Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(7):1959-1967. doi:10.7150/jca.39087 This issue Cite
Research Paper
High Expression of Long Noncoding RNA PCNA-AS1 Promotes Non-Small-Cell Lung Cancer Cell Proliferation and Oncogenic Activity via Upregulating CCND1
1. Department of Laboratory Medicine, Shanghai Chest Hospital, Shanghai Jiao Tong University, Shanghai 200030, China.
2. Department of Laboratory Medicine, Yueyang Hospital of Integrated Traditional Chinese and Western Medicine, Shanghai University of Traditional Chinese Medicine, Shanghai 200437, China.
3. Department of Anatomy and Physiology, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.
4. Department of orthopedics, Hospital of No.83 army, Xinxiang, 453000, China.
*These authors contributed equally to this work.
Received 2019-10-8; Accepted 2019-12-1; Published 2020-1-29
Abstract
Accumulating evidences showed that aberrantly expressed long noncoding RNAs (lncRNAs) have critical roles in many cancers. However, the expression and roles of a poorly studied lncRNA PCNA-AS1 in non-small-cell lung cancer (NSCLC) remain unknown. In this study, we investigated the expression, clinical significance, biological roles, and functional mechanism of PCNA-AS1 in NSCLC. Our results showed that PCNA-AS1 was upregulated in NSCLC tissues and cell lines, and correlated with TNM stages. Functional experiments showed that overexpression of PCNA-AS1 promoted NSCLC cell proliferation and cell cycle progression. Depletion of PCNA-AS1 inhibited NSCLC cell proliferation and cell cycle progression, and also inhibited NSCLC tumor growth in vivo. Mechanistically, we found that PCNA-AS1 upregulated CCND1 expression. The expression of PCNA-AS1 was positively correlated with that of CCND1 in NSCLC tissues. Moreover, depletion of CCND1 abrogated the oncogenic roles of PCNA-AS1 in NSCLC. In conclusion, highly expressed PCNA-AS1 promotes NSCLC cell proliferation and oncogenic activity via upregulating CCND1. Our results imply that PCNA-AS1 might serve as a therapeutic target for NSCLC.
Keywords: long noncoding RNA, PCNA-AS1, non-small-cell lung cancer, proliferation, cell cycle, CCND1
Introduction
Lung cancer is the most common human malignancy and the primary cause of cancer-related death worldwide [1]. Non-small-cell lung cancer (NSCLC) accounts for approximate 80% of all lung cancer, which include adenocarcinoma, squamous cell carcinoma, adenosquamous cell carcinoma, and large cell carcinoma [2, 3]. Despite great advances in therapeutic methods, including radical resection, chemotherapy, and molecular targeted therapy, the prognosis of NSCLC is still very poor [4, 5]. Therefore, it is imperative to elucidate the molecular mechanisms underlying tumorigenesis and progression of NSCLC, and identify novel therapeutic targets for NSCLC.
Long noncoding RNA (lncRNA) is a novel type of noncoding transcript with more than 200 nucleotides in length [6, 7]. Accumulating evidences suggest that lncRNAs are involved in various pathophysiological processes, particular in cancers [8-11]. Dysregulated expression of lncRNAs have been revealed in several cancers, including lung cancer, hepatocellular carcinoma, gastric cancer, melanoma, and et al. [12-16]. LncRNAs also have been found to play critical roles in various cancers, including tumor initiation, progression, and metastasis [17-19]. In our previous study, we found that lncRNA GPC3-AS1 is upregulated in hepatocellular carcinoma, associated with poor prognosis, and promotes hepatocellular carcinoma cell proliferation and metastasis [20]. The aberrant expressions and roles of several lncRNAs in NSCLC have also been reported, such as PVT1, BCYRN1, MALAT1, HOTAIR [21-24]. These studies demonstrated the importance of lncRNAs in cancers, which has previous been recognized as transcriptional noise.
Transcriptome RNA sequencing results revealed that more than 50,000 lncRNAs were identified [25]. But the clinical significances and functions of most of these lncRNAs are still unknown. PCNA-AS1 is a recently identified and poorly understood lncRNA, which is reported to be upregulated in hepatocellular carcinoma and promote hepatocellular carcinoma cell growth [26]. The gene coding PCNA-AS1 is located at chromosome 20p12.3. PCNA-AS1 has only one exon and is transcribed in antisense orientation with PCNA. However, the expression, clinical significance, and biological roles of PCNA-AS1 in lung cancer are still unknown.
In this study, we measured the expression of PCNA-AS1 in NSCLC tissues and cell lines, and investigated its correlation with clinicopathological characteristics. Using gain-of-function and loss-of-function assays, we explored the biological roles of PCNA-AS1 in NSCLC. Furthermore, the molecular mechanisms underlying the roles of PCNA-AS1 in NSCLC are also explored.
Materials and Methods
Tissues samples
Eighty-two NSCLC tissues and paired adjacent non-tumor tissues were excised from patients who were diagnosed with NSCLC and underwent surgery before radiation or chemotherapy at Shanghai Chest Hospital (Shanghai, China). All the tissues were diagnosed by histopathological examination. The tissues were immediately frozen in liquid nitrogen and stored at -80 °C until use. The Research Ethics Committee of Shanghai Chest Hospital approved this study and informed consents were obtained from all patients.
Cell lines and cell culture
The human normal bronchial epithelial cell line 16HBE and human NSCLC cell lines A549, H460 and H1299 were obtained from the cell bank of the Chinese Academy of Sciences (Shanghai, China). 16HBE cells were cultured in DMEM medium (Invitrogen, Carlsbad, CA, USA). A549, H460 and H1299 cells were cultured in RPMI-1640 medium (Invitrogen). All the medium were supplemented with 10% fetal bovine serum (Invitrogen), and all the cells were maintained in a humidified incubator with 5% CO2 at 37 °C.
RNA isolation and quantitative real-time polymerase chain reaction (qRT-PCR) assays
Total RNA was extracted from tissues or cells with TRIzol reagent (Invitrogen) according to the manufacturer's instruction. First-strand cDNA was generated using the M-MLV Reverse Transcriptase (Invitrogen) and either random primers or gene-specific primers. Quantitative real-time polymerase chain reaction (qRT-PCR) was carried out using the SYBR® Premix Ex Taq™ II kit (Takara, Dalian, China) on ABI StepOnePlus system (Applied Biosystems, Foster City, CA, USA). GAPDH was used as internal normalized reference. The primer sequences used in this study were as follows: PCNA-AS1: 5'-TTGTTGCCACTCCGCCAC-3' (reverse transcription), 5'-TTTGGACATACTGGTGAGG-3' (forward) and 5'-AAGGTGTTGGAGGCACTC-3' (reverse); PCNA: 5'-GCTGTTGTAATTTCCTGTGC-3' (forward) and 5'-CATACTGAGTGTCACCGTTG-3' (reverse); CCND1: 5'-TTCCTGTCCTACTACCGC-3' (forward) and 5'-CTCCTCCTCTTCCTCCTC-3' (reverse); GAPDH: 5'-GGAGCGAGATCCCTCCAAAAT-3' (forward) and 5'-GGCTGTTGTCATACTTCTCATGG-3' (reverse). The relative expression of RNAs was calculated by the comparative Ct method.
Vectors construction, small interfering RNA (siRNA) synthesis and transfection
PCNA-AS1 full-length transcript was PCR amplified by the TaKaRa Ex Taq® Hot Start Version (Takara) from cDNA which is synthesized from RNA of NSCLC tissues, and subcloned into the Hind III and BamH I sites of pcDNA3.1(+) (Invitrogen), named pcDNA3.1-PCNA-AS1. The primer sequences used were as follows: 5'-CCCAAGCTTCTTCAAATACTAGCGCCAAGGTAT-3' (forward) and 5'-CGGGATCCTTTTTTCGCAACGCGGCGCAGG-3' (reverse). The oligonucleotides for shRNA specifically targeting PCNA-AS1 were synthesized by GenePharma (Shanghai, China). The shRNA target sequence was: 5'-GCTGGAGCTAATATCCCAGCA-3'. After annealing, the oligonucleotides were inserted into the shRNA expression vector pGPU6/Neo (GenePharma). CCND1 specific siRNA was designed and synthesized by Invitrogen. Transfection was carried out using Lipofectamine 3000 (Invitrogen) following the manufacturer's protocol.
Stable cell lines construction
To constructing PCNA-AS1 stably overexpressed A549 and H1299 cells, pcDNA3.1-PCNA-AS1 or pcDNA3.1 was transfected into A549 and H1299 cells, and selected with neomycin (1000 µg/ml) for four weeks. To constructing PCNA-AS1 stably depleted A549 and H1299 cells, PCNA-AS1 or scrambled shRNAs expression vectors were transfected into A549 and H1299 cells, and selected with neomycin (1000 µg/ml) for four weeks.
Cell proliferation assays
Cell proliferation was assessed using Cell Counting Kit-8 (CCK-8) assays and Ethynyl deoxyuridine (EdU) incorporation assays. For CCK-8 assays, a total of 3000 cells were plated in each well of 96-well plate. Cell proliferation was measured using the CCK-8 kit (Dojindo, Kumamoto, Japan) at indicated time through collecting the absorbance at 450nm according to the manufacturer's protocol. EdU incorporation assays were performed using the EdU kit (Roche, Mannheim, Germany) following the manufacturer's instruction. The results were collected and quantified using the Zeiss photomicroscope (Carl Zeiss, Oberkochen, Germany) and Image-Pro plus 6.0 software.
Cell cycle analysis
The cell cycles of indicated NSCLC cells were analyzed using the Cell Cycle Analysis Kit (Beyotime, Jiangsu, China) on FACSCalibur flow cytometer following the manufacturer's instruction.
Animal experiments
The Research Ethics Committee of Shanghai Chest Hospital approved the animal experiments. Briefly, 3×106 indicted NSCLC cells were subcutaneously injected into five-week-old athymic BALB/c nude mice purchased from the Shanghai Experimental Animal Center of Chinese Academy of Sciences (Shanghai, China). Subcutaneous tumors growth was measured every 7 days with a caliper, and the tumor volume was calculated as 1/2 a×b2 (a, long axes; b, short axes).
Immunohistochemistry assays
Immunohistochemistry was carried out with the standard approach as previously described [27]. Briefly, formalin-fixed, paraffin-embedded subcutaneous tumors were cut into 4 µm sections. The sections were incubated with a primary antibody against Ki67 (Abcam, Hong Kong, China). After being washed, the sections were incubated with horseradish peroxidase-conjugated second antibody (Abcam) and visualized using DAB Horseradish Peroxidase Color Development Kit (Beyotime).
Western blot analysis
Proteins were extracted, quantified and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and then being transferred to PVDF membrane. After being blocked using 5% not-fat milk, the blots were incubated with antibodies for CCND1 (Abcam) or β-actin (Abcam) at 4 °C overnight. After being washed, the blots were incubated with fluorescence-labeled secondary antibodies, and detected using an Odyssey infrared scanner (Li-Cor, Lincoln, NE, USA).
Statistical analysis
All data are shown as the mean ± standard deviation (SD) of at least three independent experiments. Comparisons between groups were performed using the SPSS 18.0 software package (SPSS, Chicago, IL, USA) with Wilcoxon signed-rank test, Mann-Whitney test, Student's t-test, Pearson correlation analysis, and one-way ANOVA followed by Dunnett's multiple comparisons test as indicated. P values < 0.05 were defined as statistically significant.
Results
PCNA-AS1 is upregulated in NSCLC tissues and cell lines
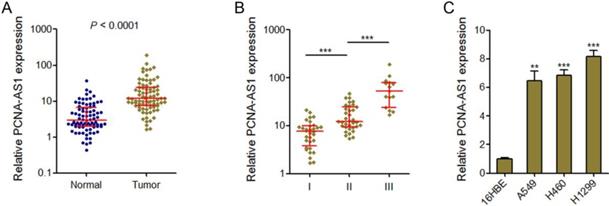
PCNA-AS1 expression levels were measured in 82 pairs of NSCLC tissues and adjacent non-tumor tissues by qRT-PCR assays. The results showed that PCNA-AS1 expression was significantly upregulated in NSCLC tissues compared with adjacent normal tissues (Figure 1A). Next, we investigated the correlation between PCNA-AS1 expression levels and TNM stages. As shown in Figure 1B, PCNA-AS1 expression was significantly upregulated in NSCLC samples with advanced TNM stages. Furthermore, we measured PCNA-AS1 expression in NSCLC cell lines (A549, H460 and H1299) and normal bronchial epithelial cell line (16HBE). As shown in Figure 1C, PCNA-AS1 expression was significantly upregulated in NSCLC cell lines compared with normal bronchial epithelial cell line. Collectively, these results suggested that PCNA-AS1 is upregulated in NSCLC.
Ectopic expression of PCNA-AS1 promotes NSCLC cell proliferation
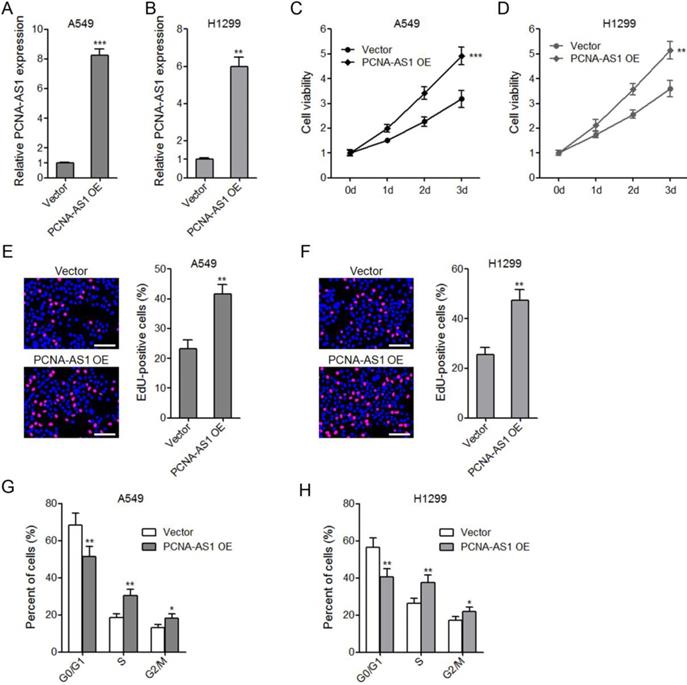
To explore the biological roles of PCNA-AS1 on NSCLC, we constructed PCNA-AS1 stably overexpressed A549 and H1299 cells (Figure 2A and 2B). CCK-8 assays showed that ectopic expression of PCNA-AS1 promoted A549 and H1299 cell proliferation (Figure 2C and 2D). EdU incorporation assays further verified the pro-proliferative roles of PCNA-AS1 on A549 and H1299 cells (Figure 2E and 2F). Furthermore, flow cytometric analysis revealed that ectopic expression of PCNA-AS1 in both A549 and H1299 cells decreased the percentage of cells at G0/G1 phase (Figure 2G and 2H), implying that PCNA-AS1 promotes NSCLC cell cycle progression.
PCNA-AS1 is highly expressed in NSCLC tissues and cell lines. (A) PCNA-AS1 expression in 82 pairs of NSCLC tissues and adjacent non-tumor tissues was measured by qRT-PCR. P < 0.0001 by Wilcoxon signed-rank test. (B) PCNA-AS1 expression in NSCLC tissues with different clinical stages. ***P < 0.001 by Mann-Whitney test. (C) PCNA-AS1 expression in normal bronchial epithelial cell line (16HBE) and NSCLC cell lines (A549, H460 and H1299) was measured by qRT-PCR. **P < 0.01, ***P < 0.001 by Student's t-test.
Ectopic expression of PCNA-AS1 promotes NSCLC cell proliferation and cell cycle progression. (A and B) PCNA-AS1 expression in PCNA-AS1 stably overexpressed and control A549 and H1299 cells was measured by qRT-PCR. (C and D) Cell proliferation of PCNA-AS1 stably overexpressed and control A549 and H1299 cells was detected by CCK-8 assays, and the relative number of cells to 0 day is shown. (E and F) Cell proliferation of PCNA-AS1 stably overexpressed and control A549 and H1299 cells was detected by EdU incorporation assays. The red color represents EdU-positive nuclei. Scale bars, 100 μm. (G and H) Cell cycle distribution of PCNA-AS1 stably overexpressed and control A549 and H1299 cells was analyzed by flow cytometry. *P < 0.05, **P < 0.01, ***P < 0.001 by Student's t-test.
Depletion of PCNA-AS1 suppresses NSCLC cell proliferation
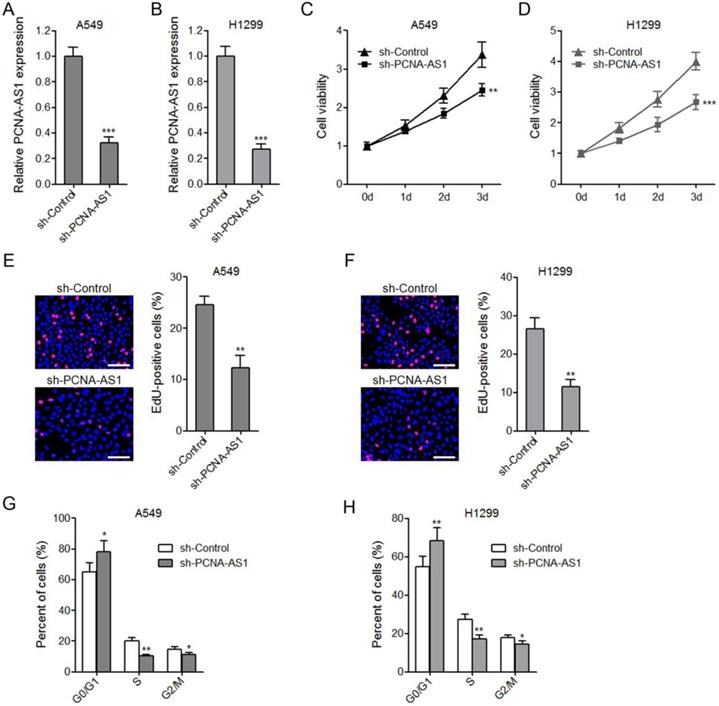
To further confirm the biological roles of PCNA-AS1 on NSCLC, we constructed PCNA-AS1 stably depleted A549 and H1299 cells by transfecting PCNA-AS1-specific shRNA (Figure 3A and 3B). CCK-8 assays showed that depletion of PCNA-AS1 significantly inhibited A549 and H1299 cell proliferation (Figure 3C and 3D). EdU incorporation assays further verified the proliferation inhibitory roles of PCNA-AS1 depletion on A549 and H1299 cells (Figure 3E and 3F). Flow cytometric analysis revealed that depletion of PCNA-AS1 in both A549 and H1299 cells increased the percentage of cells at G0/G1 phase (Figure 3G and 3H), implying that depletion of PCNA-AS1 inhibits NSCLC cell cycle progression.
Depletion of PCNA-AS1 inhibits NSCLC xenograft tumor growth in vivo
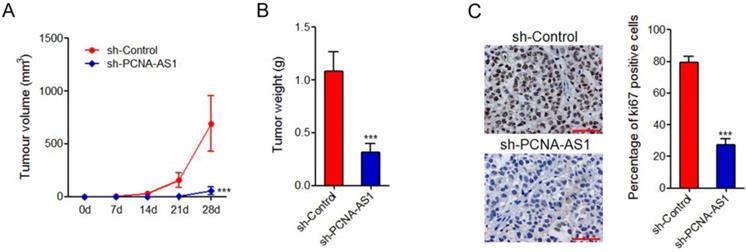
To further investigate the biological roles of PCNA-AS1 on NSCLC, PCNA-AS1 stably depleted and control H1299 cells were injected subcutaneously into nude mice. Tumor growth rates were measured every 7 days for 28 days, and then the tumors were excised and weighed. The results showed that depletion of PCNA-AS1 significantly inhibited xenograft tumor growth in vivo (Figure 4A and 4B). Ki67 staining of xenograft further showed that depletion of PCNA-AS1 inhibited H1299 cell proliferation in vivo (Figure 4C).
Depletion of PCNA-AS1 inhibits NSCLC cell proliferation and cell cycle progression. (A and B) PCNA-AS1 expression in PCNA-AS1 stably depleted and control A549 and H1299 cells was measured by qRT-PCR. (C and D) Cell proliferation of PCNA-AS1 stably depleted and control A549 and H1299 cells was detected by CCK-8 assays, and the relative number of cells to 0 day is shown. (E and F) Cell proliferation of PCNA-AS1 stably depleted and control A549 and H1299 cells was detected by EdU incorporation assays. The red color represents EdU-positive nuclei. Scale bars, 100 μm. (G and H) Cell cycle distribution of PCNA-AS1 stably depleted and control A549 and H1299 cells was analyzed by flow cytometry. *P < 0.05, **P < 0.01, ***P < 0.001 by Student's t-test.
Depletion of PCNA-AS1 inhibits NSCLC tumor growth in vivo. (A) Tumor volumes of subcutaneous tumors derived from PCNA-AS1 stably depleted and control H1299 cells were measured every 7 days. (B) Tumor weights of subcutaneous tumors derived from PCNA-AS1 stably depleted and control H1299 cells at 28 day after injection. (C) In vivo cell proliferation of PCNA-AS1 stably depleted and control H1299 cells was assessed by Ki67 immunohistochemistry staining of subcutaneous tumors. Scale bars, 50 µm. ***P < 0.001 by Mann-Whitney test.
PCNA-AS1 upregulates CCND1 expression. (A) PCNA and CCND1 mRNA level in PCNA-AS1 stably overexpressed and control A549 cells was measured by qRT-PCR. (B) PCNA and CCND1 mRNA level in PCNA-AS1 stably overexpressed and control H1299 cells was measured by qRT-PCR. (C and D) CCND1 protein level in PCNA-AS1 stably overexpressed and control A549 and H1299 cells was measured by western blot. (E) PCNA and CCND1 mRNA level in PCNA-AS1 stably depleted and control A549 cells was measured by qRT-PCR. (F) PCNA and CCND1 mRNA level in PCNA-AS1 stably depleted and control H1299 cells was measured by qRT-PCR. (G and H) CCND1 protein level in PCNA-AS1 stably depleted and control A549 and H1299 cells was measured by western blot. **P < 0.01, ***P < 0.001, ns, not significant, by Student's t-test.
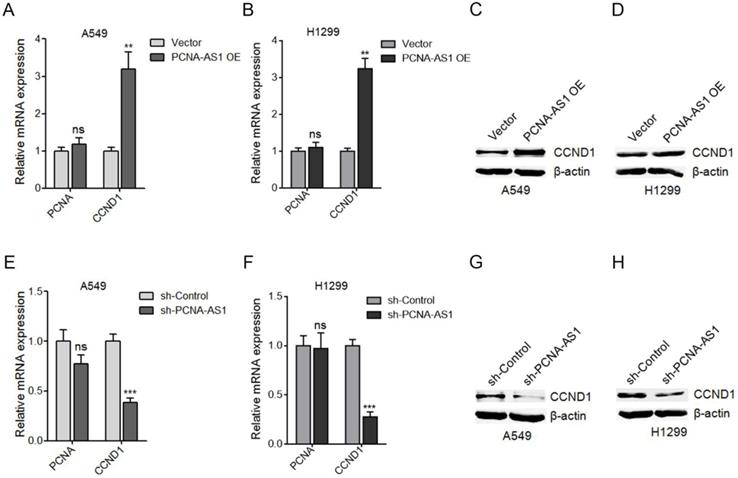
PCNA-AS1 upregulates CCND1 expression
To uncover the molecular mechanisms underlying the pro-proliferative roles of PCNA-AS1 on NSCLC, we first measured the effects of PCNA-AS1 on PCNA, which is transcribed in antisense orientation with PCNA-AS1. However, our results showed that ectopic expression of PCNA-AS1 did not regulate PCNA expression (Figure 5A). Due to the significant effects of PCNA-AS1 on G0/G1 phase cell cycle arrest, we next detected the effects of PCNA-AS1 on CCND1 (cyclin D1), whose activity is required for G1/S transition [28]. As shown in Figure 5A-D, ectopic expression of PCNA-AS1 significantly upregulated CCND1 mRNA and protein levels in both A549 and H1299 cells. In addition, although depletion of PCNA-AS1 did not regulate PCNA expression, depletion of PCNA-AS1 significantly decreased CCND1 mRNA and protein levels in both A549 and H1299 cells (Figure 5E-H). These results suggested that PCNA-AS1 upregulates CCND1 expression in NSCLC cells.
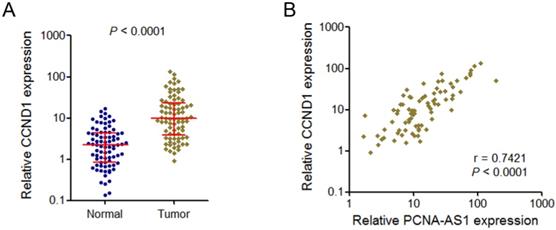
PCNA-AS1 transcript level was positively correlated with CCND1 mRNA level in NSCLC tissues
Because PCNA-AS1 could upregulate CCND1 expression, we next investigate whether a correlation exists between PCNA-AS1expression level and CCND1 in human NSCLC tissues. We examined CCND1 mRNA level in the same set of 82 pairs of NSCLC tissues and adjacent normal tissues shown in Figure 1A. The results showed that CCND1 is also significantly upregulated in NSCLC tissues compared with adjacent normal tissues (Figure 6A). Correlation analysis showed that PCNA-AS1 expression level was positively correlated with CCND1 mRNA level in these 82 NSCLC tissues (Figure 6B).
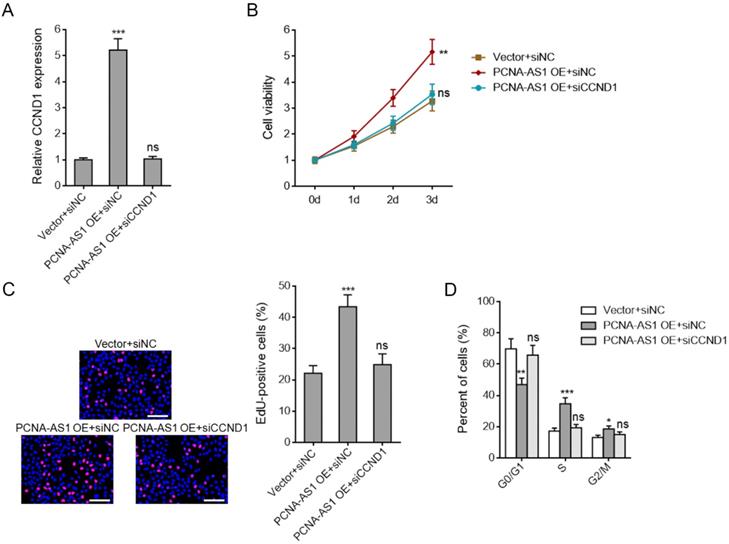
Depletion of CCND1 abolished the pro-proliferative roles of PCNA-AS1 on NSCLC cells
To investigate whether PCNA-AS1 promotes NSCLC cell proliferation through upregulating CCND1 expression, we silenced CCND1 expression in PCNA-AS1 stably overexpressed A549 cells (Figure 7A). CCK-8 assays showed that CCND1 depletion abolished the pro-proliferative roles of PCNA-AS1 on A549 cells (Figure 7B). EdU incorporation assays further verified that CCND1 depletion could abolish the pro-proliferative roles of PCNA-AS1 on A549 cells (Figure 7C). Flow cytometric analysis revealed that CCND1 depletion reversed the down-regulation of percentage of cells at G0/G1 phase caused by PCNA-AS1 overexpression (Figure 7D), implying that CCND1 depletion reversed NSCLC cell cycle progression caused by PCNA-AS1 overexpression. Collectively, these results suggested that the roles of PCNA-AS1 on NSCLC cell proliferation and cell cycle are dependent on CCND1.
Discussion
With the advances of oncological researches, many critical oncogenes or tumor suppressors in NSCLC were identified, such as p53, EGFR, PD-1/PD-L1 [29-31]. Several molecular targeted therapies have been developed, such as EGFR-tyrosine kinase inhibitor and PD-1/PD-L1 antibodies [32, 33]. However, many NSCLC patients are not sensitive to these therapies [34]. This also implies that the carcinogenesis and progression of NSCLC are very complex and many genes participate in these processes.
Recently, the crucial roles of lncRNAs in cancers have been revealed [35]. MALAT1, which is a well-known lncRNA, promotes lung cancer metastasis [23]. PVT1, another well studied lncRNA, promotes NSCLC tumorigenesis and cell proliferation [21]. Several transcriptome RNA sequencing results demonstrated that the number of lncRNAs is very larger than that of mRNAs, and over 68% of all transcripts could be classified as lncRNAs [25]. Among these many lncRNAs, only a few of them has been studied. Many other lncRNAs may also have important roles in cancers. In this study, we investigated the expression and functions of a previously poorly understood lncRNA PCNA-AS1 in NSCLC. Our results showed that PCNA-AS1 is upregulated in NSCLC tissues and cell lines and positively associated with advanced TNM stages. Gain-of-function experiments showed that overexpression of PCNA-AS1 promotes NSCLC cell proliferation and cell cycle progression. Loss-of-function experiments showed that depletion of PCNA-AS1 inhibits NSCLC cell proliferation, cell cycle progression, and in vivo tumor growth. These data demonstrate that the upregulated PCNA-AS1 functions as an oncogene in NSCLC and targeting PCNA-AS1 may be a potential therapeutic strategy for NSCLC.
PCNA-AS1 transcript level is positively correlated with CCND1 mRNA level in NSCLC tissues. (A) CCND1 mRNA level in 82 pairs of NSCLC tissues and adjacent non-tumor tissues was measured by qRT-PCR. P < 0.0001 by Wilcoxon signed-rank test. (B) The correlation between PCNA-AS1 transcript level and CCND1 mRNA level was measured in 82 NSCLC tissues. P < 0.0001 by Pearson correlation analysis.
Depletion of CCND1 abrogates the biological roles of PCNA-AS1 on NSCLC cell proliferation and cell cycle progression. (A) CCND1 expression in PCNA-AS1 stably overexpressed and control A549 cells after transfection with CCND1 specific siRNA was measured by qRT-PCR. (B) Cell proliferation of PCNA-AS1 stably overexpressed and control A549 cells after transfection with CCND1 specific siRNA was detected by CCK-8 assays, and the relative number of cells to 0 day is shown. (C) Cell proliferation of PCNA-AS1 stably overexpressed and control A549 cells after transfection with CCND1 specific siRNA was detected by EdU incorporation assays. The red color represents EdU-positive nuclei. Scale bars, 100 μm. (D) Cell cycle distribution of PCNA-AS1 stably overexpressed and control A549 cells after transfection with CCND1 specific siRNA was analyzed by flow cytometry. *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant, by one-way ANOVA followed by Dunnett's multiple comparisons test.
PCNA-AS1 is oriented in antisense direction with respect to PCNA, which has critical roles in DNA replication and cell proliferation [36]. Several antisense lncRNAs are reported to regulate the genes of the opposite strand [20, 37]. In this study, we also investigated the effects of PCNA-AS1 on PCNA. However, our results showed that neither overexpression nor depletion of PCNA-AS1 changed PCNA expression. As PCNA-AS1 had significant effects on G0/G1 phase cell cycle arrest, we investigated the regulation of PCNA-AS1 on CCND1 (cyclin D1), which is crucial for G1/S transition. Our results showed that overexpression of PCNA-AS1 upregulates CCND1 expression, and while depletion of PCNA-AS1 decreases CCND1 expression. Moreover, the expression of PCNA-AS1 is positively associated with that of CCND1 in clinical NSCLC tissues, supporting the regulation of CCND1 by PCNA-AS1. Functional experiments showed that depletion of CCND1 abolished the pro-oncogenic roles of PCNA-AS1 on NSCLC. These data suggest that PCNA-AS1 promotes NSCLC cell proliferation and oncogenic activity via upregulating CCND1. Some nuclear lncRNAs could epigenetically modulate the expression of their target genes via binding and recruiting DNA or histone modification enzymes. Some cytoplasmic lncRNAs could directly bind proteins, mRNAs, and/or microRNAs, and further change the location, expression, roles of the interacted partners. The potential molecular mechanism underlying the regulation of CCND1 by PCNA-AS1 needs further investigation.
In conclusion, we found that PCNA-AS1 is increased in NSCLC, promotes NSCLC cell proliferation and oncogenic activity via upregulating CCND1. Our results suggest that PCNA-AS1 would be a potential therapeutic target for NSCLC.
Acknowledgements
This work was supported by grants from The Innovation Group Project of Shanghai Municipal Health Commission (2019CXJQ03), Multidisciplinary Collaboration Project of Shanghai Chest Hospital (YJXT20190201) and Shanghai Outstanding Youth Clinical Medical (Medical Laboratory Scientists) Training Funding (HYWJ201605).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65:87-108
2. Ballatori E, Fatigoni S, Roila F. Treatment of lung cancer. The New England journal of medicine. 2009;361:2485 author reply 6-7
3. Li Y, Xu Y, Ye K, Wu N, Li J, Liu N. et al. Knockdown of Tubulin Polymerization Promoting Protein Family Member 3 Suppresses Proliferation and Induces Apoptosis in Non-Small-Cell Lung Cancer. Journal of Cancer. 2016;7:1189-96
4. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA: a cancer journal for clinicians. 2017;67:7-30
5. Torre LA, Siegel RL, Jemal A. Lung Cancer Statistics. Advances in experimental medicine and biology. 2016;893:1-19
6. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nature reviews Genetics. 2014;15:7-21
7. Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD. et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer cell. 2015;28:529-40
8. Atanasovska B, Rensen SS, van der Sijde MR, Marsman G, Kumar V, Jonkers I. et al. A liver-specific long non-coding RNA with a role in cell viability is elevated in human non-alcoholic steatohepatitis. Hepatology. 2017
9. Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer cell. 2016;29:452-63
10. Xu D, Yang F, Yuan JH, Zhang L, Bi HS, Zhou CC. et al. Long noncoding RNAs associated with liver regeneration 1 accelerates hepatocyte proliferation during liver regeneration by activating Wnt/beta-catenin signaling. Hepatology. 2013;58:739-51
11. Liu X, Xiao ZD, Han L, Zhang J, Lee SW, Wang W. et al. LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nature cell biology. 2016;18:431-42
12. Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X. et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer cell. 2015;27:370-81
13. Yang F, Xue X, Zheng L, Bi J, Zhou Y, Zhi K. et al. Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. The FEBS journal. 2014;281:802-13
14. Chen X, Dong H, Liu S, Yu L, Yan D, Yao X. et al. Long noncoding RNA MHENCR promotes melanoma progression via regulating miR-425/489-mediated PI3K-Akt pathway. American journal of translational research. 2017;9:90-102
15. Liu C, Lin J. Long noncoding RNA ZEB1-AS1 acts as an oncogene in osteosarcoma by epigenetically activating ZEB1. American journal of translational research. 2016;8:4095-105
16. Wang G, Chen H, Liu J. The long noncoding RNA LINC01207 promotes proliferation of lung adenocarcinoma. American journal of cancer research. 2015;5:3162-73
17. Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF. et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer cell. 2014;25:666-81
18. Takahashi K, Yan I, Haga H, Patel T. Long noncoding RNA in liver diseases. Hepatology. 2014;60:744-53
19. Lin A, Li C, Xing Z, Hu Q, Liang K, Han L. et al. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nature cell biology. 2016;18:213-24
20. Zhu XT, Yuan JH, Zhu TT, Li YY, Cheng XY. Long noncoding RNA glypican 3 (GPC3) antisense transcript 1 promotes hepatocellular carcinoma progression via epigenetically activating GPC3. The FEBS journal. 2016;283:3739-54
21. Wan L, Sun M, Liu GJ, Wei CC, Zhang EB, Kong R. et al. Long Noncoding RNA PVT1 Promotes Non-Small Cell Lung Cancer Cell Proliferation through Epigenetically Regulating LATS2 Expression. Molecular cancer therapeutics. 2016;15:1082-94
22. Hu T, Lu YR. BCYRN1, a c-MYC-activated long non-coding RNA, regulates cell metastasis of non-small-cell lung cancer. Cancer cell international. 2015;15:36
23. Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G. et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer research. 2013;73:1180-9
24. Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Functions of lncRNA HOTAIR in lung cancer. Journal of hematology & oncology. 2014;7:90
25. Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y. et al. The landscape of long noncoding RNAs in the human transcriptome. Nature genetics. 2015;47:199-208
26. Yuan SX, Tao QF, Wang J, Yang F, Liu L, Wang LL. et al. Antisense long non-coding RNA PCNA-AS1 promotes tumor growth by regulating proliferating cell nuclear antigen in hepatocellular carcinoma. Cancer letters. 2014;349:87-94
27. Jiang S, Zhang LF, Zhang HW, Hu S, Lu MH, Liang S. et al. A novel miR-155/miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. The EMBO journal. 2012;31:1985-98
28. Li Y, Li D, Yang W, Fu H, Liu Y, Li Y. Overexpression of the transcription factor FOXP3 in lung adenocarcinoma sustains malignant character by promoting G1/S transition gene CCND1. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37:7395-404
29. Ali A, Bhatti MZ, Shah AS, Duong HQ, Alkreathy HM, Mohammad SF. et al. Tumor-suppressive p53 Signaling Empowers Metastatic Inhibitor KLF17-dependent Transcription to Overcome Tumorigenesis in Non-small Cell Lung Cancer. The Journal of biological chemistry. 2015;290:21336-51
30. Trusolino L. Oncogenic MET as an Effective Therapeutic Target in Non-Small Cell Lung Cancer Resistant to EGFR Inhibitors: The Rise of the Phoenix. Cancer discovery. 2016;6:1306-8
31. Ji M, Liu Y, Li Q, Li X, Ning Z, Zhao W. et al. PD-1/PD-L1 expression in non-small-cell lung cancer and its correlation with EGFR/KRAS mutations. Cancer biology & therapy. 2016;17:407-13
32. Xu J, Yang H, Jin B, Lou Y, Zhang Y, Zhang X. et al. EGFR tyrosine kinase inhibitors versus chemotherapy as first-line therapy for non-small cell lung cancer patients with the L858R point mutation. Scientific reports. 2016;6:36371
33. Nicolazzo C, Raimondi C, Mancini M, Caponnetto S, Gradilone A, Gandini O. et al. Monitoring PD-L1 positive circulating tumor cells in non-small cell lung cancer patients treated with the PD-1 inhibitor Nivolumab. Scientific reports. 2016;6:31726
34. Gainor JF, Shaw AT, Sequist LV, Fu X, Azzoli CG, Piotrowska Z. et al. EGFR Mutations and ALK Rearrangements Are Associated with Low Response Rates to PD-1 Pathway Blockade in Non-Small Cell Lung Cancer: A Retrospective Analysis. Clinical cancer research: an official journal of the American Association for Cancer Research. 2016;22:4585-93
35. Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. British journal of cancer. 2013;108:2419-25
36. Nguyen VN, Mirejovsky P, Mirejovsky T, Melinova L, Mandys V. Expression of cyclin D1, Ki-67 and PCNA in non-small cell lung cancer: prognostic significance and comparison with p53 and bcl-2. Acta histochemica. 2000;102:323-38
37. Sun J, Wang X, Fu C, Wang X, Zou J, Hua H. et al. Long noncoding RNA FGFR3-AS1 promotes osteosarcoma growth through regulating its natural antisense transcript FGFR3. Molecular biology reports. 2016;43:427-36
Author contact
![]() Corresponding author: Jiatao Lou, Department of Laboratory Medicine, Shanghai Chest Hospital, Shanghai Jiao Tong University, Shanghai 200030, China; Phone: 86-21-22200000-1601; Fax: 86-21-22200000-1601; E-mail: loujiataocom.
Corresponding author: Jiatao Lou, Department of Laboratory Medicine, Shanghai Chest Hospital, Shanghai Jiao Tong University, Shanghai 200030, China; Phone: 86-21-22200000-1601; Fax: 86-21-22200000-1601; E-mail: loujiataocom.

 Global reach, higher impact
Global reach, higher impact