):653-8
10. Miyazaki I, Kawai T, Harada Y, Moriyasu F. A predictive factor for the response to S-1 plus cisplatin in gastric cancer. World J Gastroenterol. 2010;16(36):4575-82
11. Koizumi W, Tanabe S, Azuma M, Ishido K, Nishimura K, Sasaki T. et al. Impacts of fluorouracil-metabolizing enzymes on the outcomes of patients treated with S-1 alone or S-1 plus cisplatin for first-line treatment of advanced gastric cancer. Int J Cancer. 2010;126(1):162-70
12. Jeung HC, Rha SY, Shin SJ, Lim SJ, Roh JK, Noh SH. et al. Predictive values of 5-fluorouracil pathway genes for S-1 treatment in patients with advanced gastric cancer. Anticancer Drugs. 2011;22(8):801-10
13. Ichikawa W, Takahashi T, Suto K, Shirota Y, Nihei Z, Shimizu M. et al. Simple combinations of 5-FU pathway genes predict the outcome of metastatic gastric cancer patients treated by S-1. Int J Cancer. 2006;119(8):1927-33
14. Hashiguchi K, Kitajima Y, Kai K, Hiraki M, Nakamura J, Tokunaga O. et al. A quantitative evaluation of the determinant proteins for S-1 responsiveness in a biopsy specimen assists in patient selection to neoadjuvant therapy in cases of advanced gastric cancer. Int J Oncol. 2010;37(2):257-64
15. Choi IS, Lee HS, Lee KW, Kim H, Kim KH, Kim YJ. et al. Biomarker analysis in patients with advanced gastric cancer treated with S-1 plus cisplatin chemotherapy: orotate phosphoribosyltransferase expression is associated with treatment outcomes. Med Oncol. 2011;28(4):991-8
16. Kodera Y, Ito S, Fujiwara M, Mochizuki Y, Nakayama G, Ohashi N. et al. Gene expression of 5-fluorouracil metabolic enzymes in primary gastric cancer: correlation with drug sensitivity against 5-fluorouracil. Cancer Lett. 2007;252(2):307-13
17. Matsubara J, Nishina T, Yamada Y, Moriwaki T, Shimoda T, Kajiwara T. et al. Impacts of excision repair cross-complementing gene 1 (ERCC1), dihydropyrimidine dehydrogenase, and epidermal growth factor receptor on the outcomes of patients with advanced gastric cancer. Br J Cancer. 2008;98(4):832-9
18. Van Triest B, Peters GJ. Thymidylate synthase: a target for combination therapy and determinant of chemotherapeutic response in colorectal cancer. Oncology. 1999;57(3):179-94
19. Park SR, Kong SY, Nam BH, Choi IJ, Kim CG, Lee JY. et al. CYP2A6 and ERCC1 polymorphisms correlate with efficacy of S-1 plus cisplatin in metastatic gastric cancer patients. Br J Cancer. 2011;104(7):1126-34
20. Yamada Y, Boku N, Nishina T, Yamaguchi K, Denda T, Tsuji A. et al. Impact of excision repair cross-complementing gene 1 (ERCC1) on the outcomes of patients with advanced gastric cancer: correlative study in Japan Clinical Oncology Group Trial JCOG9912. Ann Oncol. 2013;24(10):2560-5
21. Terashima M, Kitada K, Ochiai A, Ichikawa W, Kurahashi I, Sakuramoto S. et al. Impact of expression of human epidermal growth factor receptors EGFR and ERBB2 on survival in stage II/III gastric cancer. Clin Cancer Res. 2012;18(21):5992-6000
22. Boku N, Ohtsu A, Nagashima F, Shirao K, Koizumi W. Relationship between expression of vascular endothelial growth factor in tumor tissue from gastric cancers and chemotherapy effects: comparison between S-1 alone and the combination of S-1 plus CDDP. Jpn J Clin Oncol. 2007;37(7):509-14
23. Sobin LH, Compton CC. TNM seventh edition: what's new, what's changed: communication from the International Union Against Cancer and the American Joint Committee on Cancer. Cancer. 2010;116(22):5336-9
24. Japanese classification of gastric carcinoma. 3rd English edition. Gastric Cancer. 2011;14(2):101-12
25. van Grieken NC, Aoyama T, Chambers PA, Bottomley D, Ward LC, Inam I. et al. KRAS and BRAF mutations are rare and related to DNA mismatch repair deficiency in gastric cancer from the East and the West: results from a large international multicentre study. Br J Cancer. 2013;108(7):1495-501
26. Loh M, Chua D, Yao Y, Soo RA, Garrett K, Zeps N. et al. Can population differences in chemotherapy outcomes be inferred from differences in pharmacogenetic frequencies? The pharmacogenomics journal. 2013;13(5):423-9
27. Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet. 2009 Chapter 2:Unit 2 12
28. Tan IB, Ivanova T, Lim KH, Ong CW, Deng N, Lee J. et al. Intrinsic subtypes of gastric cancer, based on gene expression pattern, predict survival and respond differently to chemotherapy. Gastroenterology. 2011;141(2):476-85 85 e1-11
29. Altman DG, Lausen B, Sauerbrei W, Schumacher M. Dangers of using "optimal" cutpoints in the evaluation of prognostic factors. J Natl Cancer Inst. 1994;86(11):829-35
30. Mazumdar M, Smith A, Bacik J. Methods for categorizing a prognostic variable in a multivariable setting. Stat Med. 2003;22(4):559-71
31. S. S, editor. Statistical Analysis of Epidemiologic Data. New York: Oxford University Press. 1996
32. Amatori F, Di Paolo A, Del Tacca M, Fontanini G, Vannozzi F, Boldrini L. et al. Thymidylate synthase, dihydropyrimidine dehydrogenase and thymidine phosphorylase expression in colorectal cancer and normal mucosa in patients. Pharmacogenet Genomics. 2006;16(11):809-16
33. Hur H, Kang J, Kim NK, Min BS, Lee KY, Shin SJ. et al. Thymidylate synthase gene polymorphism affects the response to preoperative 5-fluorouracil chemoradiation therapy in patients with rectal cancer. Int J Radiat Oncol Biol Phys. 2011;81(3):669-76
34. Li HY, Ge X, Huang GM, Li KY, Zhao JQ, Yu XM. et al. GSTP1, ERCC1 and ERCC2 polymorphisms, expression and clinical outcome of oxaliplatin-based adjuvant chemotherapy in colorectal cancer in Chinese population. Asian Pac J Cancer Prev. 2012;13(7):3465-9
35. Zaanan A, Dalban C, Emile JF, Blons H, Flejou JF, Goumard C. et al. ERCC1, XRCC1 and GSTP1 Single Nucleotide Polymorphisms and Survival of Patients with Colon Cancer Receiving Oxaliplatin-Based Adjuvant Chemotherapy. J Cancer. 2014;5(6):425-32
36. Fox KM, Maley F, Garibian A, Changchien LM, Van Roey P. Crystal structure of thymidylate synthase A from Bacillus subtilis. Protein Sci. 1999;8(3):538-44
37. Kawakami K, Salonga D, Park JM, Danenberg KD, Uetake H, Brabender J. et al. Different lengths of a polymorphic repeat sequence in the thymidylate synthase gene affect translational efficiency but not its gene expression. Clin Cancer Res. 2001;7(12):4096-101
38. Kawakami K, Watanabe G. Identification and functional analysis of single nucleotide polymorphism in the tandem repeat sequence of thymidylate synthase gene. Cancer Res. 2003;63(18):6004-7
39. Ajani JA, Rodriquez W, Bodoky G, Moiseyenko V, Lichinister M, Gorbunova V. et al. Multicenter phase III comparison of cisplatin/S-1 (CS) with cisplatin/5-FU (CF) as first-line therapy in patients with advanced gastric cancer (FLAGS): Secondary and subset analyses. Journal of clinical oncology. 2009;27(suppl):15s (abstract 4511)
40. Lee KH, Chang HJ, Han SW, Oh DY, Im SA, Bang YJ. et al. Pharmacogenetic analysis of adjuvant FOLFOX for Korean patients with colon cancer. Cancer Chemother Pharmacol. 2013;71(4):843-51
41. Suh KW, Kim JH, Kim do Y, Kim YB, Lee C, Choi S. Which gene is a dominant predictor of response during FOLFOX chemotherapy for the treatment of metastatic colorectal cancer, the MTHFR or XRCC1 gene? Ann Surg Oncol. 2006;13(11):1379-85
42. Tatewaki N, Maekawa K, Katori N, Kurose K, Kaniwa N, Yamamoto N. et al. Genetic variations and haplotype structures of the glutathione S-transferase genes, GSTT1 and GSTM1, in a Japanese patient population. Drug Metab Pharmacokinet. 2009;24(1):118-26
43. Shiraishi K, Kohno T, Tanai C, Goto Y, Kuchiba A, Yamamoto S. et al. Association of DNA repair gene polymorphisms with response to platinum-based doublet chemotherapy in patients with non-small-cell lung cancer. J Clin Oncol. 2010;28(33):4945-52
44. Yamada Y, Boku N, Nishina T, Yamaguchi K, Denda T, Tsuji A. et al. Impact of excision repair cross-complementing gene 1 (ERCC1) on the outcomes of patients with advanced gastric cancer: correlative study in Japan Clinical Oncology Group Trial JCOG9912. Ann Oncol. 2013;24(10):2560-5
45. Ma GF, Liu YM, Gao H, Miao Q, Luo TC, Zeng XQ. et al. HER2 mRNA status contributes to the discrepancy between gene amplification and protein overexpression in gastric cancer. Dig Dis Sci. 2014;59(2):328-35
Author contact
![]() Corresponding authors: Heike I Grabsch, Maastricht University Medical Center, Department of Pathology, P. Debyelaan 25, 6229 HX Maastricht, The Netherlands; H.Grabschnl and Patrick Tan, Duke-NUS Graduate Medical School, 11 Hospital Drive, Singapore 169610, Singapore; gmstanpedu.sg
Corresponding authors: Heike I Grabsch, Maastricht University Medical Center, Department of Pathology, P. Debyelaan 25, 6229 HX Maastricht, The Netherlands; H.Grabschnl and Patrick Tan, Duke-NUS Graduate Medical School, 11 Hospital Drive, Singapore 169610, Singapore; gmstanpedu.sg

 Global reach, higher impact
Global reach, higher impact