Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(11):3387-3399. doi:10.7150/jca.40186 This issue Cite
Research Paper
Transcriptional Regulation of Latency-Associated Transcripts (LATs) of Herpes Simplex Viruses
1. The First School of Clinical Medicine, Health Science Center, Yangtze University, Nanhuan Road, Jingzhou, Hubei 434023, China.
2. Laboratory of Oncology, Center for Molecular Medicine, School of Basic Medicine, Health Science Center, Yangtze University, 1 Nanhuan Road, Jingzhou, Hubei 434023, China
3. Department of Biochemistry and Molecular Biology, School of Basic Medicine, Health Science Center, Yangtze University, Jingzhou, Hubei 434023, China.
4. Clinical Medical Research Center, Affiliated Hospital of Inner Mongolia Medical University, Hohhot 010050, China.
5. Department of Neural Surgery, People's Hospital of Dongsheng District of Erdos City, Erdos, Inner Mongolia, 017000, China.
6. State Key Laboratory of Respiratory Disease, Affiliated Cancer Hospital Institute of Guangzhou Medical University, Guangzhou 510095, China.
7. Department of Pathophysiology, School of Basic Medicine, Health Science Center, Yangtze University, Jingzhou, Hubei 434023, China.
8. Department of Laboratory Medicine, School of Basic Medicine, Health Science Center, Yangtze University, 1 Nanhuan Road, Jingzhou, Hubei 434023, China.
*Authors contributed equally
Received 2019-9-11; Accepted 2020-2-6; Published 2020-3-5
Abstract
Herpes simplex viruses (HSVs) cause cold sores and genital herpes and can establish lifelong latent infection in neurons. An engineered oncolytic HSV (oHSV) has recently been approved to treat tumors in clinics. HSV latency-associated transcripts (LATs) are associated with the latent infection, but LAT transcriptional regulation was seldom reported. For a better treatment of HSV infection and tumors, here we sequenced the LAT encoding DNA and LAT transcription regulatory region of our recently isolated new strain HSV-1-LXMW and did comparative analysis of the sequences together with those of other four HSV-1 and two HSV-2 strains. Phylogenetic analysis of LATs revealed that HSV-1-LXMW is evolutionarily close to HSV-1-17 from MRC University, Glasgow, UK. For the first time, Using a weight matrix-based program Match and multi-sequences alignment of the 6 HSV strains, we identified HSV LAT transcription regulatory sequences that bind to 9 transcription factors: AP-1, C-REL, Comp1, E2F, Hairy, HFH-3, Kr, TCF11/MAFG, v-Myb. Interestingly, these transcription regulatory sequences and factors are either conserved or unique among LATs of HSV-1 and HSV-2, suggesting they are potentially functional. Furthermore, literature analysis found that the transcription factors v-myb and AP-1 family member JunD are functional in regulating HSV gene transcription, including LAT transcription. For the first time, we discovered seven novel transcription factors and their corresponding transcription regulatory sequences of HSV LATs. Based on our findings and other reports, we proposed potential mechanisms of the initiation and maintenance of HSV latent infection. Our findings may have significant implication in our understanding of HSV latency and engineering of better oncolytic HSVs.
Keywords: oncolytic Herpes Simplex Virus (oHSV), Latency-Associated Transcripts (LATs), transcription regulatory regions (TRRs), Transcription Regulatory Sequences (TRSs), Transcription Factors (TFs), latent neural infection
Introduction
Up to now, tumor treatments are still a big challenge. Although there are many methods to treat tumors [1-3], tumor recurrence rate is high and the metastasis is irreversible, resulting in poor clinical efficacy [4]. Nowadays, there are mainly eight types of herpes virus, including herpes simplex virus type 1 and type 2 (HSV-1 and 2), varicella zoster virus (VZV), cytomegalovirus (CMV), Epstein-Barr virus (EBV), human herpes simplex virus types 6,7,8. Some herpes viruses cause tumors. For example, 27% of children with retinoblastoma are diagnosed with CMV infection [5]. In addition, EBV is associated with nasopharyngeal carcinoma. However there is no direct evidence that herpes simplex virus causes tumors, so that HSV can be engineered as oncolytic HSV (oHSV) to combat cancer [6-9]. In oHSVs, HSV can infect a variety of host cells to meet the needs of oncolytic virus therapy, and various forms of transgenic vectors have been developed for cancer therapy [10, 11]. For example, the most advanced oHSVs T-Vec (Talimogene laherparepvec), G207, 1716, G47Δ and HF10 have been evaluated in clinical trials for their benefits in treating advanced cancers such as melanoma, glioma, head and neck cancer and breast cancer [12]. Furthermore, a novel oHSV Ld0-GFP (which was derived from the oncolytic ICP0-null HSV) targeting hepatocellular carcinoma (HCC) has recently been reported [12]. In 2016, the oHSV T-VEC has been approved for the treatment of melanoma [13,14].
Herpes viruses are classified into α, β and γ three genera [15]. Among them γ herpes viruses include HSV-1 and 2 [16], and are able to build lifelong latent infections in neuron [15]. HSVs have a wide host range, and specific therapeutic agents (acyclovir). Initially, a HSV effectively infects epithelial cells or tissues (lytic infections) and then slowly spreads further around, stopping to form latent infections in the nucleus of a sensory neuron [17]. HSVs express immediate early (IE or α), early (E or β) and late (L or γ) genes [18]. IE genes encode five infected cell polypeptide (ICP) proteins, ICP0/RL2, ICP4/RS1, ICP22/US1, ICP27/UL54, and ICP47/US12 [19]. ICP0 plays an essential role in virus replication, cell growth and apoptosis [20, 21]. ICP4 has multiple target sites recognized by CD8+ T cells and is a very important key regulator of early and late gene expression [22].
During latent infection of sensory ganglia in mammalian host [23], only the latency-associated transcripts (LATs) is the product of abundant expression of HSVs [24]. LATs play a crucial role in establishing latency [25], maintaining latency [26, 27], reactivation of the virus from latency, and protection of neurons from apoptosis [28, 29]. For understanding the function of LAT genes, we need to lean the structure of LATs and their transcriptional regulation in host cells.
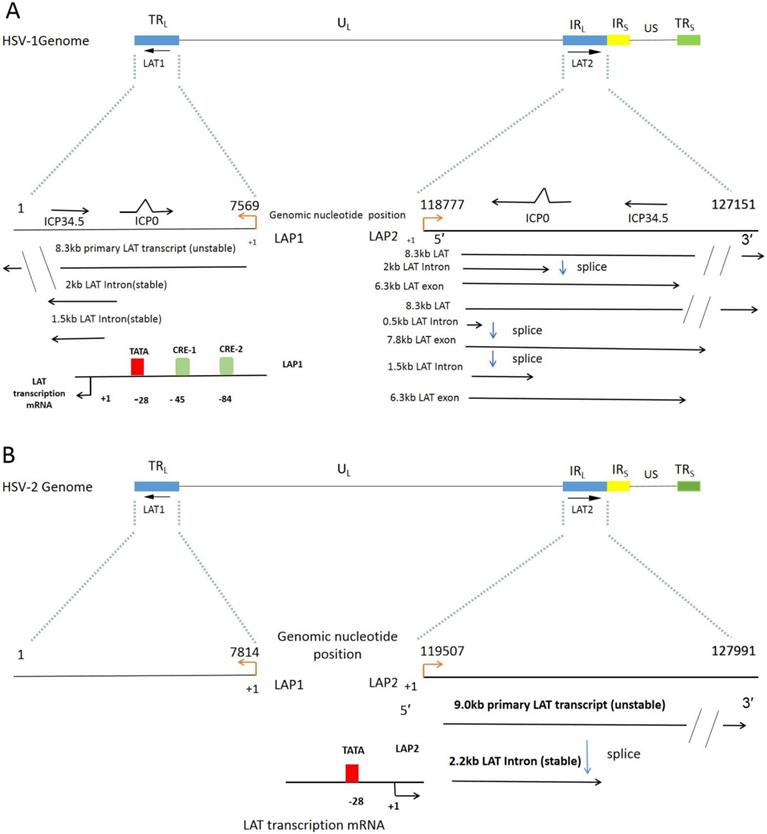
As shown in Figure 1, genes encoding LATs have two intervals, called fragment 1 (LAT1) and fragment 2 (LAT2). LATs are spliced into non-major LAT (minor LAT or primary LAT) and major LAT. HSV-1 LATs are the transcription family of a group of RNAs, consisting mainly of 8.3 kb of low-level original transcripts and 3 introns. The 2 kb intron LAT generally presents at high levels during lytic and latent infection. The 1.5 kb intron LAT is detected during latency. The 0.5 kb LAT was not detected.[30] The 6.3kb exon, which were not easily detected by Northern blot analysis [31, 32]. Translation of a protein from the LAT exon mRNA has yet to be convincingly proved [33], nevertheless, it is obvious that exon 1 region of LAT is a vital part to prevent apoptosis [28, 34]. LAT1 has two overlapping introns (2.0kb/1.5kb) produced by LAT transcripts, named to as double introns. Likewise, HSV-2 LAT gene was transcribed to generate about a 9.0 kb of non-primary LAT, and a stable 2.2 kb of main LAT. However, Transcripts of HSV-2 LAT1 are rarely reported. Both HSV-1 and HSV-2 have LAT promoter 1 (LAP1) and LAT promoter 2 (LAP2) [35]. LAP1 is one of the most critical promoters for the efficient activation of LAT1s expression, which has been demonstrated in the incubation period of HSV infection. The same LAP2 has also been shown to have promoter activity, and the sequences in this region have enhanced HSV-1 LAP1 activity during a latent infection [36].
HSV latency in neurons is a major issue in the treatment of HSV infection and engineering of effective and safe oHSVs. Here we sequenced the LAT encoding DNA and LAT transcription regulatory region of our recently isolated new strain HSV-1-LXMW, and identified HSV LAT TRSs that bind to 9 transcription factors. Our results further illustrated HSV LAT biology and provided new options for better HSV treatment and oHSVs in the future.
Materials and Methods
LAT genomic DNA sequencing of our new strain HSV-1-LXMW
Our laboratory isolated a new HSV strain named HSV-1-LXMW from a male patient with oral herpes in Beijing, China [13]. LAT DNA sequencing was carried out by Beijing institute of genomics and these sequences were analyzed using Burrows-Wheeler Aligner (BWA) software as described earlier [13]. The genome sequences of the other 5 strains of HSVs were obtained from the NCBI reference database and summarized in table 1. Interestingly, we found that HSV-1 Strain Macl and HSV-2 strain H1226 did not have LAT1.
Phylogenetic analysis of HSV LATs
The online MEGA7 software was used for phylogenetic analysis of TRRs of the LAT genes in 6 HSV strains. The evolution history of LAT is explored on the basis of time model by means of self-extracting uniform tree and maximum likelihood method. In 50% of boot replicates, the branch corresponding to the replicated partition forms a fold. Then, the original homologous tree of LAT can be obtained by combining the neighborhood join algorithm and BioNJ algorithm to estimate the distance matrix. Finally, the suitable topology of logarithmic likelihood value is selected.
The LAT and its LAT Twintron processing. A. The linear HSV-1 (strain 17) genome contains with the Unique Long (UL) and Unique Short (US) regions flanked by the terminal and internal Repeats (TRL, IRL, IRS and TRS). LAT genes are distributed in two intervals, the first interval LAT1 is located in TRL and UL, and the second interval LAT2 is located in the region where IRL connects with UL [37]. A fragment of IRL was expanded to reveal the location of LAT1 gene, overlapping ICP0 and ICP34.5, 8.3 kb original LAT (unstable), 2 kb and 1.5 kb LAT intron (stable). The direction of transcription is indicated by the black arrowhead. TATA indicates the location (in the genomic DNA) of the LAT1 promoter TATA box. The start of LAT1 transcription is indicated by +1, corresponding to nt 7569 of the genome. CRE-1 and CRE-2 denote the locations of the binding sites of the cAMP response elements 1 and 2 identified by Leib et al and Kenny et al [38, 39]. LAT2 locus overlaps with the ICP0 and ICP34.5. The 8.3 kb primary LAT2 is spliced to an unusually stable 2.0 kb intron and a 6.3 kb short lived unconfirmed mRNA. Alternatively the 8.3 kb transcript can be spliced as Twintron introns to a 0.5 kb unstable intron and a 7.8 kb RNA (1.5 kb plus 6.3 kb) [40, 41]. B. HSV-2 genome contains the LAT locus similar to HSV-1 in Figure 1A [37]. LAT2 transcripts: 9.0 kb primary LAT (unstable), 2.2 kb major LAT (stable). The start of LAT2 transcription is indicated by +1, corresponding to nt 119507 of the genome.
HSV LAT genomic DNA sequencing.
| HSV strain | Gene Bank ID | Tax-ID | Sub-Date | LAT DNA sequence | Start site of transcription (+1) | University, Country |
|---|---|---|---|---|---|---|
| HSV-1 strain LXMW | LAT1 (1—7589) LAT2(118783-127138) | LAT1 (7589→1) LAT2(118812-127138) | Yangtze University, Jingzhou, China | |||
| HSV-1 strain 17 | JN555585.1 | 10299 | 2011-08-02 | LAT1 (1--7569) LAT2(118777-127151) | LAT1 (7569→1) LAT2(118805→127151) | MRC University, Glasgow, UK |
| HSV-1 isolate SC16 | KX946970.1 | 10309 | 2016-10-30 | LAT1(1-8251) LAT2 (118713-126490) | LAT1(8223→1) LAT2 (119234-126490) | SeveroOchoa, Spain |
| HSV-1 Strain Macl | KM222720.1 | 10298 | 2014-7-21 | LAT2 (118188-126929) | LAT2(118216-126929) | Pennslyvania State University, USA |
| HSV-2 strain HG52 | JN561323.2 | 10315 | 2011-08-05 | LAT1 (1-7814) LAT2 (119507-127991) | LAT1 (7767→1) LAT2 (119554→127991) | University of Glasgow, UK |
| HSV-2 strain H1226 | KY922720.1 | 10310 | 2017-4-06 | LAT2 (119356-128342) | LAT2 (119403→128342) | Pennslyvania State University, USA |
Genomic DNA sequencing of the transcription regulatory region (TRR) of our new strain HSV-1-LXMW
To determine the transcriptional regulation sequence of LAT gene, we extended by 2kb from the upstream of LAT gene as its TRR. For instance, taking the standard strain HSV-1-17 as an example, LAT1 is a reverse gene whose gene position is 1-7569bp, and 7569 is set as the transcription initiation site (represented by +1). TRR1 was chosen from 7569 to 9569 bp. LAT2 is a forward gene, whose position is 118777-127151bp. TRR2 was chosen from 116777 to 118777bp. Similarly, the TRRs of other HSV LAT genes were chosen (Table 2). LAT TRR genomic DNA sequencing of our original strain HSV-1-LXMW was performed as described earlier and above [13].
Prediction of LAT transcription regulatory sequences (TRSs) and factors (TFs)
To predict LAT TRSs, TFs, we used the online program Matching. Match (http://gene-regulation.com/pub/programs.html) is designed to predict DNA transcription factor binding sites (TFBS) using a location-weight matrix library from TRANSFAC Public 6.0. The prediction was performed when all parameters were set to the strict default condition.
Alignment of LAT TRRs of HSV-1 and HSV-2 strains
To know if there are conservative sequences among the LAT TRRs, we used the software ApE (http://www.bio-soft.net/plasmid/ApE.htm) to compare the 10 TRRs of LATs from 6 HSV strains at a setting of > 5 base pairs. The alignment parameters were set at default except blocks 10, mismatch penalty 0, gap penalty 0, gap ext penalty 0.
Results
LAT DNA sequences of our new strain HSV-1-LXMW and other HSV strains
The sequences of HSV-1-LXMW LAT were determined as follows: LAT1 from 1 to 7589bp, LAT2 from 118783 to 127151bp (see supplementary results 1, 2). Blast analysis showed that our HSV-1-LXMW LAT and HSV-1-17 LAT strains are highly similar, but our HSV-1-LXMW sequences are very different that of HSV-2 strains. Therefore, our data validated that the new strain of HSV-1-LXMW belongs to HSV-1 (Table 1).
LAT phylogenetic analysis showed HSV-1-LXMW is close to strain HSV-1-17
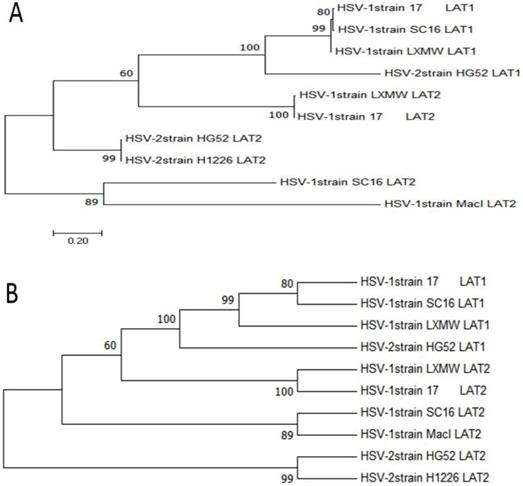
To study the evolutionary relationships of our new strain HSV-1-LXMW with other HSV-1 and HSV-2 strains, phylogenetic analysis was performed using MEGA7 software (http://www.liangchan.net/ liangchan/9113.html).The sequences of LAT1 and LAT2 of HSV-1-LXMW and 5 strains were analyzed. They include HSV-1-LXMW LAT1 and LAT2, HSV-1-17 LAT1 and LAT2, HSV-1-SC16 LAT1 and LAT2, HSV-1-Macl LAT2, HSV-2-HG52 LAT1 and LAT2, HSV-2-H1226 LAT2. Both the phylogenetic tree data (Figure 2) and neighbor network data showed the presence of four groups of clustering structures. Our new strain HSV-1-LXMW isolated in Beijing, China is close to the strain HSV-1-17 from MRC University, Glasgow, UK and far from the strain HSV-2-HG52 in University of Glasgow, UK and the strain HSV-2-H1226 in Pennslyvania State University, USA. The data showed a mean distance of approximately 52.7% among the strains tested collectively.
The sequences of LAT transcription regulatory regions (TRRs) of our new strain HSV-1-LXMW and other strains
The sequences of LAT TRRs of new HSV-1- LXMW strain are 7589-9589 bp for TRR1, and 116797- 118797 bp for TRR2 (See Table 2, and supplementary results 3 and 4). The sequences of the LAT TRRs of the other 5 HSV strains studied in this article are listed in Table 2. It was found that the TRRs of LAT1 overlapped with part or all of the UL1 gene (envelope glycoprotein L), while the TRRs of LAT2 overlapped with part or all of the UL56 gene. Interestingly, the HSV-1-macl LAT2 transcriptional regulatory region overlaps with part of the RS1 gene (ICP4) as shown in Table 2.
Potential LAT TRSs and TFs were identified
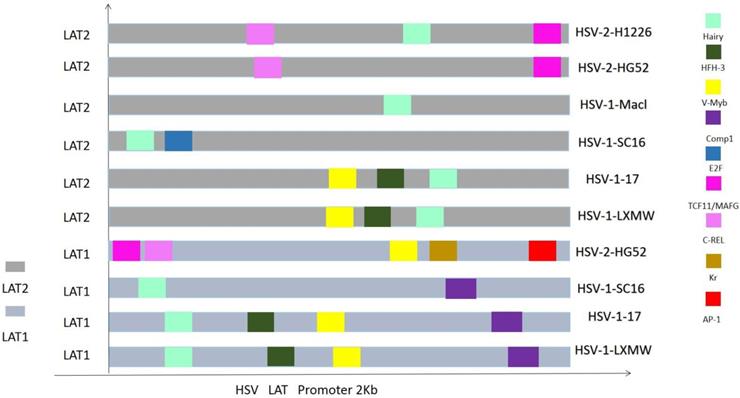
Discovery of TRSs and TFs of LAT is vital to understand the transcriptional regulation of LAT expression, HSV latency and oHSV engineering. Using the online program Matching, we found 9 potential TFs and their corresponding transcription regulatory sequences for the 6 strains of HSV LATs (Table 3). The nine transcription factors are Hairy, HFH-3, V-Myb, COMP1, E2F, TCF11/MafG, C-REL, Kr, AP-1.
The LAT TRSs and TFs are conserved or unique
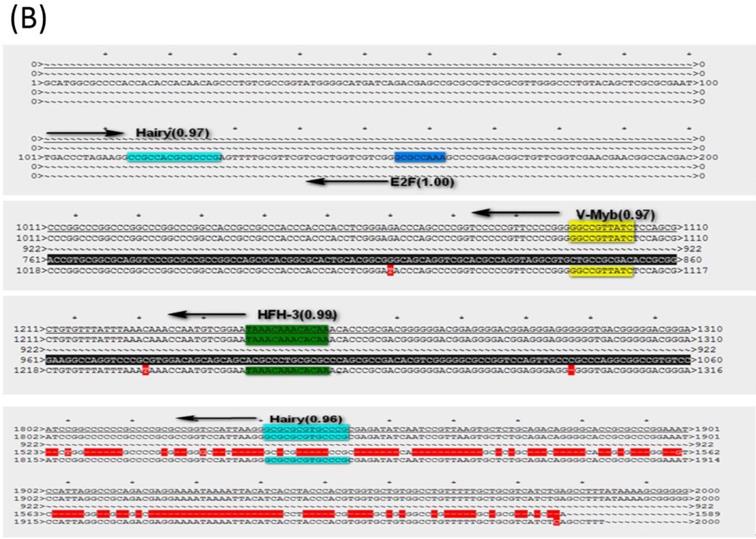
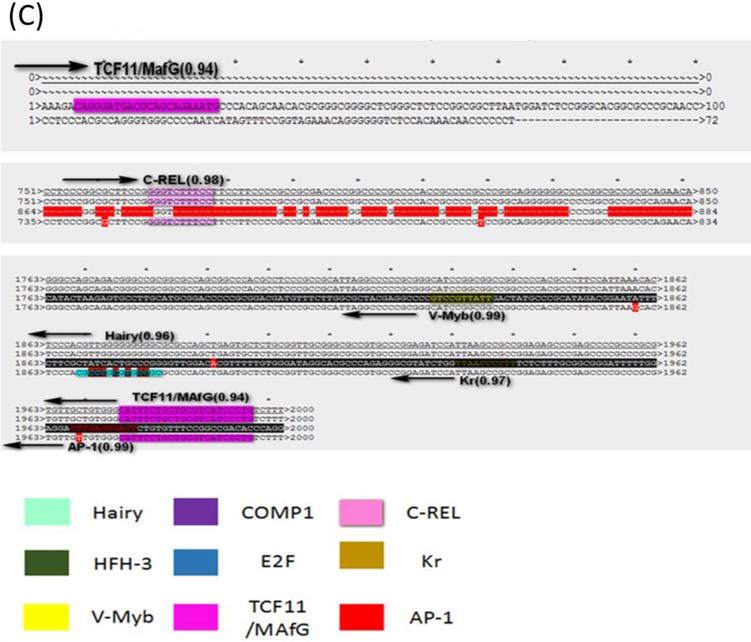
To know if the predicted LAT TRSs and TFs are conserved among HSVs and thus likely to be biologically functional, we did sequence alignment of the TRRs of LAT1 and LAT2 of HSV-1 and HSV-2 genomes (Figure 3 and 4). The identified transcription factors and their relative positions in the TRRs of LAT1 and LAT2 of HSV-1 and HSV-2 genomes are shown in Figure 3. Among all transcription factors, Hairy is shared for all LATs of HSV1 and HSV-2 except the strain HSV-2-HG52. Furthermore, HFH-3, V-Myb, Comp1, Kr and E2F are found only in HSV-1 strains; while C-REL, TCF11/MafG and AP-1 are found only in HSV-2 strains. In addition, Comp1, Kr and AP-1 are found only for LAT1; while E2F is found only for LAT2.
The sequences of the LAT TRRs of our new strain HSV-1-LXMW and other strains.
| HSV Strain | Gene (bp) | The direction of the promoter | transcriptional regulation sequence(2000bp) | The closest gene to LAT | Overlap sequence(bp) |
|---|---|---|---|---|---|
| HSV-1--LXMW | LAT1 | → | 7589-9589 | none | none |
| LAT2 | ← | 118797-116797 | none | none | |
| HSV-1-17 | LAT1 | → | 7569-9569 | UL1 (9338-10949) | 9338-10949 (Overlap 1611) |
| LAT2 | ← | 118777-116777 | UL56 ( 116197-116926) | 116926-116777 (Overlap 729) | |
| HSV-1-SC16 | LAT1 | → | 8251-10251 | UL1 (9989-11600) | 9989-10251 (Overlap 262) |
| LAT2 | ← | 118713-116713 | UL56 (116630-117359) | 116713-117359 (Overlap 646) | |
| HSV-1-Macl | LAT2 | → | 126929-128929 | RS1 (126951-131250) | 126951-128929 (Overlap 1978) |
| HSV-2-HG52 | LAT1 | → | 7814-9814 | UL1 (9463-11060) | 9463-9814 (Overlap 351) |
| LAT2 | ← | 119507-117507 | UL56 (117079-117817) | 117507-117817 (Overlap 310) | |
| HSV-2-H1226 | LAT2 | ← | 119356-117356 | UL56 (117000-117738) | 117738-117356 (Overlap 382) |
LAT transcription regulatory sequences and factors in HSV.
| HSV strain | Matrix identifier | Position on the genome | Core match | Matrix match | Sequence | Factor name | |
|---|---|---|---|---|---|---|---|
| HSV-1 strain LXMW | I$HAIRY_01 | 7759 | 1.000 | 0.967 | cgggCACGCgcgcc | Hairy | LAT1 |
| V$HFH3_01 | 8352 | 1.000 | 0.994 | ttgTGTTTgttta | HFH-3 | ||
| V$VMYB_01 | 8503 | 1.000 | 0.972 | gatAACGGcc | v-Myb | ||
| V$COMP1_01 | 9490 | 1.000 | 0.874 | gatcttGATTGgcgttacgagacc | COMP1 | ||
| V$VMYB_01 | 117689 | 1.000 | 0.972 | ggCCGTTatc | v-Myb | LAT2 | |
| V$HFH3_01 | 117541 | 1.000 | 0.994 | taaacAAACAcaa | HFH-3 | ||
| I$HAIRY_01 | 116948 | 1.00 | 0.967 | ggcgcGCGTGcccg | Hairy | ||
| HSV-1 strain 17 | I$HAIRY_01 | 7738 | 1.000 | 0.967 | cgggCACGCgcgcc | Hairy | LAT1 |
| V$HFH3_01 | 8338 | 1.000 | 0.994 | ttgTGTTTgttta | HFH-3 | ||
| V$VMYB_01 | 8489 | 1.000 | 0.972 | gatAACGGcc | v-Myb | ||
| V$COMP1_01 | 9489 | 1.000 | 0.874 | gatcttGATTGgcgttacgagacc | COMP1 | ||
| V$VMYB_01 | 117676 | 1.000 | 0.972 | ggCCGTTatc | v-Myb | LAT2 | |
| V$HFH3_01 | 117528 | 1.000 | 0.994 | taaacAAACAcaa | HFH-3 | ||
| I$HAIRY_01 | 116929 | 1.000 | 0.967 | ggcgcGCGTGcccg | Hairy | ||
| HSV-1 strain SC16 | I$HAIRY_01 | 8392 | 1.000 | 0.967 | cgggCACGCgcgcc | Hairy | LAT1 |
| V$COMP1_01 | 10140 | 1.000 | 0.866 | gatcttGATTGgcgctacgagacc | COMP1 | ||
| I$HAIRY_01 | 118827 | 1.000 | 0.973 | ccgcCACGCgcccg | Hairy | LAT2 | |
| V$E2F_02 | 118558 | 1.000 | 1.000 | gCGCCAaa | E2F | ||
| HSV-1 strain Macl | I$HAIRY_01 | 119524 | 1.000 | 0.972 | gccgCACGCggcct | Hairy | LAT2 |
| HSV-2 strain HG52 | V$TCF11MAFG_01 | 7820 | 1.000 | 0.948 | cagggATGACgcagcagaaatg | TCF11/MafG | LAT1 |
| V$CREL_01 | 6588 | 1.000 | 0.982 | GGAAAgaccc | c-Rel | ||
| V$VMYB_01 | 5988 | 1.000 | 0.990 | gtCCGTTatt | v-Myb | ||
| I$KR_01 | 5884 | 1.000 | 0.975 | ttAACCCctt | Kr | ||
| V$AP1_Q4 | 5847 | 1.000 | 0.990 | cttgAGTCAct | AP-1 | ||
| V$CREL_01 | 120274 | 1.000 | 0.982 | gggtcTTTCC | c-Rel | LAT2 | |
| V$TCF11MAFG_01 | 117532 | 1.000 | 0.948 | catttctgctgcGTCATccctg | TCF11/MafG | ||
| HSV-2 strain H1226 | V$CREL_01 | 120107 | 1.000 | 0.982 | gggtcTTTCC | c-Rel | LAT2 |
| I$HAIRY_01 | 117488 | 1.000 | 0.962 | cgcccGCGTGccgc | Hairy | ||
| V$TCF11MAFG_01 | 117381 | 1.000 | 0.948 | catttctgctgcGTCATccctg | TCF11/MafG |
Phylogenetic analysis of HSV-1-LXMW together with 5 other HSV strains. A. Phylogenetic trees take the number of substituents at each site as the length of their branches and are plotted to a certain scale. B. The taxon was presented using the bootup consistent tree method, and the evolutionary history of the evolutionary tree was analyzed using MEGA7.
The LAT TRRs and TFs in HSVs.
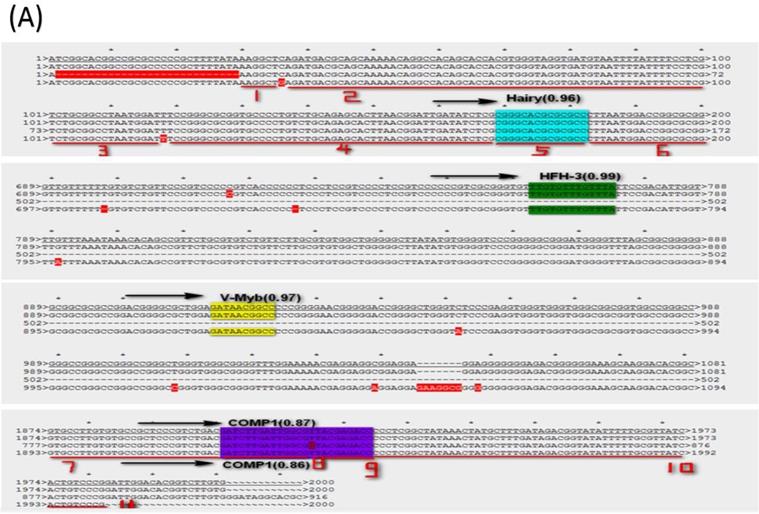
Because LAT TRRs of HSV-1 and HSV-2 are very diverse, and the two segments of LAT genes (LAT1 and LAT2) have different directions, we compared LAT TRRs in three groups (group a, b and c), using ApE Program. Group A: All HSV-1 LAT1; Group B: All HSV-1 LAT2; Group C: HSV-2 LAT1, LAT2. We found that the TRRs of our new strain HSV-1-LXMW are highly similar to the strain HSV-1-17. Our data also showed that the LAT TRRs are conserved within each group, and the TRSs identified above were mostly overlapped with the conserved sequences with certain exceptions (Figure 4).
The conservation of the identified TRSs and TFs suggests that these may have biological functions; meanwhile HSV-1 or 2 specific and LAT-1 or 2 specific sequences and factors may be related to their specific biological function. However, to validate these, further functional studies are needed.
The LAT transcription regulatory sequences and factors in HSVs. (A) HSV-1-LXMW LAT1 TRR (7589-9589, 2000bp) alignment to those of HSV-1-LXMW, HSV-1-17 and HSV-1-SC16. When aligned to HSV-1-LXMW LAT1 TRR, HSV-1-17 and HSV-1-SC16 have 1963 and 887 matched base pairs, 14 and 5 mismatched base pairs, and 48 and 1133 gaps, respectively. There were 11 conserved regions starting from the 30 bp, 37 bp, 101 bp, 119 bp, 169bp, 183 bp, 1893 bp, 1922bp, 1937bp, 1946 bp and 1993bp of the HSV-1-LXMW TRR, and four conserved TRSs binding to the four TFs of Hairy, HFH-3, V-Myb and comp1, respectively. (B) HSV-1-LXMW LAT1 TRR (116797-118797, 2000bp) alignment to those of HSV-1-LXMW, HSV-1-17, HSV-1-SC16 and Macl. When aligned to HSV-1-LXMW LAT1 TRR, HSV-1-17, HSV-1-SC16 and HSV-1-Macl have 1969, 424 and 96 matched base pairs, 14, 2 and 3 mismatched base pairs, and 36, 2071 and 162 gaps, respectively. There were four conserved TRSs binding to the four TFs of Hairy, E2F, v-myb, and HFH-3, respectively. (C) HSV-2-HG52 LAT2 TRR (117507-119507, 2000bp) alignment to those of HSV-1-HG52 LAT2, HSV-1-HG52 LAT1 and HSV-2-H1126 LAT2. When aligned to HSV-1-HG52 LAT2 TRR, HSV-1-HG52 LAT1 and HSV-2-H1126 LAT2 have 331 and 1881 matched base pairs, 19 and 13 mismatched base pairs, and 1003 and 214 gaps, respectively. There were six TRSs binding to the six TFs of TCF11/MAfG, C-REL, V-Myb, Hairy, Kr, AP-1, respectively.
The transcription factors v-myb and AP-1 family JunD are functional in regulating HSV gene transcription
| Tissue type | HSV | V-Myb | AP-1 | TF function | Ref |
|---|---|---|---|---|---|
| Brain: Human glioblastoma cells | HSV-1-F | B-Myb | --- | activate/γ34.5 | [42] |
| Urinary: Human PDAC-derived Capan-2 cells, BxPC-3, pancreatic | HSV-1-F | Myb | --- | Inhibit/ICP6 | [44] |
| Colon: Human HT29 carcinoma cells, | HSV-1-F | Myb | --- | activate/γ34.5 | [43] |
| The bone marrow: myelomonocytic hematopoietic cells, NIH 3T3 cells. | HSV-1- McKrae | v-myb | --- | activate/TK | [45] |
| Neuronal: neuroblastoma C1300 Cell | HSV-1- 17 | --- | AP-1 family JunD | activate/LAT1 | [48] |
| Genital epithelial cells HEC-1-A | HSV-2 | --- | not reported | activate/AP-1 | [52] |
The transcription factors v-myb and AP-1 family JunD are functional in regulating HSV gene transcription, including LATs transcription for HSV infection and latency
To validate if the identified TRSs and TFs are actually functional in HSV infected cells, we searched the literature that reported each of the 9 TFs in HSV infected cells, and found that the transcription factors v-myb and AP-1 are functional in regulating HSV gene transcription, including LATs transcription for HSV infection and latency (Table 4), and no report of the other 7 TFs in HSV infected cells.
In HSV-1, v-myb has been reported to activateγ34.5 [42, 43] and inhibit ICP6 [44] gene transcription. Interestingly, the gene encoding γ34.5 overlaps with the genes for LATs. Studies have also shown that v-myb activates the transcription of thymidine kinase (TK) gene [45]. TK is required for HSV DNA replication, lytic infection and reactivation from latent infection [46]. The absence of TK limits virus replication in non-dividing cells, such as ganglia neurons [47].
According to the literature, AP-1 family JunD can bind and activate the promoter of LAT1 [48]. HSV-1 LAT transcription regulatory cyclic-AMP (cAMP) response element (CRE)-like sequences, CRE-1 and CRE-2, were shown to regulate latency reactivation and support basal LAT expression in neuronal cells, respectively. Suggesting that the AP-1 family of transcription factors function in regulating CRE-dependent LAT1 transcription activation. The reaction of sensory ganglia to HSV infection is consistent with the neurobiological responses observed during nerve regeneration of peripheral processes following injury, including the alterations of neuropeptide production [49], neurite sprouting [50], and upregulation of AP-1 factors [51]. It is possible that these regenerative responses are linked to signaling pathways that regulate LAT transcription activity. In addition, HSV-2 can induce AP-1 transcriptional activation through the HSV replication affecting TLR4-MyD88/TRIF pathway in human genital epithelial cell [52].
However, these predicted transcription factors above have not been reported to regulate LAT gene transcription, which requires further study in the future.
Discussion
Recent studies have found that high expression of HSV LAT in both the human trigeminal nerve and the eye is associated with latent infection of HSV [53]. Deletion of the LAT gene can significantly reduce the latent infection rate and the activation of the virus [54]. Studies have shown that HSV-1 recombinants with deletions in the LAT promoter and portions of the 5' exon coding region lead an increase in apoptotic neurons during the acute infection [55], resulting in a 2-3 fold decrease in total HSV-1 DNA detected in the ganglia during latency in the rabbit eye [56]. The deletion of both LAP2 and LAP1 eliminated the ability to detect LAT in acute and latent infections. The deletion of LAP2 alone diminished levels of LAT expression in both acute and latent infections [57]. Thus, sequences important for wild-type-level LAT transcription during productive infections and during latency appear to reside within LAP2. To make LAP2 deletions, most of the sequences between the 5' end of the primary LAT and the 5' end of the LAT intron (positions 4081 to 4382) were deleted [58].
Interestingly, a consistent effect of the LAP2 deletion on LAT expression was observed in every experiment, including those performed during the productive infection of Vero and human neuroblastoma cells and with the latently infected ganglia [57]. More recently, studies have shown that HF10 (an oHSV with natural deletions: the UL56/IRL junction has been deleted from 116.515 bp to 120.346 bp, leading to the lack of expression of UL56 and LATs [59]. It is worth noting that the deleted sequence in HF10 overlaps with the LAT2 regulatory sequence we studied (116.797 bp to 118.797 bp) in our strain HSV-1-LXMW. This suggests that the HSV LAT sequences we studied is of significance for HSV LAT transcription and oHSVs. Actually clinical studies on HF10 have been completed in breast, head and neck, and pancreatic cancers in Japan [59, 60]. Meanwhile, in the United States, HF10 has completed phase I clinical trials for refractory superficial cancers and melanoma [59]. Importantly, HF10 has been shown to have a high safety profile and low adverse reactions in all patients treated [59]. HF10 viral replication and cytotoxicity has also been studied in human and mouse melanoma cell lines [59]. Our newly discovered transcription factors and their corresponding transcription regulatory sequences may be used to effectively regulate the expression of LAT, to control HSV latency for developing safer oHSVs in the future. In this article, for the first time, we did systematic comparative sequence analysis of LATs and their TRRs from 6 different HSV-1 and HSV-2 strains. Our important findings include the following.
Our recently isolated new strain HSV-1-LXMW was found to be closely related to HSV-1-17 from MRC University, Glasgow, UK and SC16 from the Spain, which is different from the ICP27-based results [13], and different from other methods using PCR [61], FISH [62], ISH [63], Northern blot analysis [64]. The chimeric virus HSV-1-17/LAT2, which is an HSV-1 virus engineered to express HSV-2 LAT (complementary to HSV-2 333/LAT1) preferentially establishes latent infections in KH10-positive neurons, as does wild-type HSV-2 [65]. Therefore, it is likely that our newly isolated HSV-1-LXMW strains can be used to establish a latent model and build a platform for further study on the virus latency mechanism since it's highly homologous to HSV-1-17.
We identified novel conservative HSV LAT transcription regulatory sequences and 9 potential LAT transcription factors of Hairy, HFH-3, v-Myb, Comp1, E2F, TCF11/MAFG, C-REL, Kr and AP-1. Previous studies analyzed fewer HSV strains and shorter DNA sequences (2000 vs. 900 nt) [57] [66] [67] and reported only two LAT transcription factors v-myb [42-45] and AP-1[52, 68]. For the first time, we discovered seven novel transcription factors and their corresponding transcription regulatory sequences of HSV LATs. Our findings may have significant implication in our understanding of HSV latency and engineering of better oncolytic HSVs. Among them v-Myb and AP-1 were reported to be functional in HSV infected cells, while the other 7 were reported here for the first time. Their functions in HSV1/2 tissue tropism and latency need to be further explored. We summarized the HSV-1/2 tissue tropism and the TFs expression in a variety of tissues (Table 5). The table shows in detail that most of our 7 transcription factors are moderately expressed in HSV-1 trigeminal ganglion and moderately or highly expressed in HSV-2 reproductive system. We propose that the variable expression of the transcription factors may contribute to the LAT transcription in different tissues, which may be related to the HSV latency in the proper cells.
Hairy is a developmental suppressor gene and a crucial part of normal body development and nervous system development [69]. HFH-3 is a cell-type-specific transcription factor [70]. v-Myb with DNA-binding, activation and repression domains, interacts with the CAAT-enhancer binding protein (C/EBP) family, and regulates proliferation and differentiation of hematopoietic cells [71]. The cellular transcription factor E2F, initially identified as a target for transactivation by adenovirus ElA [72, 73], is found in all kind of cell types. TCF11 and the Maf proteins -F, -G and -K/p18 are widely expressed, and interact with each other in numerous cell types [74]. C-Rel is a protooncogene product that may be involved in the transcriptional regulation of the family of genes that contain this ISRE [75]. AP-1 is a big family that includes Jun and Fos [76]. The results indicate that at least one neuronal AP-1 family member, JunD, is involved in a major fraction of the LAP1 CRE binding activity [77]. JunD has been observed to be induced by treatment of myeloid cell lines with a cAMP analog [48]. AP-1 activates caspase 8 transcription, which mediates palmitic acid induced apoptosis of human cardiac myocytes, leading to diabetic cardiomyopathy [78]. Because partial core promoter region was not included in our analysis, transcription factor AP-1 binding site was not detected in HSV-1 strains. However AP-1 family JunD was reported to bind to the CRE elements in the core promoter region.
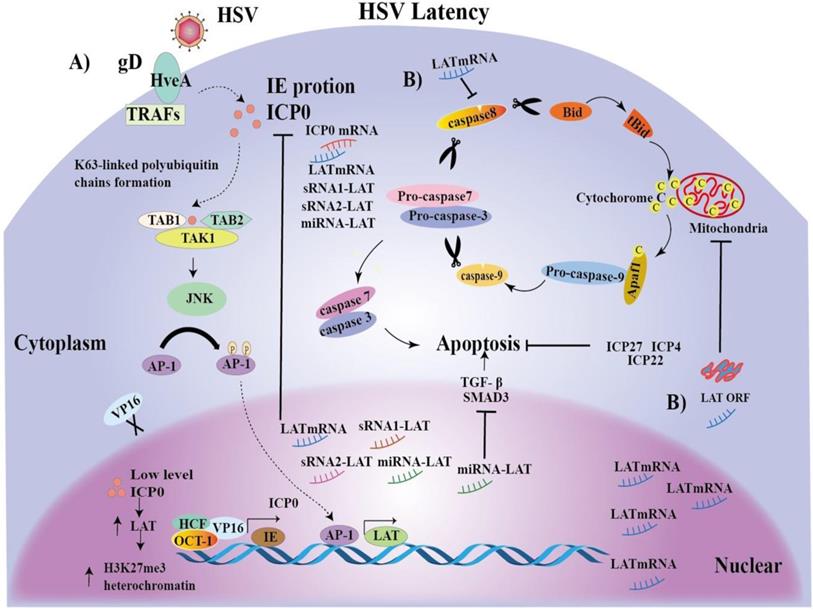
In a word, AP-1 may promote LAT transcriptional expression and latency. The mechanisms of LAT-mediated latent infection have been proposed as many hypotheses, but it is still unclear. To better understand the significance of our identification of the novel LAT TRSs and TFs, we integrated reported hypothesis and our novel findings, and proposed possible mechanisms for the initiation and maintenance of HSV latent infection (Figure 5).
The HSV-1/2 tissue tropism and the TF expression from all sorts of tissues.
| System | Cell/ tissue | HSV1 | HSV2 | Hairy | HFH-3 | v-Myb | E2F | TCF11/MAfG | C-REL | AP-1 |
|---|---|---|---|---|---|---|---|---|---|---|
| Blood system | CD34 + stem cell | + | _ | N | M | M | H | L | H | M |
| 721 B lymphoblasts | + | _ | N | L | M | M | M | H | M | |
| CD19 + B cell | + | _ | H | L | M | H | L | H | H | |
| Leukemia lymphoblastic | + | _ | M | L | H | M | L | M | L | |
| Bonemarrow | + | _ | M | M | M | L | L | H | L | |
| Pituitariy | + | _ | H | M | H | L | M | H | L | |
| Head | Prefrontal Cortex | + | _ | H | L | M | M | M | H | H |
| Pineal | + | _ | H | L | H | H | H | H | M | |
| Tongue | + | _ | M | M | H | L | M | H | L | |
| Tonsil | + | _ | M | L | M | M | L | H | L | |
| Retina | + | _ | H | M | H | M | H | H | M | |
| Trigeminal ganglion | + | _ | L | L | M | L | M | M | M | |
| Cerebellum | + | _ | M | M | M | L | L | H | L | |
| viscera | Heart | + | _ | H | M | M | L | M | H | M |
| Lung | + | _ | H | M | M | L | M | H | H | |
| Liver | + | _ | H | M | M | M | L | H | M | |
| Kidney | + | _ | H | L | M | L | M | M | L | |
| Smooth Muscles | + | _ | H | L | M | L | M | H | L | |
| Adipocyte | + | _ | H | M | M | L | M | H | L | |
| Secretory system | Adrenalgland | + | _ | H | L | M | L | M | M | M |
| Pancreaticlstet | + | _ | H | M | M | M | L | H | M | |
| Genital system | Placenta | + | + | H | L | H | M | M | H | L |
| Fetalthyroid | + | + | H | M | M | L | L | H | L | |
| Uterus | + | + | M | L | M | M | M | M | M | |
| Testis | + | + | M | M | M | M | M | M | L |
Hold increase to median fluorescence intensity on Affymetrix microarray chips from http://biogps.org: 0-2.5 (L), >2.5-<5 (M), >5 (H)
Mechanisms of the initiation and maintenance of HSV latent infection. A) The HSV envelope glycoprotein gD binds to the TNFR member HSV entry mediator A (HveA), whose cytoplasmic region binds to TRAFs [79]. Upon HSV entry, multiprotein complex VP16, Oct-1 and HCF initiate IE gene transcription, including ICP0, promoting lytic infection [80]. In turn, ICP0, together with Ubc13ev1A, catalyzes the K63-linked polyubiquitin chains, and recognizes and activates TAK1 [80]. Activated TAK1 then phosphorylates MKK6, leading to the activation of JNK kinase [80]. Activated TAK1 then phosphorylates MKK6, leading to the activation of JNK kinase [80]. Ap-1 transcription factors are phosphorylated by JNK and traffic into the nucleus to bind to the LAT transcription regulatory sequences, and, promote LAT transcription. LAT increases accumulation of H3K27me3 heterochromatin, leading to gene silencing and latency initiation [17]. Then, a large number of LAT transcripts, such as mRNA, miRNA-LAT and sRNA1-LAT or sRNA2-LAT, inhibit ICP0 gene expression by partial base complementation to maintain latency. ICP0 can also be reduced by VP16 loss. B) On the other hand, LAT expression inhibits caspase 8 and LAT ORF inhibits caspase 9, promoting cell survival [81]. Interestingly, a miRNA-LAT generated from the exon 1 region of LATs was found to exert an anti-apoptotic effect through targeting transforming growth factor beta (TGF-β) and SMAD3 expression. Finally, the regulatory proteins ICP4, ICP22 and ICP27 may indirectly inhibit apoptosis by promoting the production of anti-apoptotic viral products.
Abbreviations
HSV: Herpes simplex viruses; oHSV: oncolytic herpes simplex virus; LATs: Latency-Associated Transcripts; TRRs: transcription regulatory regions; TRSs: Transcription Regulatory Sequences; TFs: Transcription Factors; TFBS: transcription factor binding sites; LAP1: LAT promoter 1; LAP2: LAT promoter 2; UL: Unique Long; US: Unique Short; TRL: Terminal Repeat Long; IRL: Internal Repeat Long; IRS: Internal Repeat Short; TRS: Terminal Repeat Short; IE: immediate-early; HSV-1-LXMW: strain LXMW of type 1 HSV; ICP: Infected Cell Polypeptide; UL1: envelope glycoprotein L; TK: thymidine kinase; HveA: HSV entry mediator A; JNK: c-Jun N-terminal kinase; AP-1: activator protein 1; TAK1: transforming growth factor-h-activated protein kinase 1; TRAFs: TNF receptor-associated factors; TGF-β: transforming growth factor beta.
Supplementary Material
Supplementary results.
Acknowledgements
This work was partly supported by grants from the National Natural Science Foundation of China (81872412 to XHW, 81602303 to XY, 31700736 to WXW). We thank Hubei Province Natural Science Foundation of China (2016CFB180 to WXW), Foundation of Health and Family Planning Commission of Hubei Province (WJ2016-Y-02 to MZ, WJ2016Y07 to WXW), Hubei Province Scientific and Technological Research Project (Q20171306 to XWW), Jingzhou Science and Technology Development Planning Project (JZKJ15063 to WXW) and Yangtze University Fellowship to graduate student ZY.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Xin HW, Ambe CM, Hari DM, Wiegand GW, Miller TC, Chen JQ. et al. Label-retaining liver cancer cells are relatively resistant to sorafenib. Gut. 2013;62:1777-86
2. Xin HW, Ambe CM, Miller TC, Chen JQ, Wiegand GW, Anderson AJ. et al. Liver Label Retaining Cancer Cells Are Relatively Resistant to the Reported Anti-Cancer Stem Cell Drug Metformin. Journal of Cancer. 2016;7:1142-51
3. Xin HW, Ambe CM, Ray S, Kim BK, Koizumi T, Wiegand GW. et al. Wnt and the cancer niche: paracrine interactions with gastrointestinal cancer cells undergoing asymmetric cell division. Journal of Cancer. 2013;4:447-57
4. Xin HW, Hari DM, Mullinax JE, Ambe CM, Koizumi T, Ray S. et al. Tumor-Initiating Label-Retaining Cancer Cells in Human Gastrointestinal Cancers Undergo Asymmetric Cell Division. Stem Cells. 2012;30:591-8
5. Han MS, Choi EH, Lee HJ, Yun KW, Kang HJ, Hong KT. et al. Cytomegalovirus disease in a retinoblastoma cohort: The role of preemptive screening. Pediatric blood & cancer. 2019 e28101
6. Wang D, Wang X-W, Peng X-C, Xiang Y, Song S-B, Wang Y-Y. et al. CRISPR/Cas9 genome editing technology significantly accelerated herpes simplex virus research. Cancer Gene Therapy. 2018;25:93-105
7. Wen Z, Keli G, Qian Z, Xiufen Z, Zhenling D, Lingling L. et al. A novel oHSV-1 targeting telomerase reverse transcriptase-positive cancer cells via tumor-specific promoters regulating the expression of ICP4. Oncotarget. 2015;6:20345-55
8. Wu ZJ, Tang FR, Ma ZW, Peng XC, Xiang Y, Zhang Y. et al. Oncolytic Viruses for Tumor Precision Imaging and Radiotherapy. Human gene therapy. 2018;29:204-22
9. Watanabe D, Goshima F. Oncolytic Virotherapy by HSV. Advances in experimental medicine and biology. 2018;1045:63-84
10. Russell SJ, Peng KW, Bell JC. Oncolytic virotherapy. Nature biotechnology. 2012;30:658-70
11. Sanchala DS, Bhatt LK, Prabhavalkar KS. Oncolytic Herpes Simplex Viral Therapy: A Stride toward Selective Targeting of Cancer Cells. Frontiers in Pharmacology. 2017;8:270
12. Luo Y, Lin C, Ren W, Ju F, Xu Z, Liu H. et al. Intravenous Injections of a Rationally Selected Oncolytic Herpes Virus as a Potent Virotherapy for Hepatocellular Carcinoma. Molecular therapy oncolytics. 2019;15:153-65
13. Wang YY, Lyu YN, Xin HY, Cheng JT, Liu XQ, Wang XW. et al. Identification of Putative UL54 (ICP27) Transcription Regulatory Sequences Binding to Oct-1, v-Myb, Pax-6 and Hairy in Herpes Simplex Viruses. Journal of Cancer. 2019;10:430-40
14. Zhang T, Suryawanshi YR, Woyczesczyk HM, Essani K. Targeting Melanoma with Cancer-Killing Viruses. The open virology journal. 2017;11:28-47
15. Dai X, Zhou ZH. Structure of the herpes simplex virus 1 capsid with associated tegument protein complexes. Science. 2018;360:7298-23
16. Fujie X, Sternberg MR, Kottiri BJ, Mcquillan GM, Lee FK, Nahmias AJ. et al. Trends in herpes simplex virus type 1 and type 2 seroprevalence in the United States. Jama. 2006;296:964-73
17. Raja P, Lee JS, Pan D, Pesola JM, Coen DM, Knipe DM. A Herpesviral Lytic Protein Regulates the Structure of Latent Viral Chromatin. mBio. 2016;7:e00633-16
18. Lou W, Ji F, Fu J, Han Z, Di W, Zhang N. RETRACTED ARTICLE: Transcriptional retargeting of herpes simplex virus for cell-specific replication to control cancer. Journal of Cancer Research & Clinical Oncology. 2018;144:1-11
19. Guo Q, Weng W, Zeng F, Zheng R, Luo Y, Du J. Research of UL54-specific siRNA on herpes simplex virus type II replication. International Journal of Dermatology. 2011;50:362-6
20. Lou W, Ji F, Fu J, Han Z, Di W, Zhang N. Transcriptional retargeting of herpes simplex virus for cell-specific replication to control cancer. Journal of Cancer Research & Clinical Oncology. 2018;144:1-11
21. Everett RD. A detailed analysis of an HSV-1 early promoter: sequences involved in trans-activation by viral immediate-early gene products are not early-gene specific. Nucleic acids research. 1984;12:3037-56
22. Pereira RA, Simmons A. Cell surface expression of H2 antigens on primary sensory neurons in response to acute but not latent herpes simplex virus infection in vivo. Journal of virology. 1999;73:6484
23. Stevens JG. Human herpesviruses: a consideration of the latent state. Microbiological reviews. 1989;53:318-32
24. Sawtell NM. Comprehensive quantification of herpes simplex virus latency at the single-cell level. Journal of virology. 1997;71:5423-31
25. Margolis TP, Yumi I, Li Y, Vicky V, Krause PR. Herpes simplex virus type 2 (HSV-2) establishes latent infection in a different population of ganglionic neurons than HSV-1: role of latency-associated transcripts. Journal of virology. 2007;81:1872
26. Garber DA, Schaffer PA, Knipe DM. A LAT-associated function reduces productive-cycle gene expression during acute infection of murine sensory neurons with herpes simplex virus type 1. Journal of virology. 1997;71:5885-93
27. Mador N, Goldenberg D, Cohen O, Panet A, Steiner I. Herpes Simplex Virus Type 1 Latency-Associated Transcripts Suppress Viral Replication and Reduce Immediate-Early Gene mRNA Levels in a Neuronal Cell Line. Journal of virology. 1998;72:5067
28. Inman M, Perng GC, Henderson G, Ghiasi H, Nesburn AB, Wechsler SL. et al. Region of herpes simplex virus type 1 latency-associated transcript sufficient for wild-type spontaneous reactivation promotes cell survival in tissue culture. Journal of virology. 2001;75:3636-46
29. Perng GC, Jones C, Ciacci-Zanella J, Stone M, Henderson G, Yukht A. et al. Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript. Science. 2000;287:1500-3
30. Farrell MJ, Dobson AT, Feldman LT. Herpes simplex virus latency-associated transcript is a stable intron. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:790-4
31. Spivack JG, Fraser NW. Detection of herpes simplex virus type 1 transcripts during latent infection in mice. Journal of virology. 1987;61:3841
32. Zabolotny JM, Krummenacher C, Fraser NW. The herpes simplex virus type 1 2.0-kilobase latency-associated transcript is a stable intron which branches at a guanosine. Journal of virology. 1997;71:4199-208
33. Henderson G, Jaber T, Carpenter D, Wechsler SL, Jones C. Identification of herpes simplex virus type 1 proteins encoded within the first 1.5 kb of the latency-associated transcript. Journal of neurovirology. 2009;15:439
34. Ahmed M, Lock M, Miller CG, Fraser NW. Regions of the herpes simplex virus type 1 latency-associated transcript that protect cells from apoptosis in vitro and protect neuronal cells in vivo. Journal of virology. 2002;76:717-29
35. Chen X, Schmidt MC, Goins WF, Glorioso JC. Two herpes simplex virus type 1 latency-active promoters differ in their contributions to latency-associated transcript expression during lytic and latent infections. Journal of virology. 1995;69:7899-908
36. Lokensgard JR, Berthomme H, Feldman LT. The latency-associated promoter of herpes simplex virus type 1 requires a region downstream of the transcription start site for long-term expression during latency. Journal of virology. 1997;71:6714-9
37. Rajčáni J, Andrea V, Ingeborg R. Peculiarities of Herpes Simplex Virus (HSV) Transcription: An overview. Virus Genes. 2004;28:293
38. Leib DA, Nadeau KC, Rundle SA, Schaffer PA. The promoter of the latency-associated transcripts of herpes simplex virus type 1 contains a functional cAMP-response element: role of the latency-associated transcripts and cAMP in reactivation of viral latency. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:48-52
39. Kenny JJ, Krebs FC, Hartle HT, Gartner AE, Chatton B, Leiden JM. et al. Identification of a Second ATF/CREB-like Element in the Herpes Simplex Virus Type 1 (HSV-1) Latency-Associated Transcript (LAT) Promoter. Virology. 1994;200:220-35
40. Sumit B, Wong AC, Steitz JA. Drosophila hnRNP A1 homologs Hrp36/Hrp38 enhance U2-type versus U12-type splicing to regulate alternative splicing of the prospero twintron. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2577-82
41. Copertino DW, Hallick RB. Group II twintron: an intron within an intron in a chloroplast cytochrome b-559 gene. Embo Journal. 1991;10:433-42
42. Chung RY, Saeki SY, Chiocca EA. B-myb promoter retargeting of herpes simplex virus gamma34.5 gene-mediated virulence toward tumor and cycling cells. Journal of Virology. 1999;73:7556-64
43. Nakamura H. et al. Regulation of herpes simplex virus gamma(1)34.5 expression and oncolysis of diffuse liver metastases by Myb34.5. J Clin Invest. 2002;109:871-82
44. Gayral M, Lulka H, Hanoun N, Biollay C, Selves J, Vignolle-Vidoni A. et al. Targeted oncolytic herpes simplex virus type 1 eradicates experimental pancreatic tumors. Human gene therapy. 2015;26:104-13
45. Bortner DM, Ostrowski MC. Analysis of the v-myb structural components important for transactivation of gene expression. Nucleic acids research. 1991;19:1533-9
46. Jacobson JG, Ruffner KL, Koszvnenchak M, Hwang CB, Wobbe KK, Knipe DM. et al. Herpes simplex virus thymidine kinase and specific stages of latency in murine trigeminal ganglia. Journal of virology. 1993;67:6903-8
47. Leist TP, Sandri-Goldin RM, Stevens JG. Latent infections in spinal ganglia with thymidine kinase-deficient herpes simplex virus. Journal of virology. 1989;63:4976
48. Millhouse S, Kenny JJ, Quinn PG. et al. ATF/CREB Elements in the Herpes Simplex Virus Type 1 Latency-Associated Transcript Promoter Interact with Members of the ATF/CREB and AP-1 Transcription Factor Families. Journal of Biomedical Science. 1998;5:451-64
49. Henken DB, Martin JR. The proportion of galanin-immunoreactive neurons in mouse trigeminal ganglia is transiently increased following corneal inoculation of herpes simplex virus type-1. Neuroscience Letters. 1992;140:177-80
50. Martin RE, Henken DB, Hill JM. Altered expression and changing distribution of the nerve growth associated protein GAP-43 during ocular HSV-1 infection in the rabbit. Journal of neurovirology. 1996;2:127-35
51. Valyi-Nagy T DSL, Dillner A. et al. Induction of cellular transcription factors in trigeminal ganglia of mice by corneal scarification, herpes simplex virus type 1 infection, and explantation of trigeminal ganglia[J]. Journal of Virology. 1991;65:4142-52
52. Lv X, Wang H, Su A, Xu S, Chu Y. Herpes simplex virus type 2 infection triggers AP-1 transcription activity through TLR4 signaling in genital epithelial cells. Virology journal. 2018;15:173
53. Stevens JG, Haarr L, Porter DD, Cook ML, Wagner EK. Prominence of the herpes simplex virus latency-associated transcript in trigeminal ganglia from seropositive humans. The Journal of infectious diseases. 1988;158:117-23
54. Lee JS, Raja P, Pan D, Pesola JM, Coen DM, Knipe DM. et al. CCCTC-Binding Factor Acts as a Heterochromatin Barrier on Herpes Simplex Viral Latent Chromatin and Contributes to Poised Latent Infection. Mbio. 2018;9:e02372-17
55. Henderson G, Peng W, Jin L, Perng G-C, Nesburn A, Wechsler S. et al. Regulation of Caspase 8- and Caspase 9-Induced Apoptosis by the Herpes Simplex Virus Type 1 Latency-Associated Transcript. Journal of neurovirology. 2002;8:103-11
56. Watson ZL, Washington SD, Phelan DM, Lewin AS, Tuli SS, Schultz GS. et al. In Vivo Knockdown of the Herpes Simplex Virus 1 Latency-Associated Transcript Reduces Reactivation from Latency. Journal of virology. 2018;92:e00812-18
57. Yoshikawa T, Stanberry LR, Bourne N, Krause PR. Downstream regulatory elements increase acute and latent herpes simplex virus type 2 latency-associated transcript expression but do not influence recurrence phenotype or establishment of latency. Journal of virology. 1996;70:1535-41
58. Krause PR, Stanberry LR, Bourne N. et al. Expression of the herpes simplex virus type 2 latency-associated transcript enhances spontaneous reactivation of genital herpes in latently infected guinea pigs. J Exp Med. 1995;181:297-306
59. Eissa IR, Naoe Y, Bustos-Villalobos I, Ichinose T, Tanaka M, Zhiwen W. et al. Genomic Signature of the Natural Oncolytic Herpes Simplex Virus HF10 and Its Therapeutic Role in Preclinical and Clinical Trials. Frontiers in oncology. 2017;7:149
60. Hirooka Y, Kasuya H, Ishikawa T, Kawashima H, Ohno E, Villalobos IB. et al. A Phase I clinical trial of EUS-guided intratumoral injection of the oncolytic virus, HF10 for unresectable locally advanced pancreatic cancer. BMC cancer. 2018;18:596
61. Nicoll MP, Efstathiou S. Expression of the herpes simplex virus type 1 latency-associated transcripts does not influence latency establishment of virus mutants deficient for neuronal replication. The Journal of general virology. 2013;94:2489-94
62. TG E, DC B. In VitroLund Human Mesencephalic (LUHMES) Neuronal Cell Line Supports Herpes Simplex Virus 1 Latency. Journal of virology. 2019;93:e02210-18
63. XP C, M M, M K, JC G, DJ F. The relationship of herpes simplex virus latency associated transcript expression to genome copy number: a quantitative study using laser capture microdissection. Journal of neurovirology. 2002;8:204-10
64. AS B, A P, Y I, K A, TP M, PR K. Latency-associated transcript (LAT) exon 1 controls herpes simplex virus species-specific phenotypes: reactivation in the guinea pig genital model and neuron subtype-specific latent expression of LAT. Journal of virology. 2009;83:10007-15
65. Imai Y, Apakupakul K, Krause PR, Halford WP, Margolis TP. Investigation of the mechanism by which herpes simplex virus type 1 LAT sequences modulate preferential establishment of latent infection in mouse trigeminal ganglia. Journal of virology. 2009;83:7873-82
66. Diao L, Zhang B, Xuan C, Sun S, Yang K, Tang Y. et al. Activation of c-Jun N-terminal kinase (JNK) pathway by HSV-1 immediate early protein ICP0. Experimental cell research. 2005;308:196-210
67. DJ M, C C, G M, A D. Comparative sequence analysis of the long repeat regions and adjoining parts of the long unique regions in the genomes of herpes simplex viruses types 1 and 2. The Journal of general virology. 1991;72:3057-75
68. Millhouse S, Kenny JJ, Quinn PG. et al. ATF/CREB Elements in the Herpes Simplex Virus Type 1 Latency-Associated Transcript Promoter Interact with Members of the ATF/CREB and AP-1 Transcription Factor Families. Journal of Biomedical Science. 1998;5:451-64
69. Bianchi-Frias D, Orian A, Delrow JJ, Vazquez J, Rosales-Nieves AE, Parkhurst SM. Hairy Transcriptional Repression Targets and Cofactor Recruitment in Drosophila. Plos Biology. 2004;2:E178
70. Clevidence DE, Overdier DG, Tao W, Qian X, Pani L, Lai E. et al. Identification of nine tissue-specific transcription factors of the hepatocyte nuclear factor 3/forkhead DNA-binding-domain family. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:3948-52
71. Tahirov T H SK, Ichikawa-Iwata E. et al. Mechanism of c-Myb-C/EBPβ Cooperation from Separated Sites on a Promoter[J]. Cell. 2002;108:57-70
72. Yee AS, Raychaudhuri P, Jakoi L, Nevins JR. The adenovirus-inducible factor E2F stimulates transcription after specific DNA binding. Molecular and cellular biology. 1989;9:578-85
73. Kovesdi I, Reichel R, Nevins JR. Identification of a cellular transcription factor involved in E1A trans-activation. Cell. 1986;45:219-28
74. Johnsen O, Murphy P, Prydz H, Kolsto AB. Interaction of the CNC-bZIP factor TCF11/LCR-F1/Nrf1 with MafG: binding-site selection and regulation of transcription. Nucleic acids research. 1998;26:512-20
75. Sica A, Tan TH, Rice N. et al. The c-rel protooncogene product c-Rel but not NF-kappa B binds to the intronic region of the human interferon-gamma gene at a site related to an interferon-stimulable response element. Proceedings of the National Academy of Sciences. 1992;89:1740-4
76. Sassone-Corsi P, Ransone LJ, Verma IM. Cross-talk in signal transduction: TPA-inducible factor jun/AP-1 activates cAMP-responsive enhancer elements. Oncogene. 1990;5:427-31
77. Millhouse S, Kenny JJ, Quinn PG. et al. ATF/CREB Elements in the Herpes Simplex Virus Type 1 Latency-Associated Transcript Promoter Interact with Members of the ATF/CREB and AP-1 Transcription Factor Families. Journal of Biomedical Science. 1998;5:451-64
78. Liedtke C, Lambertz D, Schnepel N, Trautwein C. Molecular mechanism of Mitomycin C-dependent caspase-8 regulation: implications for apoptosis and synergism with interferon-alpha signalling. Apoptosis: an international journal on programmed cell death. 2007;12:2259-70
79. Liu X-Q, Xin H-Y, Lyu Y-N, Ma Z-W, Peng X-C, Xiang Y. et al. Oncolytic herpes simplex virus tumor targeting and neutralization escape by engineering viral envelope glycoproteins. Drug Delivery. 2018;25:1950-62
80. Diao L, Zhang B, Xuan C, Sun S, Yang K, Tang Y. et al. Activation of c-Jun N-terminal kinase (JNK) pathway by HSV-1 immediate early protein ICP0. Experimental Cell Research. 2005;308:196-210
81. Mott KR, Osorio N, Jin L, Brick DJ, Naito J, Cooper J. et al. The bovine herpesvirus-1 LR ORF2 is critical for this gene's ability to restore the high wild-type reactivation phenotype to a herpes simplex virus-1 LAT null mutant. The Journal of general virology. 2003;84:2975-85
Author contact
![]() Corresponding authors: Ying Xiang, PhD, MD, Center for Molecular Medicine, School of Medicine, Yangtze University, 1 Nanhuan Road, Jingzhou, Hubei 434023, China. Phone: +86 15107212530; Fax: 0716-8062633; Email: xying316com. Xiu-Lan Su, MD, Professor and Director, Clinical Medical Research Center, Affiliated Hospital of Inner Mongolia Medical University, Hohhot 010050, China. Phone: 13904710692; Email: xlsucom. Hong-Wu Xin, PhD, MD, Professor and Chief, Laboratory of Oncology, Center for Molecular Medicine, School of Basic Medicine, Health Science Center, Yangtze University, 1 Nanhuan Road, Jingzhou, Hubei 434023, China. Phone: +86 13311055391; Fax: 0716-8062633; Email: hongwu_xincom.
Corresponding authors: Ying Xiang, PhD, MD, Center for Molecular Medicine, School of Medicine, Yangtze University, 1 Nanhuan Road, Jingzhou, Hubei 434023, China. Phone: +86 15107212530; Fax: 0716-8062633; Email: xying316com. Xiu-Lan Su, MD, Professor and Director, Clinical Medical Research Center, Affiliated Hospital of Inner Mongolia Medical University, Hohhot 010050, China. Phone: 13904710692; Email: xlsucom. Hong-Wu Xin, PhD, MD, Professor and Chief, Laboratory of Oncology, Center for Molecular Medicine, School of Basic Medicine, Health Science Center, Yangtze University, 1 Nanhuan Road, Jingzhou, Hubei 434023, China. Phone: +86 13311055391; Fax: 0716-8062633; Email: hongwu_xincom.

 Global reach, higher impact
Global reach, higher impact