Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(16):4652-4661. doi:10.7150/jca.42669 This issue Cite
Review
CYP1B1 gene: Implications in glaucoma and cancer
1. Biochemisty, King Saud University, Saudi Arabia.
2. King Abdullah International Medical Research Center (KAIMRC)/ King Saud Bin Abdulaziz University for Health Sciences (KSAU-HS), MNGHA, Saudi Arabia.
Received 2019-12-2; Accepted 2020-3-6; Published 2020-5-19
Abstract
Glaucoma is a serious disease that can lead to irreversible loss of vision. Patients with primary congenital glaucoma may have elevated intraocular pressure. Hypertension causes damages to intraocular structures and affects the Schlemm's canal, collector channels, trabecular meshwork, and optic nerve's molecular structures. An important gene that is defective in patients with glaucoma is CYP1B1, a gene associated with optic nerve deterioration. CYP1B1is a key enzyme involved in the metabolism of exogenous and endogenous compounds. Also, it is critical in the detoxification of pre-carcinogens, such as polycyclic aromatic hydrocarbons and estrogen. It catalyzes their conversion into metabolites subsequently eliminated from the body. In malignant tumors, the CYP1B1 promoter is hypomethylated. CYP1B1 overexpression results in the conversion of estrogens to quinone forms, which bind with DNA and create a predisposition for cancer in several organs, such as the brain, breast, and ovary. Increased cytokine interleukin-6 and leptin lead to elevated CYP1B1 activity, which possibly causes cancer. In addition, the expression of aromatic hydrocarbon receptors is increased in tumor tissues, and it elevates oxidative stress and cell growth. TCGA database analysis showed increased survival at bladder and renal carcinoma when CYP1B1 expression is low. Therefore, alteration of CYP1B1 expression may suggest a therapeutic benefit for multiple diseases such as glaucoma and cancer.
Keywords: Glaucoma, CYP1B1, cancer, optic nerve, carcinogens, estrogens
Introduction
Metabolism of endogenous and exogenous compounds depends on cytochrome P450 activity [1,2]. The CYP protein was previously called 'cytochrome P450' following the discovery of a colored pigment in the cell that had a 450 nm wavelength spectrum when reduced and bound with carbon monoxide. The name continues to be misused as these proteins are not true cytochromes. They are more appropriately called 'heme-thiolate monooxygenases' [3]. CYP enzymes are stimulated by aromatic hydrocarbon receptors (AhRs), and they consist of a large number of proteins. They are involved in the metabolism of xenobiotics and endogenous compounds such as oestradiol [4]. They are also responsible for the biotransformation of most foreign substances and drug detoxification [5]. CYP expression is affected by many factors, such as sex, age, and genetic polymorphisms. The expression can also be influenced by stimulation from xenobiotics, through modulation by cytokines and hormones, and during disease states [6].
CYP1B1
Cytochrome P450 (CYPs) are a group of enzymes consist of a large heme prosthetic group catalyzing the oxidation reactions of a wide range of organic substrates. P450 proteins were the first group of enzymes to be classified as a “superfamily” and the total number of enzymes belong to this family is more than 2100 members [7]. The cytochrome P450 CYP1 family (CYP1) is responsible for the phase I metabolism of endogenous and exogenous substrates and they are involved in the oxidative metabolism of endogenous materials, such as bile acids, steroid hormones, and lipids. Furthermore, CYP1 proteins metabolize potential carcinogens such as aryl hydrocarbons, aromatic amines, heterocyclic aromatic amines, and heterocyclic amines. CYP1 family has an essential role in procarcinogen activation, catalyzing the metabolism of ~ 66% of potential carcinogens. The biotransformation of procarcinogens causes the creation of mutagenic materials, which form adducts with nucleic bases, responsible for the beginning of carcinogenesis [8].
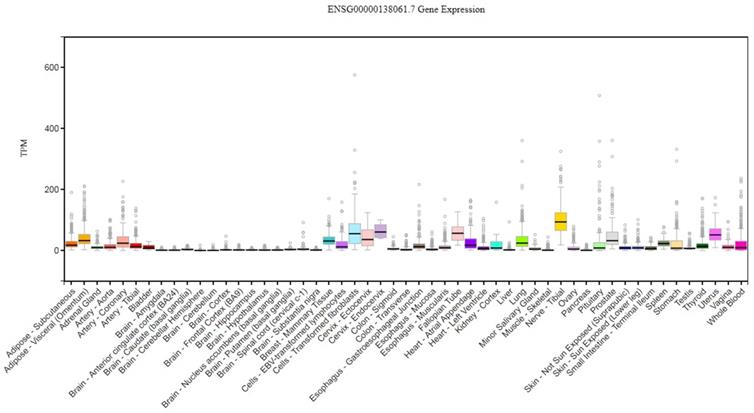
The cytochrome P450 CYP1 family consists of the CYP1A, CYP1B, CYP1C and CYP1D subfamilies [9-11]. Among the human P450 subfamilies, the CYP1B1 gene is the largest known in terms of mRNA size and number of amino acids; it is also the simplest in terms of its structure [9]. It is found on chromosome 2p21-22. Its length is approximately 8.5 kilobases (kb) of DNA, consisting of three exons and two introns. The size of its mRNA is 5.1 kb [12,13]. It has an open reading frame (ORF) starts at the 5' end of the second exon and encodes a predicted protein of 543 amino acids (Figure 1) [12]. In humans, CYP1B1 is expressed constitutively in healthy cells including eye, lungs, heart, kidney, prostate, brain, liver, uterus, ovaries and placenta [14-17]. It is also expressed in cells of the immune system and estrogen target tissues, such as mammary glands [17,18]. CYP1B1 expression in most organs was reported in the GTEx database as of October 21st, 2019 using GTEx Analysis version 7 (Figure 2). Tissues are known to accumulate melatonin or 6-hydroxymelatonin also has high expression levels of the CYP1B1 protein [19]. CYP1B1 is one of the most important enzymes involved in the metabolic activation of pre-carcinogens [1], such as polycyclic aromatic hydrocarbons [20], oxides, amines, and estrogens (Figure 3A) [21]. Moreover, CYP1B1 polymorphisms have been incriminated as risk factors in different types of cancers and glaucoma [22]. Here, we focus on the correlation between CYP1B1 and the development of glaucoma and cancer.
CYP1B1 in Glaucoma
Glaucoma is a heterogeneous disease that represents the second primary cause of irreversible loss of eyesight worldwide [23, 24]. Globally, ~64.3 million individuals are diagnosed with glaucoma. This prevalence is anticipated to increase to 76 million in 2020 [25] and 111.8 million in 2040 [26]. Glaucoma is a nonuniform group of neurodegenerative ocular disorders [27] characterized by several clinical features that include visual field defects, retinal ganglion cell death, and progressive degeneration of the optic nerve [23]. The primary source of the progressive degeneration of the optic nerve is the occurrence of a lesion in the axons of the retinal ganglion cells [28]. International Classification (CGRN/WGA) of Childhood Glaucoma divided glaucoma into two types: Primary childhood glaucoma and Secondary childhood glaucoma. The primary childhood glaucoma is classified into Primary congenital glaucoma (PCG; OMIM 231300) and Juvenile open-angle glaucoma (JOAG; OMIM 137750). The Secondary childhood glaucoma is classified into four categories: glaucoma associated with nonacquired ocular anomalies, glaucoma associated with nonacquired systemic anomalies, glaucoma associated with the acquired condition, and glaucoma following cataract surgery [29]. In children, the most common form is PCG, with a prevalence rate of 1:10,000 to 18,000 live births worldwide [30,31]. The frequency of CYP1B1 mutations in people with PCG depends on the population being considered. For example, in Saudi Arabian and Slovakian Roma residents, CYP1B1mutations were detected nearly in all cases of PCG [32]. Additionally, the prevalence of mutations in Brazil is 50%, in Indonesia it is 30%, and in Japan, it is 20%. Therefore, variability in prevalence indicates that other main genetic factors for PCG have not been identified yet. PCG is usually inherited as an autosomal recessive disease with incomplete penetrance [33]. This form is considered a rare form of glaucoma. It is characterised by isolated angle anomalies, possibly involve mild congenital iris anomalies, ocular hypertension, buphthalmos (enlargement of the eye), oedema, corneal opacification (scarring or clouding of the cornea), descemet's membrane herniation, lower density of the anterior sclera, iris atrophy and increased depth of the anterior chamber. Patients with PCG manifest variable symptoms include photophobia (intolerance to visual observation of light), blepharospasm (involuntary closing of the eyelids), and increased lacrimation (tear discharge) [33]. Optic neuropathy is not the sole contributor to PCG; glial cells are also considered major players in disease pathogenicity [34]. The development of ocular structures also involves the CYP1B1 enzyme [12], as confirmed by the detection of the CYP1B1 protein in several ocular cells of normal human fetal and adult eyes via immunolocalization assays. The intensity of CYP1B1 expression in fetal compared to adult human eyes indicates much more intense CYP1B1 immunolabeling. Anti-CYP1B1 immunoreactivity was evident in the primitive ciliary epithelium, non-pigmented ciliary epithelium, corneal epithelium, and keratocytes but was absent in the trabecular meshwork in all samples [16]. Notably, CYP1B1 was expressed in the human fetal nasal mucosa [35].
Although the diagnosis of PCG depends on clinical findings, the genetic tests could confirm the diagnosis. Several efforts have been initiated to understand the underlying mechanisms of the disease by using genome-wide association studies (GWAS) and genome sequencing technologies. Multiple PCG loci have been mapped; however, only CYP1B1 has been identified to date as the leading cause of PCG. Interestingly, more than 150 mutations in CYP1B1, including missense, nonsense, regulatory, insertions, or deletions, have been associated with PCG [33]. In glaucoma, most scientists focus on the optic nerve. However, optic nerve supporting cells and their environments are not trivial. In glaucoma, characterized CYP1B1 mutations have not been properly investigated in astrocytes, microglia, oligodendrocytes, or other neural stromal cells. In PCG, patients usually have a global mutation that increases disease severity at various levels and in a variety of cells. Therefore, scientific awareness should be practiced in areas not covered entirely in the scientific literature, and hopefully, research will be geared towards them in the future.
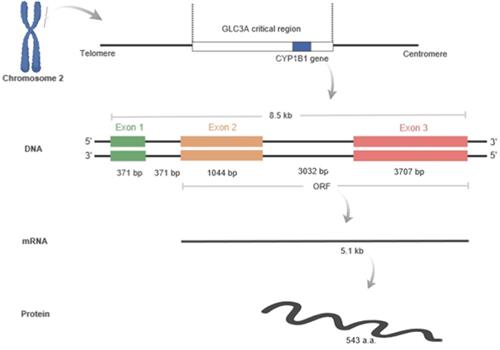
Human CYP1B1 located at 2p21-22 in the GLC3A critical region. The length of this gene is 8.5 kilobases, and it contains three exons and two introns: exon1 (371 bp), intron 1 (371 bp), exon 2 (1044 bp), intron 2 (3032 bp), and exon 3 (3707 bp). The open reading frame (ORF) starts from exon 2 and continues to exon 3. The ORF is translated into about 5.1 kb mRNA, which encodes a predicted 543 amino acids. Figure created by Biorender software.
CYP1B1 gene expression across diverse normal human tissues. The source of this figure is Genotype-Tissue expression (GTEx) project database, gtexportal.org.
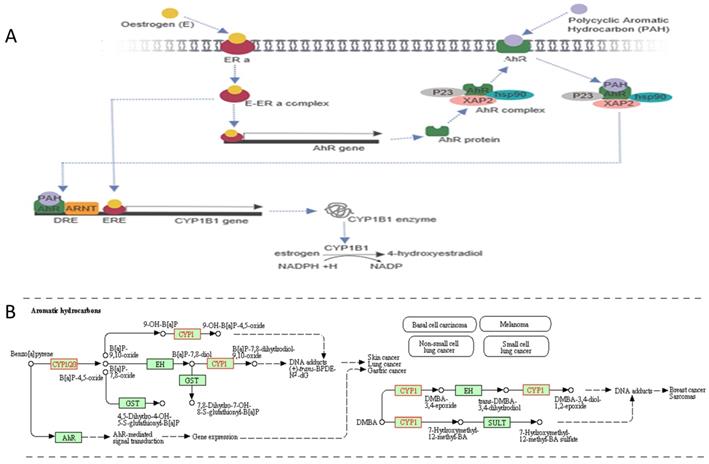
Aromatic hydrocarbon in conjunction with CYP1B1 signalling pathway. A, Oestrogen binds with the oestrogen receptor alpha (ERa) to form an oestrogen-oestrogen receptor alpha complex (E-ERa), which induces an aromatic hydrocarbon receptor (AhR) gene. The AhR protein attaches to heat shock protein-90 (hsp90), XAP2 and P23 to form an AhR complex, then transport into the cell membrane. Also, AhR binds with a polycyclic aromatic hydrocarbon (PAH) ligand to form an AhR complex/PAH complex. The AhR complex dissociates to form a heterodimer, AhR-ligand/ARNT. The heterodimer with oestrogen promotes the CYP1B1 gene. The CYP1B1 enzyme is involved in oestrogen metabolism by converting oestrogen into 4-hydroxyestradiol, which depends on NADPH. The figure created by Biorender software. B, CYP1B1 gene involved in carcinogenesis pathway using Aromatic hydrocarbons leading to different types of cancers. The figure obtained from KEGG pathways database.
CYP1B1 in Cancer
CYP1B1 belongs to a group of enzymes known as Cytochrome P450s [9]. Interestingly, CYP1B1 is one of the main enzymes that are required in the hydroxylation of estrogens and the activation of some carcinogens. CYP1B1 mediates the metabolic activity of many procarcinogens. Polycyclic aromatic hydrocarbons (PAHs) are an example of a procarcinogen in pulmonary cells that can be converted to carcinogenic metabolites capable of producing DNA adducts (Figure 3B). For instance, quinones react with the N-7 of guanine and N-3 of adenine in DNA. This reaction generates DNA adducts that are capable of producing DNA mismatches during the DNA replication process, as well as modifying promoter methylation or promoter binding, leading to an inheritable DNA mutation or irregular gene expression, and eventually tumorigenesis. The reactive metabolites of PAHs may also motivate the formation of protein adducts in cells, which may influence the normal activities of these proteins. Additionally, PAH metabolites may also increase the production of reactive oxygen species (ROS), which can affect DNA, lipids, or proteins and commence carcinogenesis [36].
Furthermore, up-regulation of CYP1B1 is an indication of cancer, since it is highly selectively expressed in cancerous rather than healthy tissues [37]. High CYP1B1 expression is found in multiple malignant tumors, including those of the brain, breast, colon, ovarian, and prostate cancers [9,38]. Immunohistochemical investigations have confirmed CYP1B1 overexpression in a majority of ovarian cancers. Most primary ovarian cancers (92%) show CYP1B1 immunoreactivity, present in the cytoplasm of the tumor cells. It is also detected in a majority (94%) of metastatic ovarian cancers. Some groups consider the CYP1B1 protein to represent an anticancer drug [39]. In a study done by Kwon and Chun on different cell lines, including MCF-7, MDA-MB-231, and HeLa cells, they reported that CYP1B1 increased invasion and inhibited cancer cell apoptosis [40]. Other studies have reported the effects of CYP1B1 knockout in vivo. When carcinogenic materials were delivered into mice with CYP1B1 knockout, decreased tumor growth, and increased DNA adduct formation was observed. These reports strongly suggest that CYP1B1 is a promising cancer biomarker and a potential target for the development of anticancer drugs [41]. However, a comprehensive molecular mechanism describing how CYP1B1 contributed to oncogenesis remains unknown [40].
Molecular mechanism of CYP1B1
Wnt/β-catenin signaling may be associated with epithelial-mesenchymal transition (EMT) because they both need β-catenin [42]. To understand CYP1B1 molecular mechanisms, multiple studies have measured key oncogenic proteins including β-catenin, c-Myc, and ZEB-2 after CYP1B1 modulation. Jung Kwon et al. explored the role of CYP1B1 in cancer development and progression, trying to find out the leading molecular players that drive CYP1B1-mediated oncogenesis. β-catenin plays a vital role in mediating the Wnt/β-catenin signaling pathway. Upon the binding of the Wnt ligand to the receptor, β-catenin escapes the proteasomal degradation and trans locates into the nucleus, where it unites with its target genes and activates various signaling pathways involved in carcinogenesis. Furthermore, Jung Kwon and colleagues measured different markers of tumor progression, including cell proliferation, invasion, and cell migration after CYP1B1 modulation, induction, or inhibition. They observed multiple target proteins, including CTNNB1, ZEB2, SNAIL1, and TWIST1, that are correlated with EMT and Wnt/β-catenin signaling were upregulated. Their findings indicate that CYP1B1 induces EMT and activates Wnt/β-catenin signaling via upregulating Sp1, a transcription factor that participates in cell proliferation and metastasis. This cascade of cellular actions inhibited E-cadherin expression and increased Wnt/β-catenin signaling upon the upregulation and nuclear translocation of β-catenin. Therefore, this was the first published study that reported the molecular mechanisms of how CYP1B mediates tumorigenesis [40]. A more detailed understanding of the underlining mechanisms of CYP1B1-mediated carcinogenesis may help in the development of new methods for cancer treatment.
Sex hormone metabolism has defected in mammary gland tumorigenesis, and variable metabolites may have different effects on mammary epithelial cells. 4-hydroxy estradiol (4-OHE2), an oxidized metabolite of estrogen, is significantly expressed in human breast cancer cells. This catechol estrogen has been proposed as an inducer of hormonal carcinogenesis, such as breast cancer. Furthermore, when transgenic mice with increased expression of CYP1B1, an essential enzyme of 4-hydroxy metabolites [1], were stimulated with estrogen, cancer cells were found in their mammary glands [43].
Hypomethylation in the promoter region of CYP1B1 regulates the overexpression of this gene, as determined in clinical prostate tissues and prostate cancer cell lines using methylation-specific PCR and bisulfite-modified DNA sequencing [44]. Another study investigated the function of miRNA in drug-metabolizing enzymes provided another reason for elevated CYP1B1 levels. The results of luciferase assays suggested that miR-27b regulates CYP1B1 at the post-transcriptional stage. A decreased level of miR-27b was associated with elevated levels of CYP1B1 protein in cancerous tissues [45]. Recently, another miRNA, miR-187-5p, was also shown to have decreased expression in non-small lung cancer. Interestingly, CYP1B1 is a direct target gene for miR-187-5p, which is expressed during cancer cell growth [46].
Moreover, elevated levels of the pro-inflammatory cytokine interleukin-6 also have effects on CYP1B1 expression in HCT116 and SW480 colorectal cancer cell lines. This cytokine can manipulate CYP1B1 expression and can induce the activation of dietary carcinogens and DNA damage that can trigger colorectal cancer progression [47]. Additionally, the breast cancer microenvironment can be guided by leptin, which directly stimulates estrogen metabolism genes, including CYP1B1, that are involved in the oxidative metabolism (4-hydroxylation) of estrogen. Oxidative metabolism of estrogen leads to DNA adduct formation and cancer mutations [48].
Cigarette smoke carcinogens such as 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and polycyclic aromatic hydrocarbons (PAH) are known as significant causes of lung cancer, which kills on average 3000 people per day worldwide [49]. NNK and PAH upregulate CYP1B1 enzyme activity upon binding to AhR (Figure 3B). The AhR overexpression upregulated the expression of CYP1B1 in the early stage of lung adenocarcinoma, which could increase intracellular oxidative stress and promote cell growth. Therefore, the disruption of AhR expression could prevent the early development of lung adenocarcinomas [50].
Flavonoids are a group of common substances with variable phenolic structures. These compounds are found in natural products such as fruits, vegetables, grains, blossoms, tea, and wine. These particular items are well known for their beneficial impacts on wellbeing, and tremendous efforts are being made to isolate Flavonoids from these natural products. Because of Flavonoids anti-oxidative, anti-inflammatory, and anti-carcinogenic properties and due to their ability to balance main cellular enzyme functions, they are considered as an essential component in multiple nutraceuticals, pharmaceutical, medicinal, and cosmetic uses [51]. Flavonoids have a tight, rigid structure that enables them to fit precisely within the active site of the CYP1B1 enzyme [21].
Interestingly, dietary flavonoids can be metabolized by CYP1B1 to products that can inhibit cancer proliferation [52]. Furthermore, many studies recommended the use of flavonoids to treat several types of cancer, such as melanoma, hepatocellular carcinoma, and thyroid cancer. The administration of flavonoid in animal models of cancer has enhanced the tolerance to cancer treatment [51].
Natural flavonoids are the primary source of CYP1B1 inhibitors. Methoxy types of flavones and flavonols were appeared to prevent CYP1B1 activity, such as chrysoeriol and isorhamnetin specifically. Various flavonoids including quercetin, rutin, apigenin, and amentoflavone, show an inhibitory effect of CYP1B1. Some CYP1B1 inhibitors, such as kaempferol and isorhamnetin, can also antagonize the expression of AHR, which may show synergetic inhibition on the expression of CYP1B1. Interestingly, some anti-cancer drugs used in clinics are competitive inhibitors of CYP1B1, such as flutamide, paclitaxel, mitoxantrone, and docetaxel. CYP1B1 inhibitors can be utilized to understand the CYP1B1 function; thus, it might be considered as anti-cancer agents for the treatment of certain types of cancer [53].
In cancer cells, CYP1B1 is highly expressed. This observation proposes that CYP1B1 enzyme may metabolize variable anticancer drugs. Cisplatin is considered an essential chemotherapeutic agent, but it might also play a role in chemo-resistance. The presence of CYP1B1 in tumor cells decreases the sensitivity of the cells to the cytotoxic effects of Cisplatin. However, the co-administration of potent, cell permeable and water soluble CYP1B1 inhibitors, such as the synthetic pyridylchalcones 7k (DMU2105) and 6j (DMU2139), with Cisplatin can overcome CYP1B1-mediated resistance and re-sensitized cancerous cells once again to the Cisplatin [54]. These findings suggest the useful roles of CYP1B1 inhibitors in the treatment of cancer.
Prognosis and interactions of CYP1B gene
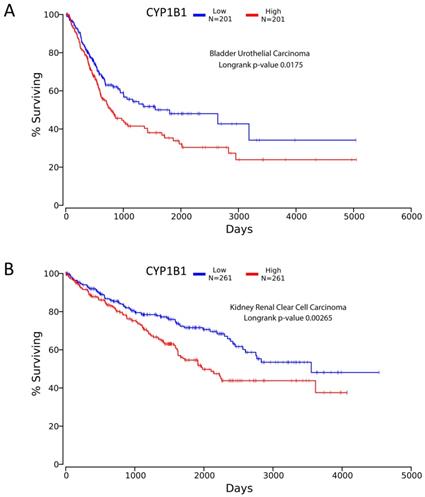
Looking at TCGA database, significance was found for CYP1B1 expression relevant to bladder and kidney carcinoma. Low expression of CYP1B1 in cancer corresponded to better survival detected by Kaplan-meier plot for kidney (renal) clear cell carcinoma and bladder urothelial carcinoma with p-value 0.00265 and 0.0175 respectively (Figure 4). The data was extracted from over 200 patients suffering from cancer in both diseases [55]. This finding opens the door for scientists to investigate possible treatment methods such as CYP inhibitors to achieve better survival in both cancers.
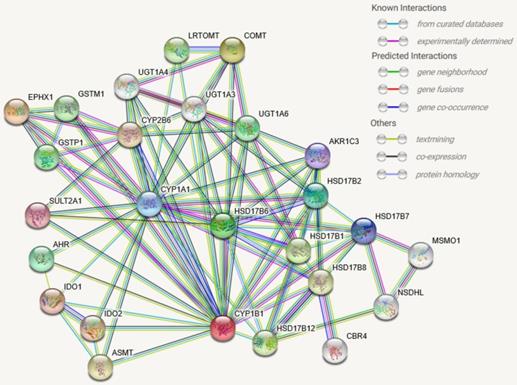
Interaction of CYP1B1 genes have been documented in many research papers through frpm STRING database. Level of confidence was assessed based on physical experiment, bioinformatics predictions, co-expressions anticipation and indirect association, See Figure 5. These interactions involve biological processes, molecular functions, and/or cellular components. The STRING database has identified 100 publications for CYP1B1 interactions and minimum 22 pathways when scale of confidence was set at high confidence. This information will enhance the scientific investigation toward validating predicted phenomenon or interaction. In addition, Table 1. lists functions of molecules that directly interact with CYP1B1.
List of direct interactive items with CYP1B1
| No. | Abbreviation | Names of Direct interactive items with CYP1B1 in Figure 5 |
|---|---|---|
| 1 | EPHX1 | Epoxide hydrolase 1; Biotransformation enzyme that catalyzes the hydrolysis of arene and aliphatic epoxides to less reactive and more water soluble dihydrodiols by the trans addition of water (By similarity). May play a role in the metabolism of endogenous lipids such as epoxide-containing fatty acids. |
| 2 | COMT | Catechol O-methyltransferase; Catalyzes the O-methylation, and thereby the inactivation, of catecholamine neurotransmitters and catechol hormones. Also shortens the biological half-lives of certain neuroactive drugs, like L-DOPA, alpha-methyl DOPA and isoproterenol; Seven-beta-strand methyltransferase motif containing. |
| 3 | HSD17B1 | Estradiol 17-beta-dehydrogenase 1; Favors the reduction of estrogens and androgens. Also has 20-alpha-HSD activity. Uses preferentially NADH; Short chain dehydrogenase/reductase superfamily. |
| 4 | HSD17B6 | 17-beta-hydroxysteroid dehydrogenase type 6; NAD-dependent oxidoreductase with broad substrate specificity that shows both oxidative and reductive activity (in vitro). Has 17-beta-hydroxysteroid dehydrogenase activity towards various steroids (in vitro). Converts 5-alpha-androstan-3- alpha,17-beta-diol to androsterone and estradiol to estrone (in vitro). Has 3-alpha-hydroxysteroid dehydrogenase activity towards androsterone (in vitro). Has retinol dehydrogenase activity towards all-trans-retinol (in vitro). |
| 5 | AHR | Aryl hydrocarbon receptor; Ligand-activated transcriptional activator. Binds to the XRE promoter region of genes it activates. Activates the expression of multiple phase I and II xenobiotic chemical metabolizing enzyme genes (such as the CYP1A1 gene). Mediates biochemical and toxic effects of halogenated aromatic hydrocarbons. Involved in cell-cycle regulation. Likely to play an important role in the development and maturation of many tissues. |
| 6 | HSD17B2 | Estradiol 17-beta-dehydrogenase 2; Capable of catalyzing the interconversion of testosterone and androstenedione, as well as estradiol and estrone. Also has 20-alpha-HSD activity. Uses NADH while EDH17B3 uses NADPH; Short chain dehydrogenase/reductase superfamily. |
| 7 | CYP1A1 | Cytochrome P450 1A1; Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. |
| 8 | HSD17B7 | 3-keto-steroid reductase; Responsible for the reduction of the keto group on the C-3 of sterols; Short chain dehydrogenase/reductase superfamily. |
| 9 | AKR1C3 | Aldo-keto reductase family 1 member C3; Catalyzes the conversion of aldehydes and ketones to alcohols. Catalyzes the reduction of prostaglandin (PG) D2, PGH2 and phenanthrenequinone (PQ) and the oxidation of 9-alpha,11-beta- PGF2 to PGD2. Functions as a bi-directional 3-alpha-, 17-beta- and 20-alpha HSD. Can interconvert active androgens, estrogens and progestins with their cognate inactive metabolites. |
| 10 | SULT2A1 | Bile salt sulfotransferase; Sulfotransferase that utilizes 3'-phospho-5'-adenylyl sulfate (PAPS) as sulfonate donor to catalyze the sulfonation of steroids and bile acids in the liver and adrenal glands; Sulfotransferases, cytosolic. |
| 11 | IDO1 | Indoleamine 2,3-dioxygenase 1; Catalyzes the first and rate limiting step of the catabolism of the essential amino acid tryptophan along the kynurenine pathway. Involved in the peripheral immune tolerance, contributing to maintain homeostasis by preventing autoimmunity or immunopathology that would result from uncontrolled and overreacting immune responses. Tryptophan shortage inhibits T lymphocytes division. |
| 12 | IDO2 | Indoleamine 2,3-dioxygenase 2; Catalyzes the first and rate limiting step of the catabolism of the essential amino acid tryptophan along the kynurenine pathway. Involved in immune regulation. May not play a significant role in tryptophan-related tumoral resistance. |
| 13 | CYP2B6 | Cytochrome P450 2B6; Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. Acts as a 1,4-cineole 2-exo-monooxygenase |
| 14 | ASMT | Acetylserotonin O-methyltransferase; Isoform 1 catalyzes the transfer of a methyl group onto N-acetylserotonin, producing melatonin (N-acetyl-5- methoxytryptamine). Isoform 2 and isoform 3 lack enzyme activity; Pseudoautosomal region 1. |
| 15 | HSD17B8 | Estradiol 17-beta-dehydrogenase 8; NAD-dependent 17-beta-hydroxysteroid dehydrogenase with highest activity towards estradiol. Has very low activity towards testosterone. The heterotetramer with CBR4 has NADH-dependent 3-ketoacyl-acyl carrier protein reductase activity, and thereby plays a role in mitochondrial fatty acid biosynthesis. |
| 16 | HSD17B12 | Very-long-chain 3-oxoacyl-CoA reductase; Catalyzes the second of the four reactions of the long- chain fatty acids elongation cycle. This endoplasmic reticulum- bound enzymatic process, allows the addition of two carbons to the chain of long- and very long-chain fatty acids/VLCFAs per cycle. |
| 17 | LRTOMT | Transmembrane O-methyltransferase; Catalyzes the O-methylation, and thereby the inactivation, of catecholamine neurotransmitters and catechol hormones (By similarity). Required for auditory function. Component of the cochlear hair cell's mechanotransduction (MET) machinery. |
| 18 | GSTM1 | Glutathione S-transferase Mu 1; Conjugation of reduced glutathione to a wide number of exogenous and endogenous hydrophobic electrophiles; Soluble glutathione S-transferases |
| 19 | GSTP1 | Glutathione S-transferase P; Conjugation of reduced glutathione to a wide number of exogenous and endogenous hydrophobic electrophiles. Regulates negatively CDK5 activity via p25/p35 translocation to prevent neurodegeneration; Soluble glutathione S-transferases |
| 20 | UGT1A6 | UDP-glucuronosyltransferase 1-6; UDPGT is of major importance in the conjugation and subsequent elimination of potentially toxic xenobiotics and endogenous compounds. This isoform has specificity for phenols. |
TCGA survival curves using Kaplan meier analysis. A, represents bladder urothelial carcinoma showing longer survival for low expression CYP1B1 patients. B, represents renal clear cell carcinoma showing extended survival for decreased expression CYP1B1 patients. Figures created from oncolnc.org website with TCGA database incorporated.
CYP1B1 protein-protein associations frpm STRING database. Colored nodes are query proteins and first shell of interactors. White nodes are second shell of interactors. filled nodes (structure inside the node) represent some 3D structure is known or predicted. Interactions include: i) known interactions from curated databases and experimentally determined, ii) predicted interactions by gene neighborhood, gene fusion, and gene co-occurrence, iii) others by text mining, co-expression, and protein homology. All interactions have been segregated by different colors.
Conclusion
High-advanced technologies, including whole-exome sequencing (WES) and whole-genome sequencing (WGS), helped to reveal the molecular mechanisms of how genetic abnormalities such as CYP1B1 mutations lead to glaucoma and cancer. Glaucoma-related abnormalities are severe and can cause irreversible blindness. The CYP1B1 gene is the most common gene mutated in PCG. Although many articles have pointed out the importance of this gene, molecular and physiological information are not completed yet. Due to the lack of research support, many mutations have been characterized, but others have been neglected. Also, some African and Middle-Eastern mutations are not characterized yet.
Furthermore, scientists are investigating either CYP1B1 mutations or CYP1B1 overexpression since they contribute to glaucoma or cancer, respectively. On the other hand, a balanced expression of CYP1B1 in either glaucoma or cancer has not been investigated. Thus, multiple questions remain unanswered, such as: does high expression in the eye cause cancer? Or does a reduction of expression in other organs, such as the liver, create difficulty? We have promising disease candidates for CYP1B1 inhibitor trials which are kidney renal clear cell carcinoma and bladder urothelial carcinoma. Additional information is needed to evaluate different treatment plans in those scenarios. With the development of new genetic engineering techniques, a possible option of treatment is the correction of the CYP1B1 abnormality via gene-editing technologies such as the CRISPR-Cas9 system.
Additionally, genome-wide association studies (GWAS) and other cellular techniques studying gene expression and regulation will undoubtedly change our understanding of how CYP1B1 mutations contribute to the pathogenesis of glaucoma and cancer. In order to achieve this, in the future, it is essential to have full access to glaucoma and cancer patient's samples. Also, it is vital to have an available platform occupied with all instrumental technologies needed to perform different research studies such as functional genomics, gene expression studies, pathway analysis, and epigenetic studies. Therefore, the ultimate knowledge we will get out from these studies will expand our understanding of the different cellular and molecular mechanisms of glaucoma and cancer, which in turn will help in the development of new therapeutic options for glaucoma and cancer patients.
Acknowledgements
We appreciate support from King Abdulaziz City for Science and Technology (KACST) and support from KAIMRC RC17/038/R. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. Some of the data used for the analyses described in this manuscript were obtained from: [insert, where appropriate] the GTEx Portal on 06/30/2019 and/or dbGaP accession number ENSG00000138061.7 on 06/30/2019. KEGG pathway database [56-58]. TCGA database and OncoLnc have been used to analyse and describe data in this manuscript [55].
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ingelman-Sundberg M. Human drug metabolising cytochrome P450 enzymes: properties and polymorphisms. Naunyn-Schmiedeberg's archives of pharmacology. 2004;369:89-104
2. Lewis DF, Gillam EM, Everett SA, Shimada T. Molecular modelling of human CYP1B1 substrate interactions and investigation of allelic variant effects on metabolism. Chem Biol Interact. 2003;145:281-95
3. Nebert DW, Dalton TP. The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nature Reviews Cancer. 2006;6:947
4. Pavek P, Dvorak Z. Xenobiotic-Induced Transcriptional Regulation of Xenobiotic Metabolizing Enzymes of the Cytochrome P450 Superfamily in Human Extrahepatic Tissues. Current Drug Metabolism. 2008;9:129-43
5. Verma S, Saxena R, Siddiqui MH, Santha K, Sethupathy S. Evaluation of CYP1B1 Expression, Oxidative Stress and Phase 2 Detoxification Enzyme Status in Oral Squamous Cell Carcinoma Patients. J Clin Diagn Res. 2017;11:BC01-BC5
6. Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacology & therapeutics. 2013;138:103-41
7. Munro AW, McLean KJ, Grant JL, Makris TM. Structure and function of the cytochrome P450 peroxygenase enzymes. Biochem Soc Trans. 2018;46:183-96
8. Dutkiewicz Z, Mikstacka R. Structure-Based Drug Design for Cytochrome P450 Family 1 Inhibitors. Bioinorg Chem Appl. 2018;2018:3924608
9. Murray GI, Melvin WT, Greenlee WF, Burke MD. Regulation, function, and tissue-specific expression of cytochrome P450 CYP1B1. Annual review of pharmacology and toxicology. 2001;41:297-316
10. Godard CA, Goldstone JV, Said MR, Dickerson RL, Woodin BR, Stegeman JJ. The new vertebrate CYP1C family: cloning of new subfamily members and phylogenetic analysis. Biochem Biophys Res Commun. 2005;331:1016-24
11. Goldstone JV, Stegeman JJ. Gene structure of the novel cytochrome P4501D1 genes in stickleback (Gasterosteus aculeatus) and medaka (Oryzias latipes). Mar Environ Res. 2008;66:19-20
12. Faiq MA, Ali M, Dada T, Dada R, Saluja D. A novel methodology for enhanced and consistent heterologous expression of unmodified human cytochrome P450 1B1 (CYP1B1). PloS one. 2014;9:e110473
13. Tang YM, Wo Y-YP, Stewart J, Hawkins AL, Griffin CA, Sutter TR. et al. Isolation and characterization of the human cytochrome P450 CYP1B1 gene. Journal of Biological Chemistry. 1996;271:28324-30
14. Martignoni M, Groothuis GM, de Kanter R. Species differences between mouse, rat, dog, monkey and human CYP-mediated drug metabolism, inhibition and induction. Expert opinion on drug metabolism & toxicology. 2006;2:875-94
15. Mahemuti L, Chen Q, Coughlan MC, Qiao C, Chepelev NL, Florian M. et al. Bisphenol A induces DSB-ATM-p53 signaling leading to cell cycle arrest, senescence, autophagy, stress response, and estrogen release in human fetal lung fibroblasts. Archives of toxicology. 2017:1-17
16. Doshi M, Marcus C, Bejjani BA, Edward DP. Immunolocalization of CYP1B1 in normal, human, fetal and adult eyes. Experimental eye research. 2006;82:24-32
17. Tsuchiya Y, Nakajima M, Yokoi T. Cytochrome P450-mediated metabolism of estrogens and its regulation in human. Cancer letters. 2005;227:115-24
18. Effner R, Hiller J, Eyerich S, Traidl-Hoffmann C, Brockow K, Triggiani M. et al. Cytochrome P450s in human immune cells regulate IL-22 and c-Kit via an AHR feedback loop. Scientific Reports. 2017;7:44005
19. Ma X, Idle JR, Krausz KW, Gonzalez FJ. Metabolism of melatonin by human cytochromes p450. Drug Metabolism and Disposition. 2005;33:489-94
20. Shimada T, Fujii-Kuriyama Y. Metabolic activation of polycyclic aromatic hydrocarbons to carcinogens by cytochromes P450 1A1 and1B1. Cancer science. 2004;95:1-6
21. Siddique MUM, McCann GJ, Sonawane VR, Horley N, Gatchie L, Joshi P. et al. Quinazoline derivatives as selective CYP1B1 inhibitors. European journal of medicinal chemistry. 2017;130:320-7
22. Vasiliou V, Gonzalez FJ. Role of CYP1B1 in Glaucoma. Annual Review of Pharmacology and Toxicology. 2008;48:333-58
23. Gong B, Qu C, Li X, Shi Y, Lin Y, Zhou Y. et al. Mutation spectrum of CYP1B1 in Chinese patients with primary open-angle glaucoma. Br J Ophthalmol. 2015;99:425-30
24. Kumar A, Basavaraj MG, Gupta SK, Qamar I, Ali AM, Bajaj V. et al. Role of CYP1B1, MYOC, OPTN, and OPTC genes in adult-onset primary open-angle glaucoma: predominance of CYP1B1 mutations in Indian patients. Mol Vis. 2007;13:667-76
25. Quigley HA. Glaucoma. Lancet. 2011;377:1367-77
26. Yih Chung Tham, Li X, Wong TY, Quigley H, Aung T, Cheng CY. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. 2014; 121: 2081-90.
27. Souzeau E, Hayes M, Zhou T, Siggs OM, Ridge B, Awadalla MS. et al. Occurrence of CYP1B1 Mutations in Juvenile Open-Angle Glaucoma With Advanced Visual Field Loss. JAMA Ophthalmol. 2015;133:826-33
28. Whitworth GB, Misaghi BC, Rosenthal DM, Mills EA, Heinen DJ, Watson AH. et al. Translational profiling of retinal ganglion cell optic nerve regeneration in Xenopus laevis. Dev Biol. 2017;426:360-73
29. Weinreb RN, Grajewski AL, Papadopoulos M, Grigg J, Freedman S. Childhood glaucoma. World glaucoma association consensus series 9. Kugler Publications. 2013 9
30. Do T, Shei W, Chau PT, Trang DL, Yong VH, Ng XY. et al. CYP1B1 and MYOC Mutations in Vietnamese Primary Congenital Glaucoma Patients. J Glaucoma. 2016;25:e491-8
31. Rulli E, Quaranta L, Riva I, Poli D, Hollander L, Galli F. et al. Visual field loss and vision-related quality of life in the Italian Primary Open Angle Glaucoma Study. Sci Rep. 2018;8:619
32. Garcia-Anton MT, Salazar JJ, de Hoz R, Rojas B, Ramirez AI, Trivino A. et al. Goniodysgenesis variability and activity of CYP1B1 genotypes in primary congenital glaucoma. PLoS One. 2017;12:e0176386
33. Chouiter L, Nadifi S. Analysis of CYP1B1 Gene Mutations in Patients with Primary Congenital Glaucoma. J Pediatr Genet. 2017;6:205-14
34. Chong RS, Martin KR. Glial cell interactions and glaucoma. Current Opinion in Ophthalmology. 2015;26:73-7
35. Zhang X, Zhang Q-Y, Liu D, Su T, Weng Y, Ling G. et al. Expression of Cytochrome P450 and other Biotransformation Genes in Fetal and Adult Human Nasal Mucosa. Drug Metabolism and Disposition. 2005;33:1423
36. Moorthy B, Chu C, Carlin DJ. Polycyclic Aromatic Hydrocarbons: From Metabolism to Lung Cancer. Toxicological Sciences. 2015;145:5-15
37. Nishida CR, Everett S, Ortiz de Montellano PR. Specificity determinants of CYP1B1 estradiol hydroxylation. Mol Pharmacol. 2013;84:451-8
38. Murray GI, Taylor MC, McFadyen MCE, McKay JA, Greenlee WF, Burke MD. et al. Tumor-specific Expression of Cytochrome P450 CYP1B1. Cancer Research. 1997;57:3026
39. McFadyen MC, Cruickshank ME, Miller ID, McLeod HL, Melvin WT, Haites NE. et al. Cytochrome P450 CYP1B1 over-expression in primary and metastatic ovarian cancer. Br J Cancer. 2001;85:242-6
40. Kwon YJ, Baek HS, Ye DJ, Shin S, Kim D, Chun YJ. CYP1B1 Enhances Cell Proliferation and Metastasis through Induction of EMT and Activation of Wnt/beta-Catenin Signaling via Sp1 Upregulation. PLoS One. 2016;11:e0151598
41. Wilsher NE, Arroo R, Matsoukas MT, Tsatsakis AM, Spandidos DA, Androutsopoulos VP. Cytochrome P450 CYP1 metabolism of hydroxylated flavones and flavonols: Selective bioactivation of luteolin in breast cancer cells. 2017; 110: 383-394.
42. Son H, Moon A. Epithelial-mesenchymal Transition and Cell Invasion. Toxicol Res. 2010;26:245-52
43. Miao S, Yang F, Wang Y, Shao C, Zava DT, Ding Q. et al. 4-Hydroxy estrogen metabolite, causing genomic instability by attenuating the function of spindle-assembly checkpoint, can serve as a biomarker for breast cancer. Am J Transl Res. 2019;11:4992-5007
44. Tokizane T, Shiina H, Igawa M, Enokida H, Urakami S, Kawakami T. et al. Cytochrome P450 1B1 is overexpressed and regulated by hypomethylation in prostate cancer. Clin Cancer Res. 2005;11:5793-801
45. Tsuchiya Y, Nakajima M, Takagi S, Taniya T, Yokoi T. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res. 2006;66:9090-8
46. Mao M, Wu Z, Chen J. MicroRNA-187-5p suppresses cancer cell progression in non-small cell lung cancer (NSCLC) through down-regulation of CYP1B1. Biochem Biophys Res Commun. 2016;478:649-55
47. Patel SA, Bhambra U, Charalambous MP, David RM, Edwards RJ, Lightfoot T. et al. Interleukin-6 mediated upregulation of CYP1B1 and CYP2E1 in colorectal cancer involves DNA methylation, miR27b and STAT3. Br J Cancer. 2014;111:2287-96
48. Shouman S, Wagih M, Kamel M. Leptin influences estrogen metabolism and increases DNA adduct formation in breast cancer cells. Cancer Biol Med. 2016;13:505-13
49. Church TR, Haznadar M, Geisser MS, Anderson KE, Caporaso NE. Interaction of CYP1B1, cigarette-smoke carcinogen metabolism, and lung cancer risk. 2010; 1: 295-309.
50. Chang JT, Chang H, Chen P-H, Lin S-L, Lin P. Requirement of aryl hydrocarbon receptor overexpression for CYP1B1 up-regulation and cell growth in human lung adenocarcinomas. Clin Cancer Res. 2007;13:38-45
51. Panche AN, Diwan AD, Chandra SR. Flavonoids: an overview. J Nutr Sci. 2016;5:e47
52. Androutsopoulos VP, Papakyriakou A, Vourloumis D, Spandidos DA. Comparative CYP1A1 and CYP1B1 substrate and inhibitor profile of dietary flavonoids. Bioorg Med Chem. 2011;19:2842-9
53. Li F, Zhu W, Gonzalez FJ. Potential role of CYP1B1 in the development and treatment of metabolic diseases. Pharmacol Ther. 2017;178:18-30
54. Horley NJ, Beresford KJ, Chawla T, McCann GJ, Ruparelia KC, Gatchie L. et al. Discovery and characterization of novel CYP1B1 inhibitors based on heterocyclic chalcones: Overcoming cisplatin resistance in CYP1B1-overexpressing lines. European journal of medicinal chemistry. 2017;129:159-74
55. Anaya J. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Computer Science. 2016;2:e67
56. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids research. 2000;28:27-30
57. Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. New approach for understanding genome variations in KEGG. Nucleic acids research. 2019;47:D590-D5
58. Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Science. 2019;28:1947-51
Author Biography
 Dr. Bahauddeen M. Alrfaei earned his Ph.D degree in Cellular and Molecular Pathology from University of Wisconsin-Madison. He later did postdoctoral fellowship at Stanford University with Prof. Samuel Cheshier. Dr. Alrfaei works as Research Scientist at King Abdullah International Medical Research Center within Cellular Therapy Department. His research is focusing on neurological disorders, stem cells biology and cancer.
Dr. Bahauddeen M. Alrfaei earned his Ph.D degree in Cellular and Molecular Pathology from University of Wisconsin-Madison. He later did postdoctoral fellowship at Stanford University with Prof. Samuel Cheshier. Dr. Alrfaei works as Research Scientist at King Abdullah International Medical Research Center within Cellular Therapy Department. His research is focusing on neurological disorders, stem cells biology and cancer.
![]() Corresponding author: Dr. Bahauddeen Alrfaei, E-mail: alrfaeibamed.sa., Cellular Therapy and Cancer Research Department, King Abdullah International Medical Research Center (KAIMRC)/ King Saud Bin Abdulaziz University for Health Sciences (KSAU-HS), Ministry of National Guard - Health Affairs, Saudi Arabia. P.O. Box 22490, Riyadh 11426 MailCode 1515.
Corresponding author: Dr. Bahauddeen Alrfaei, E-mail: alrfaeibamed.sa., Cellular Therapy and Cancer Research Department, King Abdullah International Medical Research Center (KAIMRC)/ King Saud Bin Abdulaziz University for Health Sciences (KSAU-HS), Ministry of National Guard - Health Affairs, Saudi Arabia. P.O. Box 22490, Riyadh 11426 MailCode 1515.

 Global reach, higher impact
Global reach, higher impact