Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(5):1277-1287. doi:10.7150/jca.35444 This issue Cite
Research Paper
M2 Macrophages Promote HCC Cells Invasion and Migration via miR-149-5p/MMP9 Signaling
1. Department of Biliary and Pancreatic Surgery, Xiangya Hospital, Central South University, Changsha, 410008, China.
2. Department of Urology, Shanghai Tenth People's Hospital, School of Medicine in Tongji University, Shanghai, China.
3. Department of Liver Surgery, Xiangya Hospital, Central South University, Changsha, 410008, China.
4. CITIC Xiangya Reproductive and Genetic Specialist Hospital, Changsha, 410008, China.
5. Department of Hepatobiliary and Pancreatic Surgery, Xiangya Hospital, Central South University, Changsha, 410008, China.
Received 2019-4-3; Accepted 2019-10-26; Published 2020-1-1
Abstract
The roles of M2 macrophages on promoting tumor progression and chemotherapy resistance have been well studied in many cancers, such as pancreatic cancer, kidney cancer and so on, but its linkage to HCC cells still remains unclear. Here we found that M2 macrophages could alter miR-149-5p to increase MMP9 expression in HCC cells and mechanism dissection revealed that miR-149-5p might directly target the 3'UTR of MMP9-mRNA to suppress its translation. The in vivo orthotopic xenografts mouse model with oemiR-149-5p also validated in vitro data. Together, these findings suggest that M2 macrophages may through altering the miR-149-5p to promote HCC progression and targeting the M2 macrophages/miR149-5P/MMP9 signaling may help in the development of the novel therapies to better suppress the HCC progression.
Keywords: Liver cancer, MiRNA-149-5p, MMP9, Invasion, Migration
Introduction
Hepatocellular carcinoma (HCC) is the fifth most frequently diagnosed and the second cause of cancer death in the word [1]. HCC is one of the most lethal tumors with high recurrence and metastatic rate, which made it one of the poorest-prognosis cancers across the globe. In 2018, 42,220 cases of liver and intra hepatic bile duct cancer were newly diagnosed in the US, and the incidence has almost increased by twice in the past thirty years [2]. For advanced HCC patients, sorafenib seems to be one of most useful targeted drugs, but its function is limited by many factors, and new therapies are eagerly yet to be developed.
Tumor microenvironment is very important for the growth, progression and metastasis of tumors, among which, inflammation is one of the most important factors [3-4]. Macrophage is one of the most common immune related cells in tumor microenvironment, and its functions to tumors have already been well documented in many cancers, such as pancreatic cancers [5-6], prostate cancers [7-8], lung cancers [9] and so on. Macrophages can be differentiated into classically (M1) and alternatively (M2) activated polar cells based on different environment situations. Usually, M1 macrophages can function as pro-inflammatory effect by expressing nitric oxide synthase (iNOS), whereas M2 macrophages can express anti-inflammatory cytokines, such as IL-10, to promote tumor progression and metastasis. More and more studies have showed that tumor-associated macrophages trend to M2 macrophages, can make tumor cells more invasive and aggressive [10-11].
In this study, we demonstrated that M2 macrophages can alter the expression of miR-149-5p to promote the invasion and migration of HCC by increasing MMP9 pathway, thus we delineated a novel signaling between M2 macrophages and HCC progression and also provide a new possibility for the novel therapies to better suppress the HCC progression.
Material and Methods
Cell culture
HCC SK-HEP-1, HEK 293T and human acute monocytic leukemia cell line THP-1 were purchased from the American Type Culture Collection (ATCC). HA22T cell line (BCRC No. 60168) was gifted from Prof. Yuh-Shan Jou, Academia Sinica, Taiwan. HA22T, SK-HEP-1 and HEK 293T cell lines were cultured in DMEM (Invitrogen) with 10% Fetal Bovine Serum, 1% penicillin/streptomycin and 1% glutamine. THP-1 cell line was cultured in RPMI1640, 100 ng/ml Phorbol 12-myristate 13-acetate (PMA, Sigma) was used for 48 hours to induce THP-1 differentiation into macrophages. To induce M2 polarized phenotype, 20 ng/ml of IL-4(Invitrogen) and IL-13 (R&D Systems) were used for 48 hours. All cell lines were cultured in a 5% (v/v) CO2 humidified incubator at 37 °C.
Lentivirus packaging
The pLKO.1-shMMP9#1, pLKO.1-shMMP9#2, pWPI-MMP9, pLKO.1-miR-149-5p, pLKO.1-miR-483-3p, pLKO.1-miR-6734-3p plasmids were transfected into 293T cells and cultured in virus room, the lentivirus soup was collected after 48 hours and used immediately or frozen in -80 °C for later use.
The plasmids sequences used were as follows:
pLKO.1-shMMP9#1 sequence,
Forward: 5'-CCGGCCACAACATCACCTATTGGATGGATCCATCCAATAGGTGATGTTGTGGTTTTTG-3',
Reverse: 5'-AATTCAAAAACCACAACATCACCTATTGGATGGATCCATCCAATAGGTGATGTTGTGG -3';
pLKO.1-shMMP9#2 sequence,
Forward: 5'-CCGGCAGTTTCCATTCATCTTCCAAGGATCCTTGGAAGATGAATGGAAACTGTTTTTG -3',
Reverse: 5'-AATTCAAAAAAGTTTCCATTCATCTTCCAAGGATCCTTGGAAGATGAATGGAAACTG -3';
pLKO.1-miR-149-5p sequence,
Forward: 5'-CCGGTCTGGCTCCGTGTCTTCACTCCCTTGGATCCGGGGAGTGAAGACACGGAGCCAGA TTTTTG-3',
Reverse: 5'-AATTCAAAAATCTGGCTCCGTGTCTTCACTCCCCGGATCCAAGGGAGTGAAGACACGGAGCCAGA-3';
pLKO.1-miR-483-3p sequence,
Forward: 5'-CCGGTCACTCCTCTCCTCCCGTCTTTTGGATCCGAAGACGGGAGGAGAGGAGTGA TTTTTG-3',
Reverse: 5'-AATTCAAAAATCACTCCTCTCCTCCCGTCTTCGGATCCAAAAGACGGGAGGAGAGGAGTGA-3';
pLKO.1-miR-6734-3p sequence,
Forward: 5'-CCGGCCCTTCCCTCACTCTTCTCTCAGTTGGATCCGCTGAGAGAAGAGTGAGGGAAGGG TTTTTG-3',
Reverse: 5'-AATTCAAAAACCCTTCCCTCACTCTTCTCTCAGCGGATCCAACTGAGAGAAGAGTGAGGGAAGGG-3'.
RNA extraction and qRT-PCR analysis
Trizol reagent (Invitrogen, Grand Island, NY) was used to extract total RNA, and 1-2ug of the total RNA was used to reverse transcribe into cDNA by using Superscript III transcriptase (Invitrogen, Grand Island, NY). Relative RNA expression was conducted by Quantitative real-time PCR (qRT-PCR) assay by using a Bio-Rad CFX96 system with SYBR green reagent and the results were normalized by GAPDH.
The primer sequences used were as follows:
CD206 primer,
Forward: 5'-GGGTTGCTATCACTCTCTATGC-3',
Reverse: 5'-TTTCTTGTCTGTTGCCGTAGTT-3';
CD163 primer,
Forward: 5'-TTTGTCAACTTGAGTCCCTTCAC-3',
Reverse: 5'-TCCCGCTACACTTGTTTTCAC-3';
Arginase-1 primer,
Forward: 5'-GGTTTTTGTTGTTGCGGTGTTC-3',
Reverse: 5'-CTGGGATACTGATGGTGGGATGT-3';
TGF β primer,
Forward: 5'-CAATTCCTGGCGATACCTCAG-3',
Reverse: 5'-GCACAACTCCGGTGACATCAA-3';
IL-10 primer,
Forward: 5'-GACTTTAAGGGTTACCTGGGTTG-3',
Reverse: 5'-TCACATGCGCCTTGATGTCTG-3';
MMP9 primer,
Forward: 5'- TGTACCGCTATGGTTACACTCG-3',
Reverse: 5'- GGCAGGGACAGTTGCTTCT-3';
GAPDH primer,
Forward: 5'- TGTGGGCATCAATGGATTTGG-3',
Reverse: 5'- ACACCATGTATTCCGGGTCAAT-3';
hsa-miR-1224-3p primer: CCCCACCTCCTCTCTCCTCAG;
hsa-miR-149-5p primer: TCTGGCTCCGTGTCTTCACTCCC;
hsa-miR-154-5p primer: TAGGTTATCCGTGTTGCCTTCG;
hsa-miR-183-5p primer: TATGGCACTGGTAGAATTCACT;
hsa-miR-204-5p primer: TTCCCTTTGTCATCCTATGCCT;
hsa-miR-211-5p primer: TTCCCTTTGTCATCCTTCGCCT;
hsa-miR-2355-5p primer: ATCCCCAGATACAATGGACAA;
hsa-miR-483-3p primer: TCACTCCTCTCCTCCCGTCTT;
hsa-miR-491-5p primer: AGTGGGGAACCCTTCCATGAGG;
hsa-miR-6734-3p primer: CCCTTCCCTCACTCTTCTCTCAG.
Western blot analysis
RIPA lysate was used to lyse the collected cells, and 30ug protein was electrophoresed in 10% SDS/PAGE gel and then transferred onto PVDF membranes. After that, specific primary antibodies were used to incubate the relative bands for overnight in a 4℃ room and then incubated with the secondary antibodies for 1-2 hours and visualized with ECL system (Thermo Fisher Scientific, Rochester, NY). GAPDH (6c5), MMP-9(2C3), c-Myc(6A10), ERK1(E-12), IGF-I(AT6F8), MAO-A(G-10), MMP-1(3B6) and VEGF(C1) antibodies were purchased from Santa Cruz Biotechnology.
Cell invasion assay
Cell invasion assay was performed by using 8 μm transwell chamber (Corning Life Science) in 24-well plates. 5×104 cells/well were seeded into upper chamber coated with diluted Matrigel (1:20 dilution, 100 μl/well; BD Biosciences) with serum-free medium and 750μL media with 10% FBS /well was added into lower chambers for incubation for 24 hours. The invaded cells were fixed by methanol and stained by 0.1% (w/v) crystal violet. Each sample was run in triplicate and repeated multiple times.
Wound-healing migration assay
Cells were seeded into 35-mm plates until they were confluent, and the plates were scraped using a sterile pipette tip to create a wound through the confluent monolayer, then cultured in serum-free medium for 12 hours and photographed at 0 and 12 hours, respectively. The distance of wound migration was measured for further analyses.
Luciferase assay
193bp fragment of human MMP9 3' UTR with wild or mutant miRNA-responsive elements was cloned into psiCHECK2 vector (Promaga, USA) downstream of the Renilla luciferase ORF. Cells were plated in 24-well plates and transfected the cDNA with Lipofectamine (Invitrogen) as the manufacturer's instruction. Dual-Luciferase Assay (Promega) was used to calculate luciferase activity according to the manufacturer's manual after 48 hours.
In vivo studies
24 6-8 weeks old nude mice were purchased from NCI and divided into 4 groups: 1×106 SK-HEP-1-Luc transduced with pLKO.1-vector co-injected with matrigel; SK-HEP-1-Luc transduced with pLKO.1-vector co-injected with THP-1; SK-HEP-1-Luc transduced with oemiR-149-5p co-injected with matrigel; SK-HEP-1-Luc transduced with oemiR-149-5p co-injected with THP-1, all the cells were injected into left lobes of liver capsule. IVIS was used to measure tumor growth once a week. The study was carried out under the approval of the ethics committee of Xiangya Hospital Central South University (Reference number: 2019020078) and followed the Interdisciplinary Principles and Guidelines for the Use of Animals in Research, Testing, and Education by the New York Academy of Sciences, Ad Hoc Animal Research Committee.
Statistical analysis
All statistical was analyzed by SPSS 19.0 system (SPSS Inc, Chicago, IL). The data values were presented as the mean ± SD. Differences in mean values between two groups were analyzed by two-tailed Student's t test and the mean values of more than two groups were compared with one way ANOVA. p≤0.05 was considered to be statistically significant.
Results
M2 macrophages can increase the invasion and migration capacity of HCC cells
To study the potential impact of M2 macrophages on HCC progression, we first applied the 100 ng/ml PMA to induce the THP-1 cells into macrophages, then 20 ng/ml IL-4 and IL-13 were used to induce M2 polarized phenotype and the M2 and M1 markers were measured (Fig. 1A, Figure S1 A). We then applied a co-culture system to culture M2 macrophages together with HCC cells for 48hrs (Fig. 1B). Results from invasion assay and “wound-healing” migration assay revealed that the invasion and migration capacity of HCC SK-HEP-1 and HA22T cells were increased after co-culture (Fig.1C-F).
Taken together, results from Fig. 1A-F and Figure S1A suggest that M2 macrophages can increase HCC cells invasion and migration capacity.
M2 macrophages can increase the invasion and migration capacity of HCC cells. (A) qRT-PCR was applied in macrophages using primers for M2 markers (CD206, CD163, Arginase-1,TGFβ, IL-10) after induced by IL-4 and IL-13. (B) Pattern diagram for co-culture, invasion and wound healing migration system. (C-D) Invasion assay was used to measure invasion capacity of HCC SK-HEP-1 and HA22T cells after co-cultured with M2 macrophages. (E-F) Migration capacity was measured in SK-HEP-1 and HA22T cells after co-cultured with M2 macrophages. All quantifications are mean ± SD, *p < 0.05, **p < 0.01.
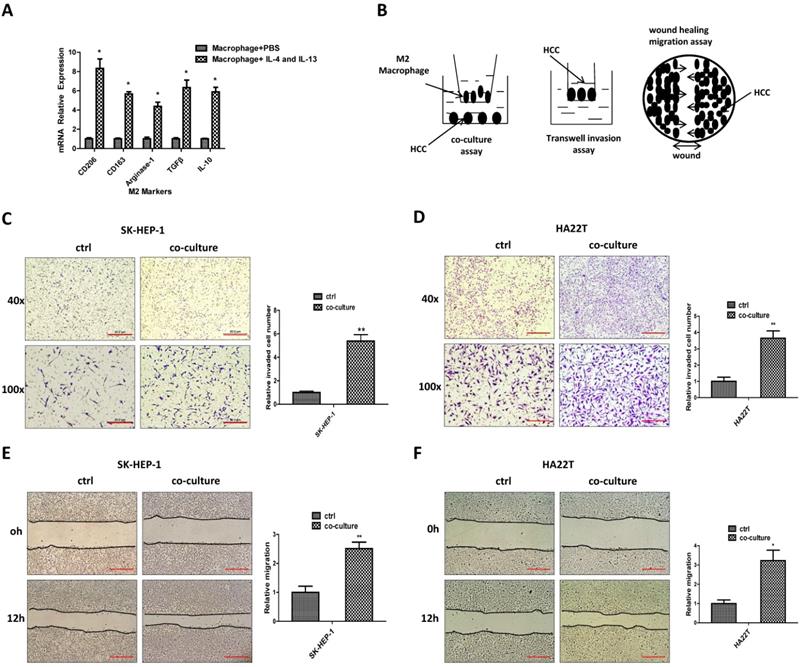
Mechanism dissection of how M2 macrophages increase the HCC cells invasion and migration: via altering the MMP9 expression
To examine the detailed mechanisms underlying M2 macrophages' role in regulating HCC cells invasion and migration, we tested the expression of proteins that have been connected with cancer cell invasion and migration [12-18] in both culture alone and in co-culture. The exploratory experiments led to the focus on MMP9 as the results showed that MMP9 protein expression was increased after co-culture, both in SK-HEP-1 and HA22T HCC cells (Fig. 2A-B).
We then examined the potential impact of altered MMP9 on the HCC cell invasion and migration, and results revealed that decreasing MMP9 via adding MMP9-shRNA into SK-HEP-1 cells could reverse M2 macrophages' function to HCC cells (Fig. 2C-E). We also used another shRNA to decrease MMP9 and the results were consistent with the previous data (Fig. 2F-H). In contrast, increasing MMP9 via adding MMP9-cDNA into HA22T cells led to increase of the invasion and migration capacity more significant (Fig. 2I-K).
Together, results from Fig. 2A-K suggest that M2 macrophages may function via increasing HCC MMP9 expression to increase the invasion and migration capacity in HCC cells.
Mechanism dissection of how M2 macrophages increase MMP9 expression of HCC cells: via altering the miR-149-5p
To study how M2 macrophages increase HCC MMP9 expression, we first tested the mRNA expression in HCC cells after co-culture and results revealed that the mRNA level of MMP9 didn't increase after co-culture (Fig. 3A).
M2 macrophages increase the HCC cells invasion and migration via altering the MMP9 expression. (A) Western bolt was used to check related genes expression in SK-HEP-1 cells after co-cultured with M2 macrophages. (B) Western blot was used to check MMP9 expression in HA22T cells after co-cultured with M2 macrophages. (C) Western blot was used to check MMP9 expression in SK-HEP-1 transfected with MMP9-shRNA#1 in co-culture system. (D-E) Invasion and migration capacity were measured in SK-HEP-1 transfected with MMP9 sh-RNA#1 in co-culture system. (F) Western blot was used to check MMP9 expression in SK-HEP-1 transfected with MMP9-shRNA#2 in co-culture system. (G-H) Invasion and migration capacity were measured in SK-HEP-1 transfected with MMP9 sh-RNA#2 in co-culture system. (I) Western blot was used to check MMP9 expression in HA22T overexpressed MMP9 in co-culture system. (J-K) Invasion and migration capacity were measured in HA22T overexpressed MMP9 in co-culture system. All quantifications are mean ± SD, *p < 0.05, **p < 0.01, ns: no significant difference.
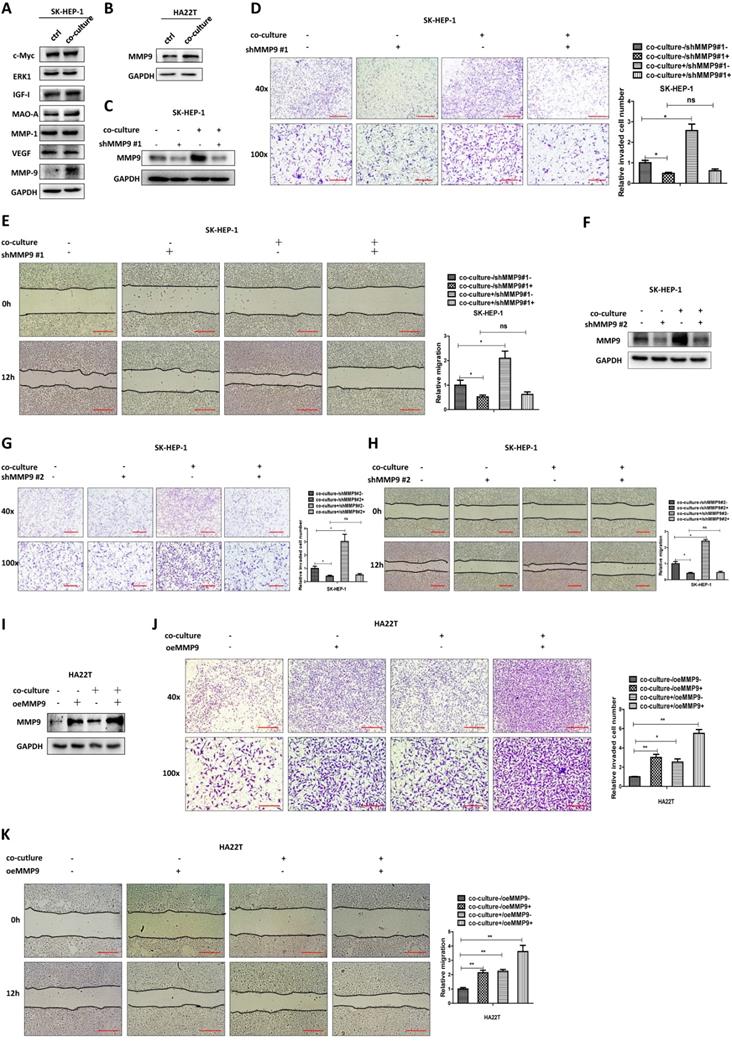
M2 macrophages increase MMP9 expression of HCC cells via altering the miR-149-5p. (A) qRT-PCR was used to check MMP9 expression in SK-HEP-1 and HA22T cells after co-cultured with M2 macrophages. (B) qRT-PCR was used to check relative miRNAs expression in SK-HEP-1 cells after co-cultured with M2 macrophages. (C) Western blot was used to check MMP9 expression after over-expression miR-149-5p/483-3p/6734-3p in SK-HEP-1 cells in co-culture system. (D) Western blot was used to check MMP9 expression after over-expression miR-149-5p in HA22T cells in co-culture system. (E-F) Invasion and migration capacity were measured after over-expression miR-149-5p in SK-HEP-1 cells in co-culture system. (G) Western blot was used to check MMP9 expression after adding miR-149-5p inhibitor in HA22T cells in co-culture system. (H-I) Invasion and migration capacity were measured after adding miR-149-5p inhibitor in HA22T cells in co-culture system. All quantifications are mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001 ns: no significant difference.
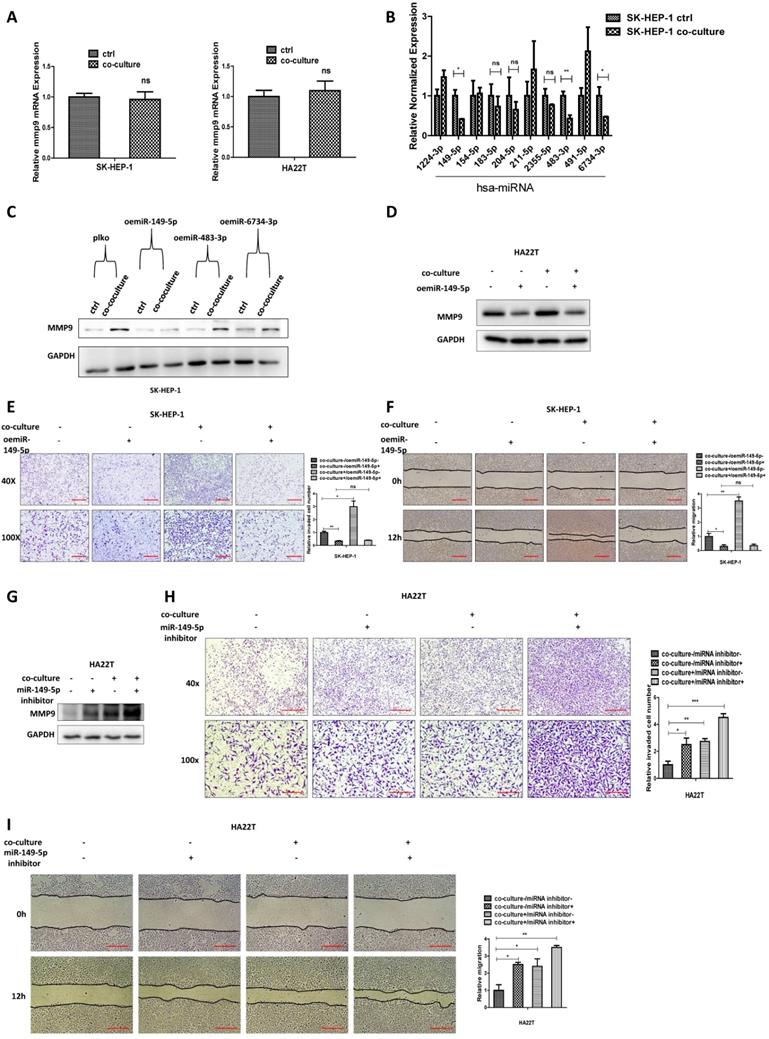
MiR-149-5p alters the MMP9 expression under the co-culture condition via directly target the 3'UTR of MMP9-mRNA. (A) Sequence alignment of the MMP9 3'UTR with wild-type versus mutant potential miR-149-5p targeting sites. (B-C) Luciferase reporter activity after transfection of wild-type or mutant MMP9 3'UTR reporter construct in SK-HEP-1 and HA22T with over expression miR-149-5p of miR-149-5p inhibitor compared to the control cells.
We also conducted the protein stability assay to check MMP9 protein stability between control and co-culture group, and the result showed no significant difference between these two groups (Figure S1B). These results suggest that M2 macrophages may regulate HCC MMP9 mRNA level through post-transcriptional regulation. So we focus on the miRNAs that can regulate MMP9. We searched the database to select out 10 miRNAs that can regulate MMP9, and then checked miRNAs expression in HCC cells after co-culture; the results revealed that miR149-5p, miR-483-3p and miR-6734-3p were decreased after co-culture (Fig. 3B). Then we over expressed the three miRNAs in HCC cells and co-cultured with M2 macrophages; the results revealed that over expressed miR-149-5p could reverse co-culture's function to MMP9, both in SK-HEP-1 and HA22T cells (Fig. 3C-D), so was the invasion and migration capacity (Fig. 3E-F). On the contrary, when we added miR-149-5p inhibitor in HCC cells, the co-culture's function to MMP9 and invasion/migration became more significant (Fig. 3G-I).
Together, results from Fig. 3A-I and Figure S1B suggest that M2 macrophages may function via decreasing miR-149-5p level of HCC cells to increase MMP9 expression.
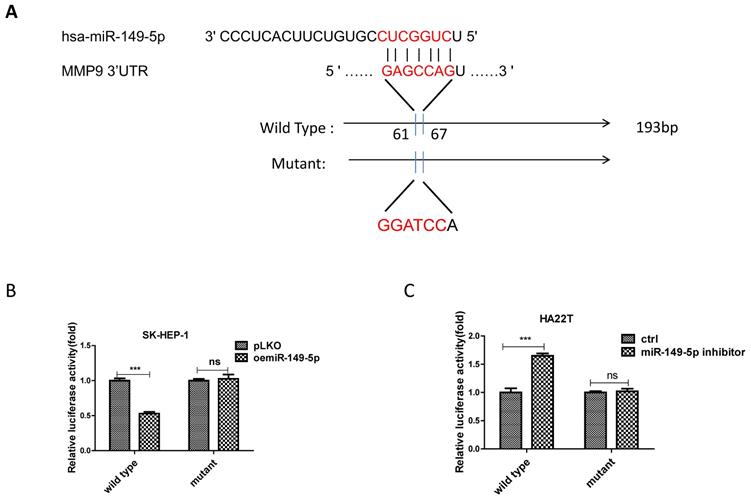
Mechanism dissection of how miR-149-5p alters the MMP9 expression under the co-culture condition: via directly target the 3'UTR of MMP9-mRNA
To dissect the mechanism of how M2 macrophage-miR-149-5p axis can alter the MMP9 expression at the molecular level, we first searched the potential miRNA targeting site located on the 3'UTR of MMP9-mRNA. We then applied the reporter assay with the psiCHECK2 vector carrying the wide-type 3'UTR and a deletion mutant without the miRNA-target site (Fig. 4A). Results from the luciferase assay revealed that addition of miR-149-5p decreased luciferase activity in SK-HEP-1 cells, while addition miR-149-5p inhibitor increased luciferase activity in HA22T cells transfected with wild type MMP9 3'UTR but not the mutant MMP9 3'UTR (Fig. 4B-C), suggesting that miR-149-5p can directly target the 3'UTR of MMP9-mRNA to suppress its protein expression.
Human Clinical study to link the MMP9 to the HCC progression
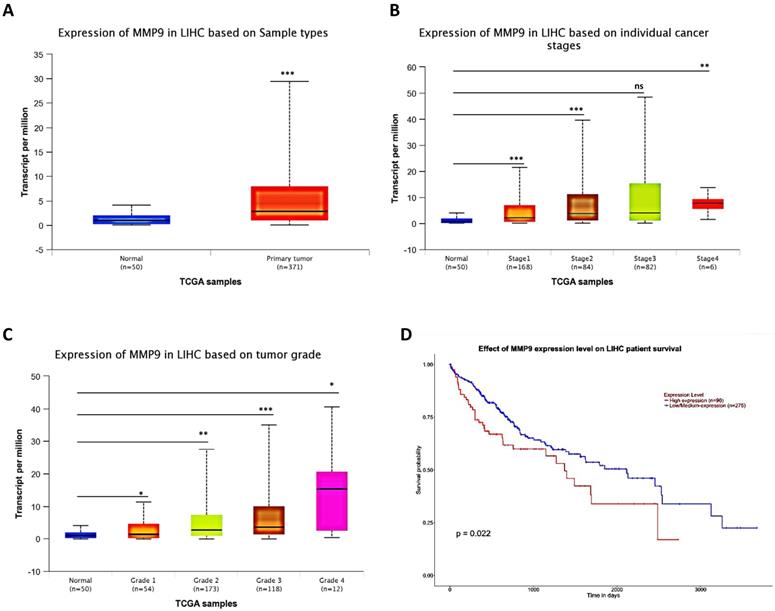
To link the above in vitro results with human HCC progression, we used UALCAN website (http://ualcan.path.uab.edu/) to analyze cancer transcriptome data in TCGA database. The results showed that MMP9 expression was higher in primary tumor samples than in normal samples (Fig. 5A), and the MMP9 expression level was relatively increased with the increased level of HCC grade and stage (Fig. 5B-C). What's more, the result also showed that high expression level of MMP9 was related to low survival probability (Fig. 5D).
Together, results of clinical data (Fig. 5A-D) from TCGA database proved that MMP9 may play important roles to promote HCC progression.
Preclinical study using in vivo mouse model to prove the roles of M2 macrophages-miR-149-5p-MMP9 axis in the HCC progression
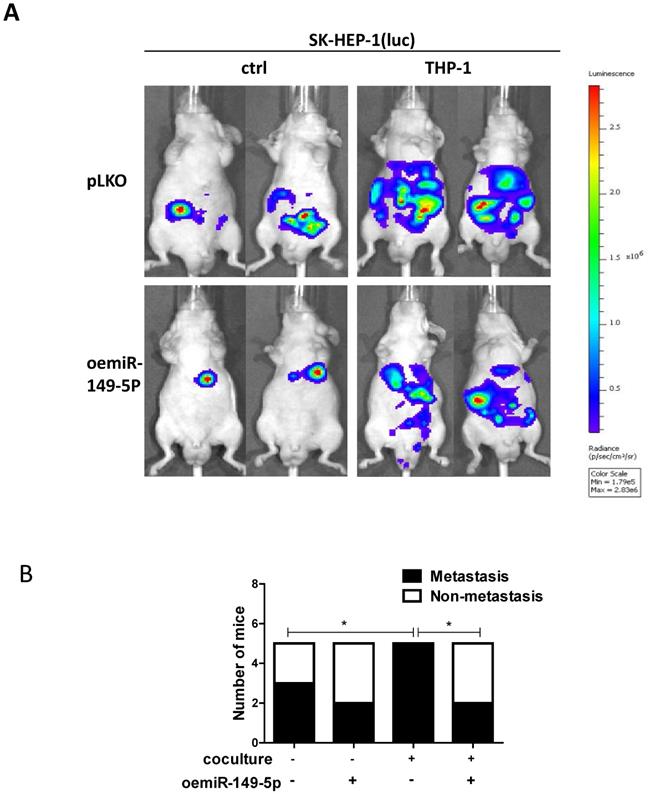
To link our in vitro studies to the clinical significance, we firstly transfected the SK-HEP-1 cells with luciferase reporter gene (for in vivo imaging system (IVIS)) to detect tumor progression inside mouse and then transfected the SK-HEP-1-Luc cells with vector control or oemiR-149-5p. The HCC cells were divided into four groups, SK-HEP-1(luc) vector control co-injected with matrigel into the left lobes of liver capsule of nude mouse(Group 1), SK-HEP-1(luc) vector control co-injected with THP-1(Group 2), SK-HEP-1 (luc) oemiR-149-5p co-injected with matrigel (Group 3), SK-HEP-1 (luc) oemiR149-5p co-injected with THP-1 (Group 4). Tumor growth and metastasis were monitored weekly via IVIS analysis.
Totally, 20 mice generated tumors (5 for each group), and our results revealed that xenografts of HCC cells with macrophages (THP-1) had more metastasis than no coculture group, and oemiR-149-5p can partly reverse in vivo function of macrophages (Fig. 6A-B), and the IHC data of MMP9 expression also confirmed our conclusions(Figure S1C).
Taken together the data from Fig. 6 and Figure S1C, suggests that M2 macrophages/miR-149-5p /MMP9 axis played a critical role to regulate HCC progression, and targeting this newly identified signaling with oemiR-149-5p led to suppression of HCC progression.
Human Clinical study to link the miR-149-5p-MMP9 axis to the HCC progression. (A) HCC patients' from TCGA database shows different expression of MMP9 in normal tissue and primary tumor tissue. (B) The TCGA data shows expression of MMP9 in HCC samples based on different individual cancer stages. (C) The TCGA data shows expression of MMP9 in HCC samples based on different tumor grade. (D) The TCGA data shows effect of MMP9 expression level on HCC patient survival. All quantifications are mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001 ns: no significant difference.
An in vivo mouse model proves the role of M2 macrophage-miR149-5p function in HCC progression. (A) SK-HEP-1-cells were transduced with Luciferase and with either pLKO or oemiR-149-5p and then co-injected with either matrigel or THP-1 into left lobe of liver and IVIS imaging was used to determine the tumor growth and metastasis in mice in each group. (B) Quantitative analysis of number of mice with metastasis in each group. *p < 0.05.
Discussion
HCC is one of the most lethal tumors across the globe. In 2018, 42,220 new cases and 30,200 new deaths of liver and intra hepatic bile duct cancer were counted in the United States [2]. High morbidity and mortality make it one of the most burdened tumors in the world. So far, we still lack effective treatments for advanced HCC. Although sorafenib sometimes works, its function is very limited. So, finding new targets and therapies seems to be urgently needed.
More and more evidence proves that the tumor microenvironment is closely related to tumor progression and metastasis, among which, hypoxia and inflammatory cells infiltration such as macrophages seem to be the most important two factors [3-4]. M2 macrophage is an alternatively activated phenotype of macrophage and can express anti-inflammatory cytokines to promote tumor progression and metastasis, which is associated with poor outcomes of tumors and has been widely reported in many cancers. Here we provide another example where M2 macrophages can function as a promoter to regulate HCC progression.
MicroRNAs belong to the endogenous small noncoding RNAs family. Usually they play key roles to regulate tumorigenesis and progression by binding to the 3'UTR of target genes mRNA to inhibitor gene expression [19-20]. MiR-149-5p is a very important member of microRNA family, which has been widely reported to be a suppressor in tumor progression. For instance, in human osteosarcoma, miR-149-5p can inhibit tumor growth by regulating TWEAK/Fn14/PI3K/AKT pathway [21]. For non-small cell lung cancer, the lncRNA-PCAT-1/miR-149-5p/LRIG2 axis plays a key role to regulate its progression [22]. Recent study showed that miR-149-5p could inhibit cell proliferation and invasion through targeting GIT1 in medullary thyroid carcinoma [23]. All this above proved the key roles of miR-149-5p on tumor progression. In our study, we found miR-149-5p decreased after co-culture, and could increase HCC cells invasion and migration by targeting 3'UTR of MMP9, what's more, by using HCC-oemiR-149-5p cells in in vitro and in vivo experiment could reverse M2 macrophages' function to HCC cells, which strongly proved miR-149-5p's negative roles on regulating tumor progression.
Matrix metalloproteinases (MMPs) family is a zinc-dependent enzyme which is involved in the degradation of the extracellular matrix both in physiological and pathological processes, such as embryonic development, tissue remodeling, arthritis and tumor progression and metastasis [24]. MMP9 is one of key member of MMPs family. Elevating MMP9 expression plays a key role in the progression of many tumors, such as bladder cancer [25], esophageal squamous cell carcinoma [26], and intrahepatic cholangiocarcinoma [27]. Our study proved that M2 macrophages can increase HCC cells invasion and migration by upregulating MMP9 expression, this conclusion was strengthened by the outcome in response to the MMP9-shRNA which could block the biochemical and cell behavior induced by M2 macrophages in the HCC tumor microenvironment.
In conclusion, M2 macrophages may increase HCC progression via altering miR-149-5p/MMP9 signaling, and miR-149-5p can target the 3'UTR of MMP9-mRNA to suppress its expression. A potential therapy to target this newly identified signaling may help improve the treatment to better suppress the HCC progression.
Supplementary Material
Supplementary figures.
Acknowledgements
This work was supported by National Natural Science Foundation of China (81903004) and The 12th Five-year Key Discipline Construction in Hunan Province (Biomedical Engineering). We thank Thelma for editing the manuscript.
Ethics approval and consent to participate
The animal study was carried out under the approval of the ethics committee of Xiangya Hospital Central South University (Reference number: 2019020078) and followed the Interdisciplinary Principles and Guidelines for the Use of Animals in Research, Testing, and Education by the New York Academy of Sciences, Ad Hoc Animal Research Committee.
Competing Interests
The authors have declared that no competing interest exists.
References
1. A. Jemal, F. Bray, M.M. Center, et al. Global cancer statistics. CA Cancer J Clin. 2011:61 (2) 69-90
2. Siegel R L, Miller K D, Jemal A. Cancer Statistics, 2018. CA Cancer J Clin. 2018;68:7-30
3. Lewis CE, Pollard JW. Distinct role of macrophages in different tumor microenvironments. Cancer Res. 2006;66:605-12
4. Ackerman D, Simon MC. Hypoxia, lipids, and cancer: surviving the harsh tumor microenvironment. Trends Cell Biol. 2014;24(8):472-8
5. Wang X, Luo G, Zhang K. et al. Hypoxic Tumor-Derived Exosomal miR-301a Mediates M2 Macrophage Polarization via PTEN/PI3Kγ to Promote Pancreatic Cancer Metastasis. Cancer Research. 2018;78(16):4586 ~4598
6. Binenbaum Y, Fridman E, Yaari Z. et al. Transfer of miRNA in Macrophage-Derived Exosomes Induces Drug Resistance in Pancreatic Adenocarcinoma. Cancer Research. 2018;78(18):5287 ~5299
7. Zheng T, Ma G, Tang M. et al. IL-8 Secreted from M2 Macrophages Promoted Prostate Tumorigenesis via STAT3/MALAT1 Pathway. International Journal of Molecular Sciences. 2019;20(1):98
8. Jones J D, Sinder B P, Paige D. et al. Trabectedin Reduces Skeletal Prostate Cancer Tumor Size in Association with Effects on M2 Macrophages and Efferocytosis. Neoplasia. 2019;21(2):172 ~184
9. Yang L, Dong Y, Li Y. et al. IL-10 derived from M2 macrophage promotes cancer stemness via JAK1/STAT1/NF-κB/Notch1 pathway in non-small cell lung cancer. International Journal of Cancer. 2019 doi:10.1002/ijc.32151
10. Talmadge JE, Donkor M, Scholar E. Inflammatory cell infiltration of tumours: Jekyll or Hyde. Cancer Metastasis Rev. 2007;26:373-400
11. Yeung O W, Lo C M, Ling C C. et al. Alternatively activated (M2) macrophages promote tumour growth and invasiveness in hepatocellular carcinoma. J Hepatol. 2015;62(3):607 ~616
12. Xiao L, Wang Y, Liang W. et al. LRH-1 drives hepatocellular carcinoma partially through induction of c-myc and cyclin E1, and suppression of p21. Cancer Management and Research. 2018;10:2389 ~2400
13. Wu T, Dong X, Yu D. et al. Natural product pectolinarigenin inhibits proliferation, induces apoptosis, and causes G2/M phase arrest of HCC via PI3K/AKT/mTOR/ERK signaling pathway. OncoTargets and Therapy. 2018;11:8633 ~8642
14. Lei T, Ling X. IGF-1 promotes the growth and metastasis of hepatocellular carcinoma via the inhibition of proteasome-mediated cathepsin B degradation. World Journal of Gastroenterology. 2015;21(35):10137 ~10149
15. Li J, Yang X, Wang Y. et al. Monoamine oxidase A suppresses hepatocellular carcinoma metastasis by inhibiting the adrenergic system and its transactivation of EGFR signaling. Journal of Hepatology. 2014;60(6):1225 ~1234
16. Kim E, Kim D, Lee J. et al. Capicua suppresses hepatocellular carcinoma progression by controlling the ETV4-MMP1 axis. Hepatology. 2018;67(6):2287 ~2301
17. Yang S, Yang C, Yu F. et al. Endoplasmic reticulum resident oxidase ERO1-Lalpha promotes hepatocellular carcinoma metastasis and angiogenesis through the S1PR1/STAT3/VEGF-A pathway. Cell Death & Disease. 2018;9(11):1105
18. Min J, Feng Q, Liao W. et al. IFITM3 promotes hepatocellular carcinoma invasion and metastasis by regulating MMP9 through p38/MAPK signaling. FEBS Open Bio. 2018;8(8):1299 ~1311
19. Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351-379
20. Li M, Li J, Ding X, He M, Cheng SY. MicroRNA and cancer. AAPS J. 2010;12:309-317
21. Xu R, Feng F, Yu X. et al. miR-149-5p inhibits cell growth by regulating TWEAK/Fn14/PI3K/AKT pathway and predicts favorable survival in human osteosarcoma. International Journal of Immunopathology and Pharmacology. 2018;32:1120487799
22. Li J, Li Y, Wang B. et al. LncRNA-PCAT-1 promotes non-small cell lung cancer progression by regulating miR-149-5p/LRIG2 axis. Journal of Cellular Biochemistry. 2018 doi:10.1002/jcb.28046
23. Ye X, Chen X. miR-149-5p inhibits cell proliferation and invasion through targeting GIT1 in medullary thyroid carcinoma. Oncology letters. 2019;17(1):372-378
24. Bonnans C, Chou J, Werb Z. Remodelling the extracellular matrix in development and disease. Nature Reviews Molecular Cell Biology. 2014;15(12):786 ~801
25. Yan H, Li J, Ying Y. et al. MIR-300 in the imprinted DLK1-DIO3 domain suppresses the migration of bladder cancer by regulating the SP1/MMP9 pathway. Cell Cycle. 2018;17(24):2790 ~2801
26. Zhou H, Yang L, Xu X. et al. miR-34a inhibits esophageal squamous cell carcinoma progression via regulation of FOXM1. Oncol Lett. 2019;17(1):706 ~712
27. Dai L, Hu W, Yang Z. et al. Upregulated expression of HOXB7 in intrahepatic cholangiocarcinoma is associated with tumor cell metastasis and poor prognosis. Laboratory Investigation. 2019 doi:10.1038/s41374-018-0150-4
Author contact
![]() Corresponding authors: Yao Xiao (Email: yaoxiaoedu.cn) and Yixiong Li (Email: liyixiong2011com).
Corresponding authors: Yao Xiao (Email: yaoxiaoedu.cn) and Yixiong Li (Email: liyixiong2011com).

 Global reach, higher impact
Global reach, higher impact