3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(21):5139-5152. doi:10.7150/jca.30816 This issue Cite
Research Paper
An NF90/long noncoding RNA-LET/miR-548k feedback amplification loop controls esophageal squamous cell carcinoma progression
Department of Surgical Oncology, the Second Affiliated Hospital of Fujian Medical University, Quanzhou 362000, Fujian, China
Jianqing Lin and Zhiyao Chen are co-first authors.
Received 2018-10-20; Accepted 2019-7-16; Published 2019-8-28
Abstract
In our previous study we have found that miR-548k has oncogenic roles in esophageal squamous cell carcinoma (ESCC) via repressing long noncoding RNA (lncRNA)-LET and further upregulating nuclear factor 90 (NF90). However, the upstream factors controlling miR-548k expression are still unknown. In this study, we found NF90 directly binds pri-miR-548k, increases the stability of pri-miR-548k, and upregulates the expression of pri-miR-548k and miR-548k. Therefore, NF90, miR-548k and lncRNA-LET forms a feedback loop. Gain-of-function and loss-of-function assays demonstrated that in accordance with the roles of miR-548k, NF90 also promotes ESCC cell proliferation and migration. Furthermore, we verified the regulatory feedback loop between NF90, miR-548k, and lncRNA-LET. We found NF90 upregulated miR-548k and downregulated lncRNA-LET. miR-548k downregulated lncRNA-LET and upregulated NF90. lncRNA-LET downregulated NF90 and miR-548k. Through the reciprocal regulations between each other, the NF90/miR-548k/lncRNA-LET feedback loop controls the expressions of NF90 targets (HIF-1α and VEGF), miR-548k targets (KLF10 and EGFR), and lncRNA-LET target (p53). Further functional assays demonstrated that activation of the NF90/miR-548k/lncRNA-LET feedback loop via simultaneously overexpressing NF90 and miR-548k and simultaneously depleting lncRNA-LET significantly promotes ESCC cell proliferation and migration in vitro and ESCC tumor growth in vivo. Targeting the NF90/miR-548k/lncRNA-LET feedback loop via simultaneously depleting NF90 and miR-548k and simultaneously overexpressing lncRNA-LET significantly inhibits ESCC cell proliferation and migration in vitro and ESCC tumor growth in vivo. In summary, our findings identified a crucial oncogenic NF90/lncRNA-LET/miR-548k feedback amplification loop, which may be promising therapeutic targets for ESCC.
Keywords: esophageal squamous cell carcinoma, NF90, miR-548k, long noncoding RNA, feedback loop
Introduction
Esophageal cancer is one of the most common cancer and the sixth leading cause of cancer-related death worldwide with an estimated 572.0 thousand new cases and 508.6 thousand deaths in 2018 [1, 2]. Esophageal squamous cell carcinoma (ESCC) is the predominant histopathological subtype of esophageal cancer [3, 4]. Although advances have been made in multiple therapeutic strategies, the overall 5-year survival rate of ESCC is still very poor, with about 10% to 25% [5]. The poor prognosis of ESCC is associated with later diagnosis of ESCC patients which are not suitable for surgical resection [6]. Except surgical resection, other therapeutic strategies are relative less efficient due to the lack of the understanding of underlying molecular mechanisms mediating the initiation and progression of ESCC [7].
Genome sequencing, exome sequencing, transcriptome sequencing, and array comparative genomic hybridization of ESCC tissues have identified many mutated genes, genomic amplified or deleted regions, and aberrantly expressed transcripts [7, 8]. Among these aberrantly expressed transcripts, not only mRNAs, but also many non-coding RNAs were identified [4, 9]. MicroRNA (miRNA) is a class of small non-coding RNA with about 21-25 nucleotides in length [10, 11]. Accumulating evidences have demonstrated that miRNAs are frequently dysregulated in many diseases, particular in cancers, and have crucial roles in various pathophysiological processes [12-17]. miRNAs induce their targets transcripts degradation and/or translation repression via base pairing with target transcripts [18-20]. In our previous study, we have found miR-548k as an oncogenic and upregulated miRNA in ESCC [21]. However, the factors regulating miR-548k expression in ESCC are still not fully understood.
Long noncoding RNA (lncRNA) is another class of non-coding RNA with more than 200 nucleotides in length and no protein coding potential [22-24]. Recent advances in genome and transcriptome sequencing have found more than 58,000 lncRNAs in human cells [25]. The number of genes coding proteins is only about 21,000 [25]. Among these lncRNAs, many are revealed to be dysregulated in cancers and implicated in almost every aspect of cancers, including cell proliferation, cell cycle, apoptosis, migration, invasion, metastasis, angiogenesis, drug-resistance, and so on [26-35]. In accordance with miRNAs, lncRNAs are also considered as important regulatory RNAs in ESCC [36, 37]. In our previous study, we have identified lncRNA-LET as a tumor suppressor in ESCC [21]. Furthermore, we found lncRNA-LET as a direct target of miR-548k [21]. Nuclear factor 90 (NF90) was identified as a downstream target of lncRNA-LET [21].
NF90 is an RNA binding protein, which has been reported to regulate the stability and translation of bound RNAs and miRNA biogenesis [38-40]. Intriguingly, in this study, we further found NF90 also bound pri-miR-548k and upregulated the expression of pri-miR-548k and miR-548k. Thus, NF90, miR-548k, lncRNA-LET forms a positive feedback loop in ESCC. We further investigated the biological roles and mechanisms of action of the NF90/miR-548k/lncRNA-LET regulatory loop in ESCC.
Materials and Methods
Cell culture and treatment
ESCC cell lines KYSE30 and Eca-109 were acquired from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). KYSE30 and Eca-109 cells were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Invitrogen) in a humidified incubator containing 5% CO2 at 37°C. Where indicated, the ESCC cells were treated with 50 μM α-amanitin (Sigma-Aldrich, Saint Louis, MO, USA) for the indicated time to inhibit RNA polymerase II-mediated transcription.
Plasmids construction and transfection
The complementary DNA (cDNA) coding NF90 was PCR-amplified with the PfuUltra II Fusion HS DNA Polymerase (Agilent Technologies, Santa Clara, CA, USA) and the primers 5'-GGGGTACCGTTGAAGTATTGATAACACCAA-3' (sense) and 5'-GGGTTTAAACACAAACCATTGAAGACACGG-3' (anti-sense). Next, the PCR products were subcloned into the Kpn I and Pme I sites of the pcDNA3.1 plasmid (Invitrogen), named as pcDNA3.1-NF90. lncRNA-LET overexpression plasmid pcDNA3.1-LET was constructed in our previous study [21]. To inhibit NF90 expression, two independent cDNA oligonucleotides specifically targeting NF90 were designed as previously described [41], synthesized by GenePharma (Shanghai, China), and inserted into the supersilencingTM shRNA expression vector pGPU6/Hygro (GenePharma), named as shRNA-NF90-1 and shRNA-NF90-2. The shRNAs target sites were 5'-GCUCAAAGCUGUGUCCGACUGGA-3' (shRNA-NF90-1) and 5'-AAGCCACUGAUGCUAUUGGGC-3' (shRNA-NF90-2). To inhibit lncRNA-LET expression, two independent cDNA oligonucleotides specifically targeting lncRNA-LET were designed as we previously described [21], synthesized by GenePharma (Shanghai, China), and inserted into the supersilencingTM shRNA expression vector pGPU6/Hygro (GenePharma), named as shRNA-LET-1 and shRNA-LET-2. The shRNAs target sites were 5'-TGGGAGTAAAGGGAAAGAGTT-3' (shRNA-LET-1) and 5'-GTGCATGTGGTAGGTTAGATT-3' (shRNA-LET-2). A scrambled shRNA was used as negative control, named as shRNA-NC. The transfection of pcDNA3.1, pcDNA3.1-NF90, pcDNA3.1- LET, shRNA-NC, shRNA-NF90-1, shRNA-NF90-2, shRNA-LET-1, and shRNA-NF90-1 were undertaken with Lipofectamine 3000 (Invitrogen) according to the protocol.
Lentivirus production and stable cell lines construction
Recombinant hsa-miR-548k overexpression lentiviruses, recombinant has-miR-548k inhibition lentiviruses, and their respective negative control lentiviruses were purchased from GenePharma. For construction of NF90 overexpressed KYSE30 cells, KYSE30 cells were transfected with pcDNA3.1-NF90 and selected with neomycin for four weeks. For construction of NF90 depleted Eca-109 cells, Eca-109 cells were transfected with shRNA-NF90-1 or shRNA-NF90-2, and selected with hygromycin for four weeks. For construction of lncRNA-LET overexpressed Eca-109 cells, Eca-109 cells were transfected with pcDNA3.1-LET and selected with neomycin for four weeks. For construction of lncRNA-LET depleted KYSE30 cells, KYSE30 cells were transfected with shRNA-LET-1 or shRNA-LET-2, and selected with hygromycin for four weeks. For construction of miR-548k overexpressed KYSE30 cells, KYSE30 cells were infected with 2×106 transducing units of miR-548k overexpression lentiviruses and selected with puromycin for four weeks. For construction of miR-548k depleted Eca-109 cells, Eca-109 cells were infected with miR-548k inhibition lentiviruses and selected with puromycin for four weeks. For construction of NF90 overexpressed and miR-548k simultaneously depleted KYSE30 cells, NF90 overexpressed KYSE30 cells were infected with miR-548k inhibition lentiviruses and selected with neomycin and puromycin for four weeks. For construction of NF90 and miR-548k simultaneously overexpressed and lncRNA-LET simultaneously depleted KYSE30 cells, NF90 overexpressed KYSE30 cells were transfected with shRNA-LET-2 and selected with neomycin and hygromycin for four weeks. Then, the cells were further infected with 2×106 transducing units of miR-548k overexpression lentiviruses and selected with puromycin, neomycin and hygromycin for four weeks. For construction of NF90 and miR-548k simultaneously depleted and lncRNA-LET simultaneously overexpressed Eca-109 cells, NF90 depleted Eca-109 cells were transfected with pcDNA3.1-LET and selected with neomycin and hygromycin for four weeks. Then, the cells were further infected with 2×106 transducing units of miR-548k inhibition lentiviruses and selected with puromycin, neomycin and hygromycin for four weeks.
Western blot
Total proteins were extracted from indicated ESCC cells with RIPA buffer (Beyotime, Jiangsu, China) added with protease inhibitors (Beyotime). Protein concentrations were determined by BCA assay using the BCA Protein Assay Kit (Beyotime). Equal amounts of proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by being transferred to nitrocellulose membrane (Millipore, Bedford, MA, USA). After being blocked, the membranes were incubated with NF90 specific primary antibody (ab131004, Abcam, Hong Kong, China) and β-actin specific primary antibody (66009-1-Ig, Proteintech, Rosemont, IL, USA). After three washes, the membranes were incubated with IRDye 700CW goat anti-mouse IgG (Li-Cor, Lincoln, NE, USA) and IRDye 800CW goat anti-rabbit IgG (Li-Cor), followed by being scanned on an Odyssey infrared scanner (Li-Cor).
RNA extraction, reverse transcription, and quantitative real time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from indicated ESCC cells with the TRIzol Regent (Invitrogen) following the protocol. The quality of RNA was assessed by formaldehyde agarose gel electrophoresis and quantified by NanoDrop UV-Vis Spectrophotometer. After being treated with DNase I (Takara, Dalian, China) to remove genomic DNA, the purified RNA was subjected to reverse transcription with the M-MLV Reverse Transcriptase (Invitrogen) following the protocol. qRT-PCR was undertaken on ABI StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The expressions of mRNAs and lncRNAs were measured using SYBR® Premix Ex Taq™ II (Takara) with the standard SYBR Green protocol. The expressions of miR-548k, pri-miR-548k, and pri-miR-21 were measured using TaqManTM MicroRNA Assay (Applied Biosystems) and TaqManTM Pri-miRNA Assay (Applied Biosystems) following the manufacturer's protocols. The sequences of the primers were as follows: for lncRNA-LET, 5'-TGAGATGCTGGAATGATG-3' (sense) and 5'-GGCTAAAGAAGGAAAAGG-3' (antisense); for HIF-1α, 5'-ATGAAGTGTACCCTAACTAGCCG-3' (sense) and 5'-CCAAGCAGGTCATAGGTGGTTTC-3' (antisense) [42]; for VEGF, 5'-CGCAAGAAATCCCGGTATAA-3' (sense) and 5'-AAATGCTTTCTCCGCTCTGA-3' (antisense) [39]; for KLF10, 5'-GGAGGAAAGAATGGAAATG-3' (sense) and 5'-GTCAGAAGGACTGTAAGG-3' (antisense); for EGFR, 5'-TTCACACATACTCCTCCTC-3' (sense) and 5'-TCTCCATCACTTATCTCCT-3' (antisense); for p53, 5'-GTGTGGTGGTGCCCTATGA-3' (sense) and 5'-GTGAGGCTCCCCTTTCTTG-3' (antisense); for 18S rRNA, 5'-ACACGGACAGGATTGACAGA-3' (sense) and 5'-GGACATCTAAGGGCATCACA-3' (antisense); for GAPDH, 5'-GGAGCGAGATCCCTCCAAAAT-3' (sense) and 5'-GGCTGTTGTCATACTTCTCATGG-3' (antisense). GAPDH was employed as endogenous control for the quantification of mRNAs, lncRNAs, and pri-miRNAs. U6 was employed as endogenous control for the quantification of miRNAs. The PCR efficiency was determined using qRT-PCR with standard curve method. The quantifications of RNAs were calculated using the comparative Ct method.
RNA Immunoprecipitation (RIP) assay
RNA Immunoprecipitation (RIP) assay was undertaken in Eca-109 cells with the EZ-Magna RIP™ RNA Binding Protein Immunoprecipitation Kit (Millipore) and NF90 specific primary antibody (ab131004, Abcam) following the protocol. The retrieved RNA was quantified using qRT-PCR as above described.
Cell proliferation assay
Cell proliferation was evaluated using Glo cell viability assay and Ethynyl deoxyuridine (EdU) incorporation assay as we previously described [21]. Briefly, for Glo cell viability assay, indicated ESCC cells were plated per well into 96-well plates and maintained for the indicated time. At indicated time points, the luminescence values were acquired with the CellTiter-Glo® Luminescent Cell Viability Assay (Promega, Madison, WI, USA) following the protocol. Then the luminescence values were used to plot the cell proliferation curves. EdU incorporation assay was undertaken using the EdU kit (Roche, Mannheim, Germany) following the protocol. The results were collected and quantified by the Zeiss fluorescence photomicroscope (Carl Zeiss, Oberkochen, Germany).
Cell migration assay
Cell migration was evaluated by transwell assay as we previously described [21]. Briefly, indicated ESCC cells re-suspended in serum-free medium with 1 μg/mL Mitomycin C to inhibit cell proliferation were seeded in the upper chamber of Millicell transwell chamber (8 μm pore size, Millipore). The lower chamber was supplemented with complete medium. After incubation for 48 hours, the cells on the upper chamber were scraped off, and the migratory cells on the bottom of the chamber were fixed, stained, and quantified using the Zeiss photomicroscope (Carl Zeiss).
Xenograft model
To construct ESCC xenograft model, 2×106 indicted ESCC cells were subcutaneously injected into 5-weeks-old male athymic BALB/c nude mice (SLRC Laboratory Animal Center, Shanghai, China). Subcutaneous tumor volumes were detected every four or seven days using a caliper and calculated by the formula V = 0.5 × L × W2 (L, length; W, width). At indicated time, the nude mice were sacrificed and subcutaneous xenografts were resected and weighted. The use of animals was reviewed and approved by the Review Board of the Second Affiliated Hospital of Fujian Medical University.
Statistical analysis
Statistical analyses were undertaken with the GraphPad Prism Software. Comparisons between groups were subjected to Student's t-test, one-way ANOVA followed by Dunnett's multiple comparison test, or Mann-Whitney U test as indicated. P < 0.05 was considered as statistically significant.
Results
NF90 upregulates miR-548k expression
To investigate the effects of NF90 on miR-548k, we overexpressed NF90 in KYSE30 cells via transfecting NF90 overexpression plasmids (Figure 1A). The expression of miR-548k in these transfected cells was measured by qRT-PCR. As displayed in Figure 1B, enhanced expression of NF90 significantly upregulated miR-548k expression. Furthermore, we inhibited NF90 in Eca-109 cells via transfecting NF90 specific shRNAs, and the results displayed that depletion of NF90 significantly downregulated miR-548k expression (Figure 1C and 1D). As an RNA binding protein, NF90 has been reported to bind mRNAs and/or pri-miRNAs and change their stabilities [38, 40]. Thus, we next investigated whether NF90 also regulates pri-miR-548k. RIP assays using NF90 specific antibody displayed that NF90 specifically interacted with pri-miR-548k, but not pri-miR-21 (Figure 1E). Next, we investigated the effects of NF90 on pri-miR-548k stability. 48 hours after transient transfection of NF90 overexpression or control plasmids into KYSE30 cells, the transfected cells were treated with 50 μM α-amanitin to block new RNA synthesis. The loss of pri-miR-548k transcript overtime was measured using qRT-PCR. As displayed in Figure 1F, enhanced expression of NF90 significantly elongated the half-life of pri-miR-548k transcript. Similarly, 48 hours after transient transfection of NF90 specific shRNAs or control shRNAs into Eca-109 cells, the transfected cells were treated with 50 μM α-amanitin and the loss of pri-miR-548k transcript overtime was measured. The results displayed that depletion of NF90 significantly shortened the half-life of pri-miR-548k transcript (Figure 1G). In addition, the effects of NF90 on pri-miR-548k expression were investigated. As displayed in Figure 1H and 1I, enhanced expression of NF90 upregulated pri-miR-548k expression, and while depletion of NF90 downregulated pri-miR-548k expression. Collectively, these data suggested that NF90 interacted with pri-miR-548k, increased pri-miR-548k stability, and upregulated the expression of pri-miR-548k and miR-548k.
NF90 promotes ESCC cell proliferation and migration
To explore the biological roles of NF90 in ESCC, we constructed NF90 overexpressed KYSE30 cells via transfecting NF90 overexpression plasmids. Glo cell viability assay displayed that enhanced expression of NF90 increased cell viability of KYSE30 cells (Figure 2A). EdU incorporation assay further displayed that enhanced expression of NF90 promoted cell proliferation of KYSE30 cells (Figure 2B). Transwell migration assay displayed that enhanced expression of NF90 promoted cell migration of KYSE30 cells (Figure 2C). In addition, we also constructed NF90 depleted Eca-109 cells via transfecting NF90 specific shRNAs. Glo cell viability assay displayed that depletion of NF90 decreased cell viability of Eca-109 cells (Figure 2D). EdU incorporation assay further displayed that depletion of NF90 inhibited cell proliferation of Eca-109 cells (Figure 2E). Transwell migration assay displayed that depletion of NF90 inhibited cell migration of Eca-109 cells (Figure 2F). Collectively, these data demonstrated that NF90 promotes ESCC cell proliferation and migration.
The regulatory roles of NF90 on miR-548k. (A) After transfection of NF90 overexpression or control plasmids into KYSE30 cells, NF90 expression were measured by western blot. (B) After transfection of NF90 overexpression or control plasmids into KYSE30 cells, miR-548k expression were measured by qRT-PCR. (C) After transfection of NF90 specific or control shRNAs into Eca-109 cells, NF90 expression were measured by western blot. (D) After transfection of NF90 specific or control shRNAs into Eca-109 cells, miR-548k expression were measured by qRT-PCR. (E) RIP assay followed by qRT-PCR was performed to detect the specific enrichment of pri-miR-548k with NF90 specific antibody compared with nonspecific IgG. pri-miR-21 was used as negative control. (F) After transfection of NF90 overexpression or control plasmids into KYSE30 cells, the stability of pri-miR-548k transcript over time was evaluated by qRT-PCR relative to time 0 after blocking new RNA synthesis with α-amanitin and normalized to 18S rRNA (transcribed by RNA polymerase I and not influenced by α-amanitin). (G) After transfection of NF90 specific or control shRNAs into Eca-109 cells, the stability of pri-miR-548k transcript over time was evaluated by qRT-PCR relative to time 0 after blocking new RNA synthesis with α-amanitin and normalized to 18S rRNA. (H) After transfection of NF90 overexpression or control plasmids into KYSE30 cells, pri-miR-548k expression was measured by qRT-PCR. (I) After transfection of NF90 specific or control shRNAs into Eca-109 cells, pri-miR-548k expression was measured by qRT-PCR. Results are presented as mean ± S.D. (n = 3). **P < 0.01, ***P < 0.001, ns, not significant, by Student's t-test (A, B, E, F and H) or one-way ANOVA followed by Dunnett's multiple comparison test (C, D, G and I).
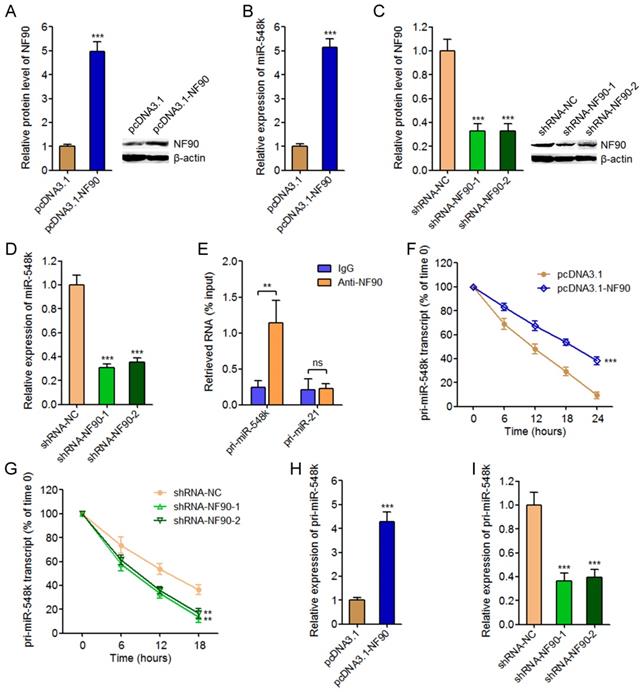
The biological roles of NF90 in ESCC cell proliferation and migration. (A) Glo cell viability assay was performed to detect cell viability of NF90 overexpressed and control KYSE30 cells. (B) EdU incorporation assay was performed to detect cell proliferation of NF90 overexpressed and control KYSE30 cells. The red color represents EdU-positive and proliferation active cells. Scale bars, 100 μm. (C) Transwell assay was performed to detect cell migration of NF90 overexpressed and control KYSE30 cells. Scale bars, 100 μm. (D) Glo cell viability assay was performed to detect cell viability of NF90 depleted and control Eca-109 cells. (E) EdU incorporation assay was performed to detect cell proliferation of NF90 depleted and control Eca-109 cells. The red color represents EdU-positive and proliferation active cells. Scale bars, 100 μm. (F) Transwell assay was performed to detect cell migration of NF90 depleted and control Eca-109 cells. Scale bars, 100 μm. Results are presented as mean ± S.D. (n = 3). **P < 0.01 by Student's t-test (A-C) or one-way ANOVA followed by Dunnett's multiple comparison test (D-F).
Inhibition of miR-548k abolishes the oncogenic roles of NF90 in ESCC
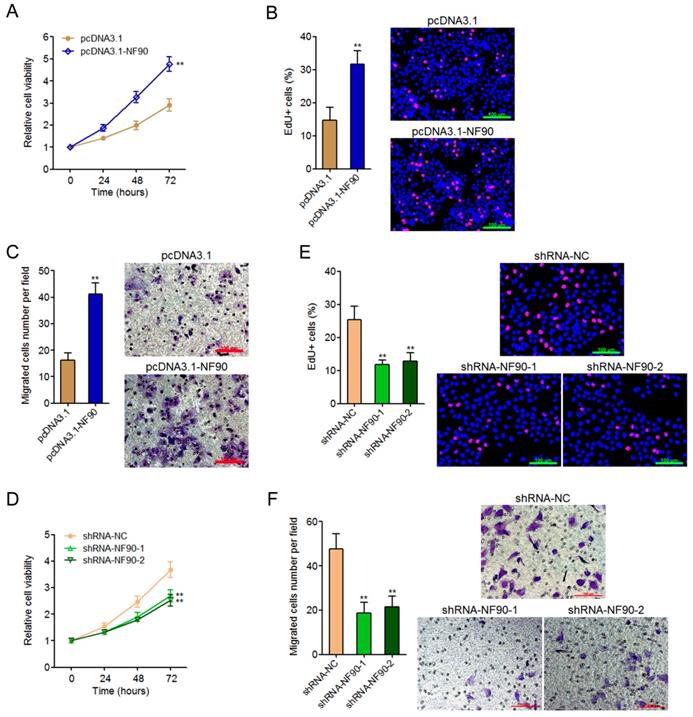
To investigate whether the oncogenic roles of NF90 in ESCC were dependent on the positive regulation of miR-548k, we inhibited miR-548k expression in NF90 overexpressed KYSE30 cells via infecting miR-548k inhibition lentiviruses (Figure 3A). Glo cell viability assay displayed that inhibition of miR-548k abolished the increasing of cell viability caused by enhanced expression of NF90 (Figure 3B). EdU incorporation assay displayed that inhibition of miR-548k abolished the pro-proliferative roles of NF90 (Figure 3C). Transwell migration assay displayed that inhibition of miR-548k abolished the pro-migratory roles of NF90 (Figure 3D). Collectively, these data demonstrated that the positive regulation of miR-548k at least partially mediates the oncogenic roles of NF90 in ESCC.
The feedback amplification loop between NF90, miR-548k, and lncRNA-LET
In our previous study, we have found miR-548k directly targets and inhibits the expression of lncRNA-LET [21]. Due to the negative regulation of NF90 by lncRNA-LET, we also found miR-548k upregulated NF90 in our previous study [39]. Combined with the positive regulation of miR-548k by NF90, we supposed a feedback regulatory loop existed between NF90, miR-548k, and lncRNA-LET (Figure 4A).
First, we investigated the effects of NF90 on miR-548k and lncRNA-LET. The above described results have demonstrated the positive regulation of miR-548k by NF90. The effects of NF90 on lncRNA-LET were investigated in NF90 overexpressed and control KYSE30 cells, and NF90 depleted and control Eca-109 cells. As displayed in Figure 4B and 4C, enhanced expression of NF90 downregulated lncRNA-LET expression, and while depletion of NF90 upregulated lncRNA-LET expression. Moreover, the expression of lncRNA-LET was evaluated in NF90 overexpressed and miR-548k simultaneously inhibited KYSE30 cells. As displayed in Figure 4D, inhibition of miR-548k abolished the downregulation of lncRNA-LET caused by enhanced expression of NF90. These data suggested that NF90 negatively regulated lncRNA-LET via positive regulation of miR-548k.
Second, our previous report has showed that miR-548k positively regulated NF90 via negative regulation of lncRNA-LET [21].
Third, we investigated the effects of lncRNA-LET on NF90 and miR-548k. Our previous report has revealed the negative regulation of NF90 by lncRNA-LET [21]. In this study, we further investigated the effects of lncRNA-LET on miR-548k. We overexpressed lncRNA-LET in Eca-109 cells via transfecting lncRNA-LET overexpression plasmids (Figure 4E). Then, the expression of miR-548k and pri-miR-548k in lncRNA-LET overexpressed and control Eca-109 cells were detected. As displayed in Figure 4F, enhanced expression of lncRNA-LET downregulated the expression of miR-548k and pri-miR-548k. Moreover, we depleted lncRNA-LET in KYSE30 cells via transfecting lncRNA-LET specific shRNAs (Figure 4G). Then, the expression of miR-548k and pri-miR-548k were detected, and the results displayed that depletion of lncRNA-LET upregulated the expression of miR-548k and pri-miR-548k (Figure 4H). Overexpression of NF90 reversed the downregulation of miR-548k and pri-miR-548k caused by enhanced expression of lncRNA-LET (Figure 4I). These data suggested that lncRNA-LET negatively regulated miR-548k via downregulation of NF90. Collectively, these results demonstrated the positive feedback loop between NF90, miR-548k, and lncRNA-LET.
The roles of NF90 in ESCC cell proliferation and migration are miR-548k dependent. (A) miR-548k expression in NF90 overexpressed and miR-548k simultaneously depleted and control KYSE30 cells was measured by qRT-PCR. (B) Glo cell viability assay was performed to detect cell viability of NF90 overexpressed and miR-548k simultaneously depleted and control KYSE30 cells. (C) EdU incorporation assay was performed to detect cell proliferation of NF90 overexpressed and miR-548k simultaneously depleted and control KYSE30 cells. The red color represents EdU-positive and proliferation active cells. Scale bars, 100 μm. (D) Transwell assay was performed to detect cell migration of NF90 overexpressed and miR-548k simultaneously depleted and control KYSE30 cells. Scale bars, 100 μm. Results are presented as mean ± S.D. (n = 3). **P < 0.01, ***P < 0.001, ns, not significant, by one-way ANOVA followed by Dunnett's multiple comparison test.
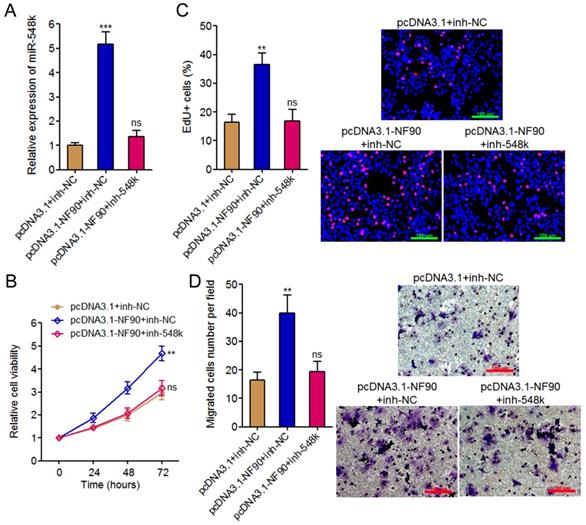
The positive feedback loop between NF90, miR-548k, and lncRNA-LET. (A) Schematic outline of the reciprocal regulation between NF90, miR-548k, and lncRNA-LET. Red arrowhead represents positive regulation, and while blue arrowhead represents negative regulation. (B) lncRNA-LET expression in NF90 overexpressed and control KYSE30 cells was measured by qRT-PCR. (C) lncRNA-LET expression in NF90 depleted and control Eca-109 cells was measured by qRT-PCR. (D) lncRNA-LET expression in NF90 overexpressed and miR-548k simultaneously depleted and control KYSE30 cells was measured by qRT-PCR. (E) lncRNA-LET expression in lncRNA-LET overexpressed and control Eca-109 cells was measured by qRT-PCR. (F) miR-548k and pri-miR-548k expressions in lncRNA-LET overexpressed and control Eca-109 cells were measured by qRT-PCR. (G) lncRNA-LET expression in lncRNA-LET depleted and control KYSE30 cells was measured by qRT-PCR. (H) miR-548k and pri-miR-548k expressions in lncRNA-LET depleted and control KYSE30 cells were measured by qRT-PCR. (I) miR-548k and pri-miR-548k expressions in lncRNA-LET and NF90 simultaneously overexpressed and control Eca-109 cells were measured by qRT-PCR. Results are presented as mean ± S.D. (n = 3). ***P < 0.001, ns, not significant, by Student's t-test (B, E and F) or one-way ANOVA followed by Dunnett's multiple comparison test (C, D, G, H and I).
The NF90/miR-548k/lncRNA-LET feedback loop controls the expression of HIF-1α, VEGF, KLF10, EGFR, and p53
NF90 has been reported to regulate hypoxia inducible factor-1α (HIF-1α) and vascular endothelial growth factor (VEGF) mRNA stability and upregulate the expression of HIF-1α and VEGF in human cancers [39, 43]. miR-548k has been reported to inhibit Kruppel like factor 10 (KLF10) and activate epidermal growth factor receptor (EGFR) in ESCC [44]. lncRNA-LET has been reported to upregulate p53 in ESCC [45]. HIF-1α, VEGF, KLF10, EGFR, and p53 are crucial factors implicated in tumorigenesis and progression. HIF-1α, VEGF, and EGFR has oncogenic roles, and while KLF10 and p53 has tumor suppressive roles. Thus, we next investigated the effects of the NF90/miR-548k/lncRNA-LET feedback loop on HIF-1α, VEGF, KLF10, EGFR, and p53 (Figure 5A).
First, we measured the expression of HIF-1α, VEGF, KLF10, EGFR, and p53 in NF90 overexpressed and control KYSE30 cells, and NF90 depleted and control Eca-109 cells. As displayed in Figure 5B and 5C, enhanced expression of NF90 upregulated HIF-1α and VEGF, downregulated KLF10, upregulated EGFR, and downregulated p53, which were consistent with the effects of miR-548k overexpression on KLF10 and EGFR, and also the effects of lncRNA-LET depletion on p53. Conversely, depletion of NF90 downregulated HIF-1α and VEGF, upregulated KLF10, downregulated EGFR, and upregulated p53, which were consistent with the effects of miR-548k depletion on KLF10 and EGFR, and also the effects of lncRNA-LET overexpression on p53.
Second, we constructed miR-548k overexpressed KYSE30 cells via infecting miR-548k overexpression lentiviruses, and also miR-548k depleted Eca-109 cells via infecting miR-548k inhibition lentiviruses (Figure 5D and 5E). Then, we measured the expression of HIF-1α, VEGF, KLF10, EGFR, and p53 in miR-548k overexpressed KYSE30 cells and miR-548k depleted Eca-109 cells. As displayed in Figure 5F and 5G, enhanced expression of miR-548k upregulated HIF-1α and VEGF, downregulated KLF10, upregulated EGFR, and downregulated p53, which were consistent with the effects of NF90 overexpression on HIF-1α and VEGF, and also the effects of lncRNA-LET depletion on p53. Conversely, depletion of miR-548k downregulated HIF-1α and VEGF, upregulated KLF10, downregulated EGFR, and upregulated p53, which were consistent with the effects of NF90 depletion on HIF-1α and VEGF, and also the effects of lncRNA-LET overexpression on p53.
The NF90/miR-548k/lncRNA-LET feedback loop regulates the expression of HIF-1α, VEGF, KLF10, EGFR, and p53. (A) Schematic outline of the regulation of HIF-1α, VEGF, KLF10, EGFR, and p53 by NF90, miR-548k, and lncRNA-LET. Red arrowhead represents positive regulation, and while blue arrowhead represents negative regulation. (B) HIF-1α, VEGF, KLF10, EGFR, and p53 expressions in NF90 overexpressed and control KYSE30 cells were measured by qRT-PCR. (C) HIF-1α, VEGF, KLF10, EGFR, and p53 expressions in NF90 depleted and control Eca-109 cells were measured by qRT-PCR. (D) miR-548k expression in miR-548k overexpressed and control KYSE30 cells was measured by qRT-PCR. (E) miR-548k expression in miR-548k depleted and control Eca-109 cells was measured by qRT-PCR. (F) HIF-1α, VEGF, KLF10, EGFR, and p53 expressions in miR-548k overexpressed and control KYSE30 cells were measured by qRT-PCR. (G) HIF-1α, VEGF, KLF10, EGFR, and p53 expressions in miR-548k depleted and control Eca-109 cells were measured by qRT-PCR. (H) HIF-1α, VEGF, KLF10, EGFR, and p53 expressions in lncRNA-LET overexpressed and control Eca-109 cells were measured by qRT-PCR. (I) HIF-1α, VEGF, KLF10, EGFR, and p53 expressions in lncRNA-LET depleted and control KYSE30 cells were measured by qRT-PCR. Results are presented as mean ± S.D. (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, by Student's t-test (B, D, E, F, G and H) or one-way ANOVA followed by Dunnett's multiple comparison test (C, I).
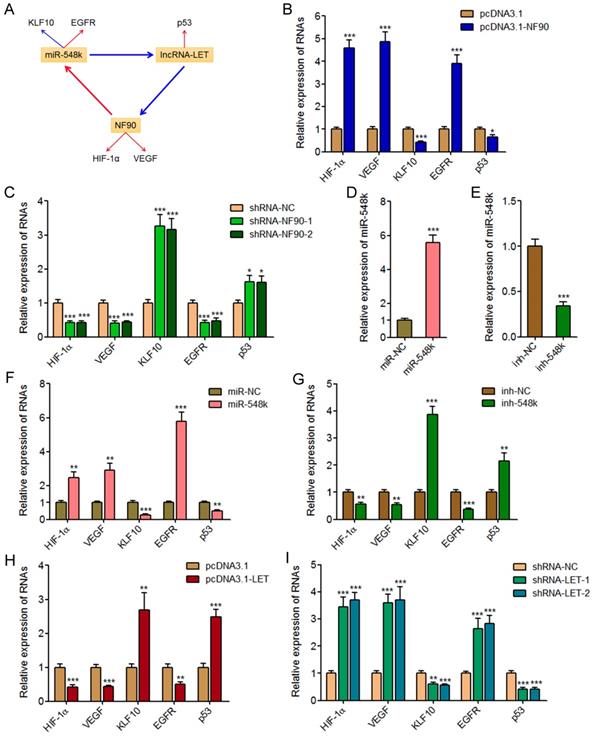
Third, we measured the expression of HIF-1α, VEGF, KLF10, EGFR, and p53 in lncRNA-LET overexpressed Eca-109 cells and lncRNA-LET depleted KYSE30 cells. As displayed in Figure 5H and 5I, enhanced expression of lncRNA-LET downregulated HIF-1α and VEGF, upregulated KLF10, downregulated EGFR, and upregulated p53, which were consistent with the effects of NF90 depletion on HIF-1α and VEGF, and also the effects of miR-548k depletion on KLF10 and EGFR. Conversely, depletion of lncRNA-LET upregulated HIF-1α and VEGF, downregulated KLF10, upregulated EGFR, and downregulated p53, which were consistent with the effects of NF90 overexpression on HIF-1α and VEGF, and also the effects of miR-548k overexpression on KLF10 and EGFR. Collectively, these data suggested that the NF90/miR-548k/lncRNA-LET feedback loop upregulates HIF-1α, VEGF, and EGFR, and while downregulates KLF10 and p53.
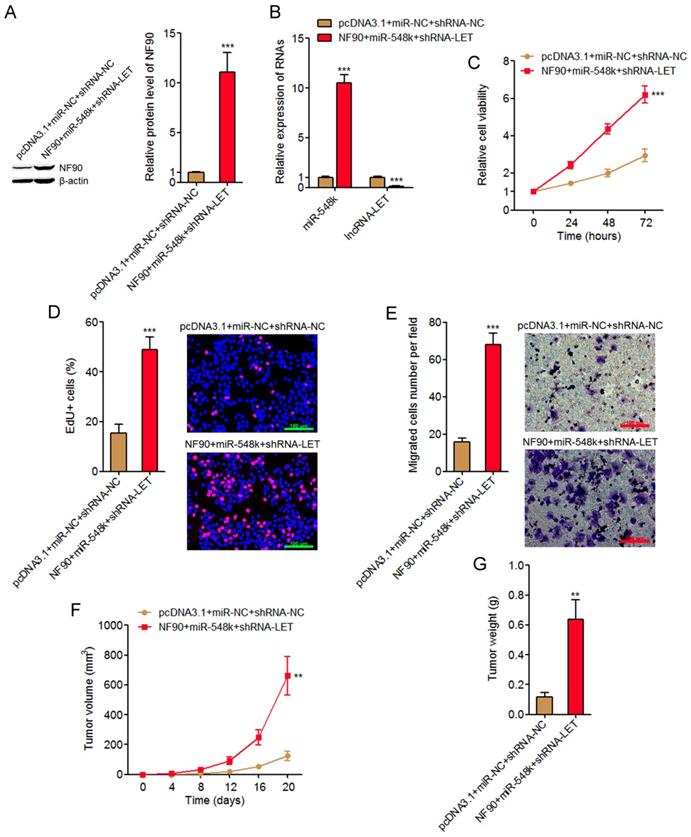
Activation of the NF90/miR-548k/lncRNA-LET feedback loop significantly promotes ESCC cell proliferation and migration in vitro and ESCC xenograft growth in vivo
The NF90/miR-548k/lncRNA-LET feedback loop upregulates HIF-1α, VEGF, and EGFR, which all have oncogenic roles. The NF90/miR-548k/lncRNA-LET feedback loop also downregulates KLF10 and p53, which both have tumor suppressive roles. Therefore, we further investigated the biological roles of the NF90/miR-548k/lncRNA-LET feedback loop in ESCC. We activated the feedback loop via constructing NF90 and miR-548k simultaneously overexpressed and lncRNA-LET simultaneously depleted KYSE30 cells (Figure 6A and 6B). Glo cell viability assay displayed that activation of the NF90/miR-548k/lncRNA-LET feedback loop markedly increased cell viability of KYSE30 cells (Figure 6C). EdU incorporation assay further displayed that activation of the NF90/miR-548k/lncRNA-LET feedback loop markedly promoted cell proliferation of KYSE30 cells (Figure 6D). Transwell migration assay displayed that activation of the NF90/miR-548k/lncRNA-LET feedback loop markedly promoted cell migration of KYSE30 cells (Figure 6E). To further explore the biological roles of the NF90/miR-548k/lncRNA-LET feedback loop in ESCC, NF90 and miR-548k simultaneously overexpressed and lncRNA-LET simultaneously depleted and control KYSE30 cells were subcutaneously injected into nude mice. Xenograft tumor growth was detected via measuring tumor volumes every four days and tumor weights at the 20th day after injection. As displayed in Figure 6F and 6G, activation of the NF90/miR-548k/lncRNA-LET feedback loop markedly promoted xenograft tumor growth in vivo. Collectively, these data supported the strongly oncogenic roles of the NF90/miR-548k/lncRNA-LET feedback loop in ESCC.
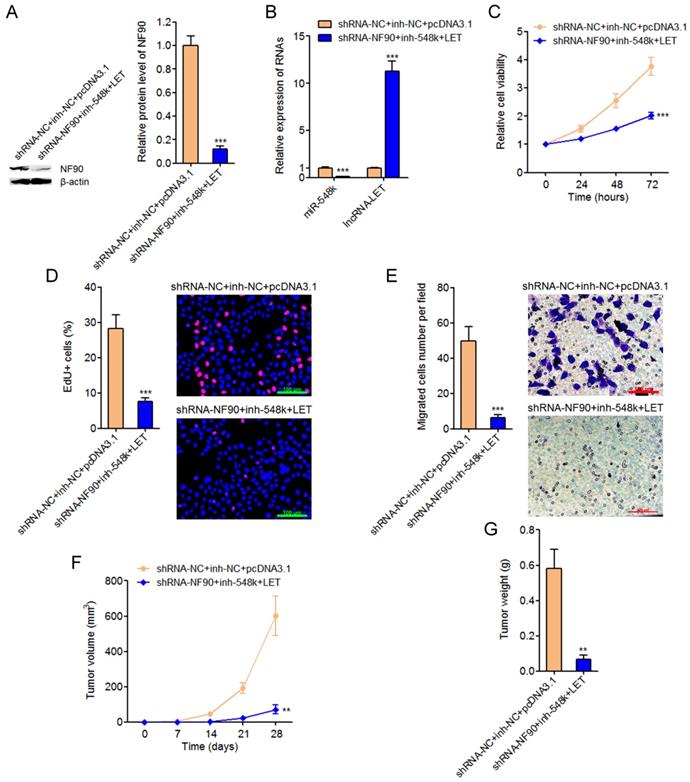
Targeting the NF90/miR-548k/lncRNA-LET feedback loop significantly represses ESCC cell proliferation and migration in vitro and ESCC xenograft growth in vivo
To explore the significances of targeting the NF90/miR-548k/lncRNA-LET feedback loop for ESCC, we constructed NF90 and miR-548k simultaneously depleted and lncRNA-LET simultaneously overexpressed Eca-109 cells (Figure 7A and 7B). Glo cell viability assay displayed that targeting the NF90/miR-548k/lncRNA-LET feedback loop markedly decreased cell viability of Eca-109 cells (Figure 7C). EdU incorporation assay further displayed that targeting the NF90/miR-548k/lncRNA-LET feedback loop markedly inhibited cell proliferation of Eca-109 cells (Figure 7D). Transwell migration assay displayed that targeting the NF90/miR-548k/lncRNA-LET feedback loop markedly inhibited cell migration of Eca-109 cells (Figure 7E). NF90 and miR-548k simultaneously depleted and lncRNA-LET simultaneously overexpressed and control Eca-109 cells were subcutaneously injected into nude mice. Xenograft tumor growth was detected via measuring tumor volumes every seven days and tumor weights at the 28th day after injection. As displayed in Figure 7F and 7G, targeting the NF90/miR-548k/lncRNA-LET feedback loop markedly repressed xenograft tumor growth in vivo. Collectively, these data demonstrated that targeted inhibition of the NF90/miR-548k/lncRNA-LET feedback loop has significantly tumor suppressive roles in ESCC.
Discussion
The underlying molecular mechanisms mediating tumorigenesis and progression of ESCC are not fully understood [46, 47]. Although many genome and transcriptome sequencings have identified many mutated genes and aberrant expressed transcripts in ESCC, the roles and regulatory mechanisms of most of these genes and transcripts in ESCC are still unclear [7, 48, 49]. Some of these genes and transcripts may be driver molecular events of ESCC. But, many others may also be passenger molecular events in ESCC. Therefore, the identification of functional molecular events is critical for providing efficient therapeutic targets for ESCC.
Activation of the NF90/miR-548k/lncRNA-LET feedback loop significantly promotes ESCC progression. (A) NF90 expressions in NF90 and miR-548k simultaneously overexpressed and lncRNA-LET simultaneously depleted KYSE30 cells were measured by western blot. (B) miR-548k and lncRNA-LET expressions in NF90 and miR-548k simultaneously overexpressed and lncRNA-LET simultaneously depleted KYSE30 cells were measured by qRT-PCR. (C) Glo cell viability assay was performed to detect cell viability of NF90 and miR-548k simultaneously overexpressed and lncRNA-LET simultaneously depleted KYSE30 cells. (D) EdU incorporation assay was performed to detect cell proliferation of NF90 and miR-548k simultaneously overexpressed and lncRNA-LET simultaneously depleted KYSE30 cells. The red color represents EdU-positive and proliferation active cells. Scale bars, 100 μm. (E) Transwell assay was performed to detect cell migration of NF90 and miR-548k simultaneously overexpressed and lncRNA-LET simultaneously depleted KYSE30 cells. Scale bars, 100 μm. For A-E, results are presented as mean ± S.D. (n = 3). ***P < 0.001 by Student's t-test. (F) NF90 and miR-548k simultaneously overexpressed and lncRNA-LET simultaneously depleted KYSE30 cells were subcutaneously injected into nude mice. Xenograft tumor volumes were detected every four days. (G) Xenograft tumor weights were detected at the 20th days after injection. For F-G, results are presented as mean ± S.D. (n = 5 mice). **P < 0.01 by Mann-Whitney U test.
In this study, we identified a positive feedback amplification loop between NF90, miR-548k, and lncRNA-LET. Functional assays demonstrated that activation of the NF90/miR-548k/lncRNA-LET feedback loop significantly promoted ESCC cell proliferation and migration in vitro, and ESCC tumor growth in vivo. Targeting the NF90/miR-548k/lncRNA-LET feedback loop significantly inhibited ESCC cell proliferation and migration in vitro, and ESCC tumor growth in vivo. Thus, our study provided a novel strategy to inhibit ESCC, which is the combined manipulation of a protein (NF90), a miRNA (miR-548k), and a lncRNA (lncRNA-LET).
Targeting the NF90/miR-548k/lncRNA-LET feedback loop significantly represses ESCC progression. (A) NF90 expressions in NF90 and miR-548k simultaneously depleted, and lncRNA-LET simultaneously overexpressed Eca-109 cells were measured by western blot. (B) miR-548k and lncRNA-LET expressions in NF90 and miR-548k simultaneously depleted, and lncRNA-LET simultaneously overexpressed Eca-109 cells were measured by qRT-PCR. (C) Glo cell viability assay was performed to detect cell viability of NF90 and miR-548k simultaneously depleted and lncRNA-LET simultaneously overexpressed Eca-109 cells. (D) EdU incorporation assay was performed to detect cell proliferation of NF90 and miR-548k simultaneously depleted and lncRNA-LET simultaneously overexpressed Eca-109 cells. The red color represents EdU-positive and proliferation active cells. Scale bars, 100 μm. (E) Transwell assay was performed to detect cell migration of NF90 and miR-548k simultaneously depleted and lncRNA-LET simultaneously overexpressed Eca-109 cells. Scale bars, 100 μm. For A-E, results are presented as mean ± S.D. (n = 3). ***P < 0.001 by Student's t-test. (F) NF90 and miR-548k simultaneously depleted and lncRNA-LET simultaneously overexpressed Eca-109 cells were subcutaneously injected into nude mice. Xenograft tumor volumes were detected every seven days. (G) Xenograft tumor weights were detected at the 28th days after injection. For F-G, results are presented as mean ± S.D. (n = 5 mice). **P < 0.01 by Mann-Whitney U test.
NF90 is an RNA binding protein, which has complex and diverse biological roles and mechanisms of action in different diseases. Zhuang et al. reported NF90 promoted gemcitabine resistance of bladder cancer via repressing miR-145 biogenesis [41]. Zhang et al. reported NF90 promoted angiogenesis of cervical cancer via regulating PI3K/Akt signaling pathway [39]. Barbier et al. reported NF90 reduced ovarian cancer proliferation and metastasis via regulating DICER expression [50]. Jiang et al. reported NF90 promoted metastasis of breast tumors via repressing p21 [51]. Zhou et al. reported NF90 promoted colorectal cancer progression via regulating VEGF [43]. However, the roles and mechanisms of action of NF90 in ESCC are still unknown. In this study, using gain-of-function and loss-of-function assays, we found NF90 promoted ESCC cell proliferation and migration. Mechanistic investigation revealed that NF90 directly bound pri-miR-548k, increased the stability of pri-miR-548k, and upregulated the expression of pri-miR-548k and miR-548k. The positive regulation of miR-548k by NF90 is different from the reported effects of NF90 on suppressing biogenesis of miR-7 and miR-145 [40, 41]. The different binding manner between NF90 and different pri-miRNAs may have different effects on the processing and/or degradation of different pri-miRNAs, which need further investigation.
In our previous study, we have found the negative regulation of lncRNA-LET by miR-548k, the negative regulation of NF90 by lncRNA-LET, and thus the positive regulation of NF90 by miR-548k via repressing lncRNA-LET [21]. Combination with the above identified positive regulation of miR-548k by NF90, NF90, miR-548k, and lncRNA-LET forms feedback amplification loop in ESCC. Indeed, we verified that NF90 upregulated miR-548k and downregulated lncRNA-LET, miR-548k downregulated lncRNA-LET and upregulated NF90, lncRNA-LET downregulated NF90 and miR-548k, which support the feedback loop between NF90, miR-548k, and lncRNA-LET. HIF-1α and VEGF are reported downstream targets of NF90 [39]. KLF10 and EGFR are reported downstream targets of miR-548k [44]. p53 is reported downstream target of lncRNA-LET [45]. Our results found that NF90, miR-548k, and lncRNA-LET all regulated HIF-1α, VEGF, KLF10, EGFR, and p53, which have important roles in tumor growth and metastasis. These results further support the feedback amplification loop between NF90, miR-548k, and lncRNA-LET. Due to the extensive and strong regulatory roles of the NF90/miR-548k/lncRNA-LET feedback loop in critical oncogenes and tumor suppressors, the NF90/miR-548k/lncRNA-LET feedback loop has strongly oncogenic roles in ESCC.
In conclusion, we identified a critical feedback amplification loop between NF90, miR-548k, and lncRNA-LET, which significantly promotes ESCC progression. Targeting the NF90/miR-548k/lncRNA-LET feedback loop significantly represses ESCC progression. Thus, our study suggests that the NF90/miR-548k/lncRNA-LET feedback loop may be promising therapeutic targets for ESCC.
Abbreviations
ESCC, esophageal squamous cell carcinoma; lncRNA, long noncoding RNA; NF90, nuclear factor 90; miRNA, microRNA; cDNA, complementary DNA; qRT-PCR, quantitative real time polymerase chain reaction; RIP, RNA Immunoprecipitation; EdU, Ethynyl deoxyuridine; HIF-1α, hypoxia inducible factor-1α; VEGF, vascular endothelial growth factor; KLF10, Kruppel like factor 10; EGFR, epidermal growth factor receptor.
Acknowledgements
This work was supported by the Natural Science Foundation of Fujian Province (2019J01050835) and Quanzhou High-level Talent Plan (2017Z008).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018
3. Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137-50
4. Wei G, Luo H, Sun Y, Li J, Tian L, Liu W. et al. Transcriptome profiling of esophageal squamous cell carcinoma reveals a long noncoding RNA acting as a tumor suppressor. Oncotarget. 2015;6:17065-80
5. Pohl H, Welch HG. The role of overdiagnosis and reclassification in the marked increase of esophageal adenocarcinoma incidence. J Natl Cancer Inst. 2005;97:142-6
6. Pennathur A, Farkas A, Krasinskas AM, Ferson PF, Gooding WE, Gibson MK. et al. Esophagectomy for T1 esophageal cancer: outcomes in 100 patients and implications for endoscopic therapy. Ann Thorac Surg. 2009;87:1048-54 discussion 54-5
7. Song Y, Li L, Ou Y, Gao Z, Li E, Li X. et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509:91-5
8. Gao YB, Chen ZL, Li JG, Hu XD, Shi XJ, Sun ZM. et al. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet. 2014;46:1097-102
9. Huang X, Zhou X, Hu Q, Sun B, Deng M, Qi X. et al. Advances in esophageal cancer: A new perspective on pathogenesis associated with long non-coding RNAs. Cancer Lett. 2018;413:94-101
10. Yan W, Wu X, Zhou W, Fong MY, Cao M, Liu J. et al. Cancer-cell-secreted exosomal miR-105 promotes tumour growth through the MYC-dependent metabolic reprogramming of stromal cells. Nat Cell Biol. 2018;20:597-609
11. Mohr S, Doebele C, Comoglio F, Berg T, Beck J, Bohnenberger H. et al. Hoxa9 and Meis1 Cooperatively Induce Addiction to Syk Signaling by Suppressing miR-146a in Acute Myeloid Leukemia. Cancer Cell. 2017;31:549-62 e11
12. Yuan JH, Yang F, Chen BF, Lu Z, Huo XS, Zhou WP. et al. The histone deacetylase 4/SP1/microrna-200a regulatory network contributes to aberrant histone acetylation in hepatocellular carcinoma. Hepatology. 2011;54:2025-35
13. Inui M, Mokuda S, Sato T, Tamano M, Takada S, Asahara H. Dissecting the roles of miR-140 and its host gene. Nat Cell Biol. 2018;20:516-8
14. Haj-Ahmad TA, Abdalla MA, Haj-Ahmad Y. Potential Urinary miRNA Biomarker Candidates for the Accurate Detection of Prostate Cancer among Benign Prostatic Hyperplasia Patients. J Cancer. 2014;5:182-91
15. Leite KR, Reis ST, Viana N, Morais DR, Moura CM, Silva IA. et al. Controlling RECK miR21 Promotes Tumor Cell Invasion and Is Related to Biochemical Recurrence in Prostate Cancer. J Cancer. 2015;6:292-301
16. Chernyy V, Pustylnyak V, Kozlov V, Gulyaeva L. Increased expression of miR-155 and miR-222 is associated with lymph node positive status. J Cancer. 2018;9:135-40
17. Ho CS, Noor SM, Nagoor NH. MiR-378 and MiR-1827 Regulate Tumor Invasion, Migration and Angiogenesis in Human Lung Adenocarcinoma by Targeting RBX1 and CRKL, Respectively. J Cancer. 2018;9:331-45
18. Bu Y, Yoshida A, Chitnis N, Altman BJ, Tameire F, Oran A. et al. A PERK-miR-211 axis suppresses circadian regulators and protein synthesis to promote cancer cell survival. Nat Cell Biol. 2018;20:104-15
19. Wu MZ, Cheng WC, Chen SF, Nieh S, O'Connor C, Liu CL. et al. miR-25/93 mediates hypoxia-induced immunosuppression by repressing cGAS. Nat Cell Biol. 2017;19:1286-96
20. Lee D, Sun S, Zhang XQ, Zhang PD, Ho AS, Kiang KM. et al. MicroRNA-210 and Endoplasmic Reticulum Chaperones in the Regulation of Chemoresistance in Glioblastoma. J Cancer. 2015;6:227-32
21. Chen Z, Lin J, Wu S, Xu C, Chen F, Huang Z. Up-regulated miR-548k promotes esophageal squamous cell carcinoma progression via targeting long noncoding RNA-LET. Exp Cell Res. 2018;362:90-101
22. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629-41
23. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7-21
24. Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF. et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666-81
25. Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y. et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199-208
26. Hu WL, Jin L, Xu A, Wang YF, Thorne RF, Zhang XD. et al. GUARDIN is a p53-responsive long non-coding RNA that is essential for genomic stability. Nat Cell Biol. 2018;20:492-502
27. Yuan JH, Liu XN, Wang TT, Pan W, Tao QF, Zhou WP. et al. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat Cell Biol. 2017;19:820-32
28. Michelini F, Pitchiaya S, Vitelli V, Sharma S, Gioia U, Pessina F. et al. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat Cell Biol. 2017;19:1400-11
29. Lin A, Hu Q, Li C, Xing Z, Ma G, Wang C. et al. The LINK-A lncRNA interacts with PtdIns(3,4,5)P3 to hyperactivate AKT and confer resistance to AKT inhibitors. Nat Cell Biol. 2017;19:238-51
30. Grelet S, Link LA, Howley B, Obellianne C, Palanisamy V, Gangaraju VK. et al. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat Cell Biol. 2017;19:1105-15
31. Wang Z, Yang B, Zhang M, Guo W, Wu Z, Wang Y. et al. lncRNA Epigenetic Landscape Analysis Identifies EPIC1 as an Oncogenic lncRNA that Interacts with MYC and Promotes Cell-Cycle Progression in Cancer. Cancer Cell. 2018;33:706-20 e9
32. Mondal T, Juvvuna PK, Kirkeby A, Mitra S, Kosalai ST, Traxler L. et al. Sense-Antisense lncRNA Pair Encoded by Locus 6p22.3 Determines Neuroblastoma Susceptibility via the USP36-CHD7-SOX9 Regulatory Axis. Cancer Cell. 2018;33:417-34 e7
33. Berger AC, Korkut A, Kanchi RS, Hegde AM, Lenoir W, Liu W. et al. A Comprehensive Pan-Cancer Molecular Study of Gynecologic and Breast Cancers. Cancer Cell. 2018;33:690-705 e9
34. Zhu XT, Yuan JH, Zhu TT, Li YY, Cheng XY. Long noncoding RNA glypican 3 (GPC3) antisense transcript 1 promotes hepatocellular carcinoma progression via epigenetically activating GPC3. FEBS J. 2016;283:3739-54
35. Li JK, Chen C, Liu JY, Shi JZ, Liu SP, Liu B. et al. Long noncoding RNA MRCCAT1 promotes metastasis of clear cell renal cell carcinoma via inhibiting NPR3 and activating p38-MAPK signaling. Mol Cancer. 2017;16:111
36. Lin C, Zhang S, Wang Y, Wang Y, Nice E, Guo C. et al. Functional Role of a Novel Long Noncoding RNA TTN-AS1 in Esophageal Squamous Cell Carcinoma Progression and Metastasis. Clin Cancer Res. 2018;24:486-98
37. Wu Y, Hu L, Liang Y, Li J, Wang K, Chen X. et al. Up-regulation of lncRNA CASC9 promotes esophageal squamous cell carcinoma growth by negatively regulating PDCD4 expression through EZH2. Mol Cancer. 2017;16:150
38. Song D, Huang H, Wang J, Zhao Y, Hu X, He F. et al. NF90 regulates PARP1 mRNA stability in hepatocellular carcinoma. Biochem Biophys Res Commun. 2017;488:211-7
39. Zhang W, Xiong Z, Wei T, Li Q, Tan Y, Ling L. et al. Nuclear factor 90 promotes angiogenesis by regulating HIF-1alpha/VEGF-A expression through the PI3K/Akt signaling pathway in human cervical cancer. Cell Death Dis. 2018;9:276
40. Higuchi T, Todaka H, Sugiyama Y, Ono M, Tamaki N, Hatano E. et al. Suppression of MicroRNA-7 (miR-7) Biogenesis by Nuclear Factor 90-Nuclear Factor 45 Complex (NF90-NF45) Controls Cell Proliferation in Hepatocellular Carcinoma. J Biol Chem. 2016;291:21074-84
41. Zhuang J, Shen L, Yang L, Huang X, Lu Q, Cui Y. et al. TGFbeta1 Promotes Gemcitabine Resistance through Regulating the LncRNA-LET/NF90/miR-145 Signaling Axis in Bladder Cancer. Theranostics. 2017;7:3053-67
42. Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F. et al. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49:1083-96
43. Zhou Q, Zhu Y, Wei X, Zhou J, Chang L, Sui H. et al. MiR-590-5p inhibits colorectal cancer angiogenesis and metastasis by regulating nuclear factor 90/vascular endothelial growth factor A axis. Cell Death Dis. 2016;7:e2413
44. Zhang W, Hong R, Li L, Wang Y, Du P, Ou Y. et al. The chromosome 11q13.3 amplification associated lymph node metastasis is driven by miR-548k through modulating tumor microenvironment. Mol Cancer. 2018;17:125
45. Wang PL, Liu B, Xia Y, Pan CF, Ma T, Chen YJ. Long non-coding RNA-Low Expression in Tumor inhibits the invasion and metastasis of esophageal squamous cell carcinoma by regulating p53 expression. Mol Med Rep. 2016;13:3074-82
46. Kashyap MK, Abdel-Rahman O. Expression, regulation and targeting of receptor tyrosine kinases in esophageal squamous cell carcinoma. Mol Cancer. 2018;17:54
47. Liu A, Zhu J, Wu G, Cao L, Tan Z, Zhang S. et al. Antagonizing miR-455-3p inhibits chemoresistance and aggressiveness in esophageal squamous cell carcinoma. Mol Cancer. 2017;16:106
48. Liu Q, Cui X, Yu X, Bian BS, Qian F, Hu XG. et al. Cripto-1 acts as a functional marker of cancer stem-like cells and predicts prognosis of the patients in esophageal squamous cell carcinoma. Mol Cancer. 2017;16:81
49. Wang W, He X, Zheng Z, Ma X, Hu X, Wu D. et al. Serum HOTAIR as a novel diagnostic biomarker for esophageal squamous cell carcinoma. Mol Cancer. 2017;16:75
50. Barbier J, Chen X, Sanchez G, Cai M, Helsmoortel M, Higuchi T. et al. An NF90/NF110-mediated feedback amplification loop regulates dicer expression and controls ovarian carcinoma progression. Cell Res. 2018;28:556-71
51. Jiang Z, Slater CM, Zhou Y, Devarajan K, Ruth KJ, Li Y. et al. LincIN, a novel NF90-binding long non-coding RNA, is overexpressed in advanced breast tumors and involved in metastasis. Breast Cancer Res. 2017;19:62
Author contact
![]() Corresponding author: Zhijun Huang, Department of Surgical Oncology, the Second Affiliated Hospital of Fujian Medical University, No.34 North Zhongshan Road, Quanzhou 362000, Fujian, China. Email: huangzjedu.cn; Phone: +86-0595-86580796; Fax: +86-0595-22195115.
Corresponding author: Zhijun Huang, Department of Surgical Oncology, the Second Affiliated Hospital of Fujian Medical University, No.34 North Zhongshan Road, Quanzhou 362000, Fujian, China. Email: huangzjedu.cn; Phone: +86-0595-86580796; Fax: +86-0595-22195115.

 Global reach, higher impact
Global reach, higher impact