3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(3):735-739. doi:10.7150/jca.50899 This issue Cite
Review
The oncogenic role of protein kinase D3 in cancer
1. The Key Laboratory of Bio-Medical Diagnostics, Suzhou Institute of Biomedical Engineering and Technology, Chinese Academy of Sciences, Suzhou 215163, P. R. China.
2. Jiangsu Key Laboratory for Molecular and Medical Biotechnology, Institute of cancer, Department of biochemistry, College of Life Science, Nanjing Normal University, Nanjing 210023, P. R. China.
3. The First Affiliated Hospital of Soochow University, Soochow University, Suzhou 215006, P. R. China.
4. School of Integrated Chinese and Western Medicine, Anhui University of Chinese Medicine, Hefei 230012, P. R.China.
5. Department of Rheumatology and Immunology, Jinling Hospital, Medical School of Nanjing University, Nanjing 210002, P. R.China.
#These authors contributed equally to this work.
Received 2020-7-20; Accepted 2020-10-30; Published 2021-1-1
Abstract
Protein kinase D3 (PRKD3), a serine/threonine kinase, belongs to protein kinase D family, which contains three members: PRKD1, PRKD2, and PRKD3. PRKD3 is activated by many stimuli including phorbol esters, and G-protein-coupled receptor agonists. PRKD3 promotes cancer cell proliferation, growth, migration, and invasion in various tumor types including colorectal, gastric, hepatic, prostate, and breast cancer. Accumulating data supports that PRKD3 is a promising therapeutic target for treatment of cancer. This review discusses the functions and mechanisms of PRKD3 in promoting tumorigenesis and tumor progression of various tumor types as well as the latest developments of small-molecule inhibitors selection for PRKD/PRKD3.
Keywords: Protein kinase D3, Cancer progression
Introduction
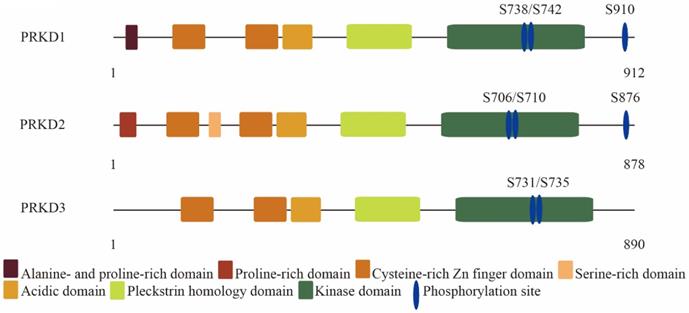
Protein kinase D (PRKD) family consists of three highly conserved members in human: PRKD1, PRKD2, and PRKD3. PRKD1 was the first identified PRKD family member [1, 2]. Two other PRKD members have since been identified, PRKD2 [3] and PRKD3 [4]. PRKD family members are effectors of diacylglycerol signaling and are activated downstream of protein kinase C by a variety of stimuli including growth factors and hormones [5]. PRKD family share similar structural features such as the highly conserved N-terminal regulatory domain containing two cysteine-rich DAG-binding C1 domains and an auto-inhibitory pleckstrin homology domain (Figure 1) [6, 7]. Despite high structural homology among the PRKD isoforms, some structural variability exists and to a certain extent can help to explain the different effects of each PRKD isoforms. For example, PRKD1 and PRKD2 contain a c-terminal PDZ binding motif, while PRKD3 does not [8]. The c-terminal PDZ binding motif allows PRKD1 and PRKD2 to regulate Kidins220 localization at the surface of neural cells and its trafficking between the plasma membrane and trans-golgi network, while PRKD3 do not have these functions [9]. In addition, PRKD1 is mainly localized within the cytosol in resting cells [10], but upon stimulation can be found in other cellular structures such as the golgi [11], nucleus [12], or mitochondria [13]. Like PRKD1, PRKD2 is mainly cytoplasmic in unstimulated cells [14]. However, PRKD3 is localized in the cytoplasm and nucleus without stimulation [15]. PRKDs have many cellular targets and have been implicated in a variety of biological effects such as cell growth [16, 17], invasion [18-20], angiogenesis [21], protein transport [22], transcriptional regulation [23], and epithelial to mesenchymal transition [24, 25]. Due to lack of an autophosphorylation site at its C terminus and the alanine- and proline-rich region at PRKD3 N terminus, PRKD3 exhibits diverse biological effects and molecular signals from other PRKD isoforms in cancer. In this review, we focused on discussing PRKD3 in the context of cancer.
The molecular architecture of protein kinase D family members: PRKD1, PRKD2 and PRKD3.
Oncogenic functions of PRKD3 in breast cancer
Breast cancer is the most heterogeneous disease in females [26]. PRKD1 is expressed and active in the normal breast ductal epithelial cells but its expression is lost during tumorigenesis. Analyses of human breast cancer specimens have shown that PRKD1 expression is completely lost in some of the most highly aggressive tumors [18, 27]. However, PRKD2 and PRKD3 are only weakly expressed in the normal breast tissues while PRKD2 is generally weakly expressed, but PRKD3 has been reported to be up-regulated in breast cancer [28, 29].
PRKD3 is involved in all aspects of oncogenic signaling. Many researches have confirmed that PRKD3 is overexpressed in invasive breast cancer cell lines [28-30]. Moreover, the mRNA and protein level of PRKD3 increased in triple negative breast cancer (TNBC) [28, 29]. PRKD3 appears to have typical oncogenic effects in breast cancer. Depletion of PRKD3 attenuated cell proliferation by up to 40% in the TNBC cell line MDA-MB-231 [31]. In other TNBC cell lines (MDA-MB-468 and HCC1806) and in the ER-/HER2+ cell line (HCC1954), the same effect on cell proliferation was also observed [28]. One of the possible mechanisms is that PRKD3 promotes TNBC cell proliferation via contributing to mammalian target of rapamycin complex 1/ribosomal protein S6 kinase B1 pathway activation [29]. Other researchers suggest that PRKD3 promotes the proliferation of breast cancer cells by activating the mitogen-activated protein kinase 3/MYC proto-oncogene axis or ELAV like RNA binding protein 1 [32, 33]. The RhoGEF GRF-H1 is claimed to activate PRKD3 for the maintenance of TNBC stem cells [34]. Besides functioning in proliferation of breast cancer cells, PRKD3 also promotes the motility, spreading, and migration of breast cancer. Basal PRKD3 activity promotes breast cancer migration via regulating cofilin phosphorylation status and activation of P21 (RAC1) activated kinase 4/LIM domain kinase 1 [35]. GIT arfGAP 1 phosphorylation on serine 46 by PRKD3 regulates paxillin trafficking and cellular protrusive activity [36]. Knockdown of PRKD3 decreased the migration of ER- breast cancer cells with increased cell spreading and altered F-actin organization [28].
PRKD3 participates in cell growth, invasion and secretion in prostate cancer
Besides breast cancer, increased levels of PRKD3 were detected in human prostate cancer specimens when compared to normal prostate specimens. In addition, there was a strong correlation between increasing prostate tumor grade and PRKD3 nuclear localization [37]. PRKD3 promotes the growth and survival of prostate cancer cells through AKT serine/threonine kinase 1 and mitogen-activated protein kinase 1 signaling pathway [37]. Interplay of PRKD3 with sterol regulatory element binding transcription factor 2 also contributes to the growth of prostate cancer cells via upregulating lipogenesis [38]. Inducible silencing of PRKD3 inhibits secretion of tumor-promoting factors (matrix metallopeptidase 9, Interleukin 6, C-X-C motif chemokine ligand 8, and C-X-C motif chemokine ligand 1) in prostate cancer [39]. Snail activated the lncRNA PCA3 expression could inhibit PRKD3 protein translation via competitive miR-1261 sponging to promote the invasion and migration of prostate cancer [40]. PRKD3 promotes the invasion of prostate cancer cells by modulating nuclear factor kappa B subunit 1- and histone deacetylase 1-mediated expression and activation of plasminogen activator, urokinase [41].
PRKD3 promotes cancer progression in other cancer type
PRKD3 acts as an important role as well as diagnostic criteria in gastric, melanoma, and hepatocellular cancer. In gastric cancer, PRKD3 promotes the development of cancer through RELA proto-oncogene, NF-KB subunit/6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 activation of glycolysis [42]. In melanoma cells, PRKD3 sensitizes RAF inhibitor RAF265 by preventing reactivation of mitogen-activated protein kinase 1 signaling [43]. The expression of PRKD3 promotes the progression of hepatocellular carcinoma and predicts a poor prognosis in the patients with hepatocellular carcinoma after hepatectomy [44].
PRKD3 inhibition in cancer therapy
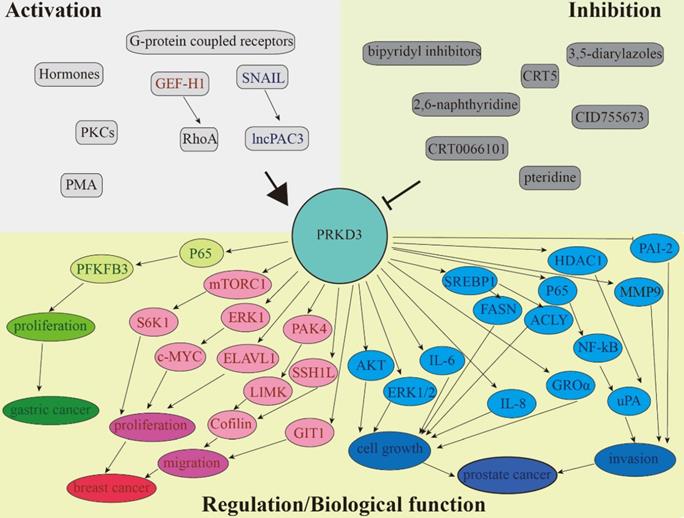
The emergence of PRKD3 as a potential therapeutic target for various cancers has encouraged the development of potent, selective, and small-molecule inhibitors. Several small-molecule inhibitors such as 2,6-naphthyridine and bipyridyl and analogs [45-47], CID755763 and analogs [48], 3,5-diarylazoles [49], pteridine [50], CRT5 [51], and CRT0066101 [52] were reported to inhibit PRKD in various cell lines. An issue with most of these chemical compounds is that although they are effective in blocking cell growth, and migration in vitro [53], they are quickly metabolized when administered to xenograft mouse models. Among all these compounds only CRT0066101 has been used successfully in xenograft mouse models of colorectal [52], pancreatic [54], and breast cancer [55]. In mice with TNBC, CRT0066101 significantly inhibited tumor growth without showing side effects [55]. More importantly, the similar results were obtained with specific inhibition of PRKD3 suggesting that PRKD3 is CRT0066101's main target in TNBC cells [28]. It is however possible, since CRT0066101 is administered orally, that some of the additional anti-cancer effects observed in the xenograft mouse model treated with CRT0066101 could be due to systemic inhibition of PRKD-mediated angiogenesis [56]. CRT0066101 seems to be a promising candidate since no harmful effects have been observed in all the tested models. For breast cancer treatments that do not express PRKD1, pan- inhibitors of PRKD could be even more effective if used in combination with PRKD2 or PRKD3's current chemotherapeutic agents have been associated to mediate multi-drug resistance [43, 57]. A potential problem with using pan-inhibitors is the management of detrimental off-target effects and to combat this problem the specificity of each compound must be fully investigated.
Alternative methods to chemical inhibition could include systemic delivery of siRNA or nucleic acid-based therapies that could allow to specifically target PRKDs [58, 59]. Such strategies have been successfully used suggesting that the therapeutic applications could be very promising in humans [60]. Last but not least, it would be promising to apply the new emerging proteolysis targeting chimera (PROTAC) for development of new drugs against PRKD3 for cancer treatment.
Conclusion and perspectives
There is a lot of evidence that PRKD3 is involved in the regulation of various signaling pathways, as well as in the integration of extracellular signals that promote migration, invasion, proliferation, and growth of cancer cells. This review summarized the various functions of PRKD3 in human tumors (Figure 2 and Table 1). Although many studies have confirmed the previously unknown mechanisms of PRKD3, it is still need for a better understanding of activation of different isoforms, isoform-specific functions, differential kinase expression and molecular cross-signaling. Delineation of potential compensatory effects between different PRKD subtypes in a specific cancer will help to improve the therapeutic prospects of PRKDs in a successful combinatorial molecular therapy approach.
The table shows the cancer-related functions of PRKD3 in a specific tumor type
| Tumor type | Cancer-related function | Activation/regulation | Reference |
|---|---|---|---|
| Breast | Proliferation | TORC-S6K1/ERK1-c-MYC | [29, 32] |
| ELAVL1 | [31, 33] | ||
| Migration | GIT1/PAK-LIMK-Cofilin | [34, 35] | |
| SSH1L-Coffilin | [36] | ||
| Prostate | Growth | AKT/ERK1/2/SREBP1-FASN | [37, 38] |
| SREBP1-ACLY/IL-6/IL-8 | [39] | ||
| GROα | |||
| Invasion | P65/NF-ΚB-uPA/HDAC1-uPA | [40, 41] | |
| MMP9/PAI-2 | |||
| Gastric | Proliferation | P65-PFKFB3 | [42] |
Acknowledgements
Authors' Contributions
Y.L, H.S, Y.Z, X.M and J.X wrote the manuscript. Z.Y and L.C critically reviewed the manuscript. All authors read and approved the final manuscript.
PRKD3 is involved in the regulation of various signaling pathways. Separate circuits show the ability of PRKD3 to promote cancer progression of gastric (green), breast (red), and prostate (blue).
Funding Sources
This study was funded by the Foundation for Young Scholars of Jiangsu Province, China (Grant No. BK20190194 awarded to Y. Liu), National Natural Science Foundation of China (Grant No.: 81974447 & 81572712 to L. Chen), Natural Science Fund for Distinguished Young Scholars of Jiangsu Province (Grant No.: SBK2020010058 to L. Chen), and Science and Technology Program of Nanjing (Grant No.: 201803052 & CLB20J023 to Z. Yu).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Johannes FJ, Prestle J, Eis S, Oberhagemann P, Pfizenmaier K. Pkcu Is a Novel, Atypical Member of the Protein-Kinase-C Family. J Biol Chem. 1994;269(8):6140-6148
2. Valverde AM, Sinnettsmith J, Vanlint J, Rozengurt E. Molecular-Cloning and Characterization of Protein-Kinase-D - a Target for Diacylglycerol and Phorbol Esters with a Distinctive Catalytic Domain. P Natl Acad Sci USA. 1994;91(18):8572-8576
3. Sturany S, Van Lint J, Muller F, Wilda M, Hameister H, Hocker M, Brey A, Gern U, Vandenheede J, Gress T. et al. Molecular cloning and characterization of the human protein kinase D2. A novel member of the protein kinase D family of serine threonine kinases. J Biol Chem. 2001;276(5):3310-3318
4. Hayashi A, Seki N, Hattori A, Kozuma S, Saito T. PKCnu, a new member of the protein kinase C family, composes a fourth subfamily with PKCmu. Biochim Biophys Acta. 1999;1450(1):99-106
5. Rozengurt E, Rey O, Waldron RT. Protein kinase D signaling. J Biol Chem. 2005;280(14):13205-13208
6. Wang QMJ. PKD at the crossroads of DAG and PKC signaling. Trends Pharmacol Sci. 2006;27(6):317-323
7. Rykx A, De Kimpe L, Mikhalap S, Vantus T, Seufferlein T, Vandenheede JR, Van Lint J. Protein kinase D: a family affair. FEBS Lett. 2003;546(1):81-86
8. Fu Y, Rubin CS. Protein kinase D: coupling extracellular stimuli to the regulation of cell physiology. EMBO Rep. 2011;12(8):785-796
9. Sanchez-Ruiloba L, Cabrera-Poch N, Rodriguez-Martinez M, Lopez-Menendez C, Jean-Mairet RM, Higuero AM, Iglesias T. Protein kinase D intracellular localization and activity control kinase D-interacting substrate of 220-kDa traffic through a postsynaptic density-95/discs large/zonula occludens-1-binding motif. J Biol Chem. 2006;281(27):18888-18900
10. Matthews SA, Iglesias T, Rozengurt E, Cantrell D. Spatial and temporal regulation of protein kinase D (PKD). EMBO J. 2000;19(12):2935-2945
11. Nhek S, Ngo M, Yang XM, Ng MM, Field SJ, Asara JM, Ridgway ND, Toker A. Regulation of Oxysterol-binding Protein Golgi Localization through Protein Kinase D-mediated Phosphorylation. Mol Biol Cell. 2010;21(13):2327-2337
12. Waldron RT, Rozengurt E. Protein kinase C phosphorylates protein kinase D activation loop Ser744 and Ser748 and releases autoinhibition by the pleckstrin homology domain. J Biol Chem. 2003;278(1):154-163
13. Storz P, Doppler H, Toker A. Protein kinase D mediates mitochondrion-to-nucleus signaling and detoxification from mitochondrial reactive oxygen species. Mol Cell Biol. 2005;25(19):8520-8530
14. Rey O, Yuan J, Rozengurt E. Intracellular redistribution of protein kinase D2 in response to G-protein-coupled receptor agonists. Biochem Biophys Res Commun. 2003;302(4):817-824
15. Rey O, Yuan J, Young SH, Rozengurt E. Protein kinase C nu/protein kinase D3 nuclear localization, catalytic activation, and intracellular redistribution in response to G protein-coupled receptor agonists. J Biol Chem. 2003;278(26):23773-23785
16. Wong C, Jin ZG. Protein kinase C-dependent protein kinase D activation modulates ERK signal pathway and endothelial cell proliferation by vascular endothelial growth factor. J Biol Chem. 2005;280(39):33262-33269
17. Zhukova E, Sinnett-Smith J, Rozengurt E. Protein kinase D potentiates DNA synthesis and cell proliferation induced by bombesin, vasopressin, or phorbol esters in Swiss 3T3 cells. J Biol Chem. 2001;276(43):40298-40305
18. Eiseler T, Doppler H, Yan IK, Goodison S, Storz P. Protein kinase D1 regulates matrix metalloproteinase expression and inhibits breast cancer cell invasion. Breast Cancer Res. 2009;11(1):R13
19. Eiseler T, Hausser A, De Kimpe L, Van Lint J, Pfizenmaier K. Protein Kinase D Controls Actin Polymerization and Cell Motility through Phosphorylation of Cortactin. J Biol Chem. 2010;285(24):18672-18683
20. Peterburs P, Heering J, Link G, Pfizenmaier K, Olayioye MA, Hausser A. Protein kinase D regulates cell migration by direct phosphorylation of the cofilin phosphatase slingshot 1 like. Cancer Res. 2009;69(14):5634-5638
21. Qin LL, Zeng HY, Zhao DZ. Requirement of protein kinase D tyrosine phosphorylation for VEGF-A(165)-induced angiogenesis through its interaction and regulation of phospholipase C gamma phosphorylation. J Biol Chem. 2006;281(43):32550-32558
22. Yeaman C, Ayala MI, Wright JR, Bard F, Bossard C, Ang A, Maeda Y, Seufferlein T, Mellman I, Nelson WJ. et al. Protein kinase D regulates basolateral membrane protein exit from trans-Golgi network. Nat Cell Biol. 2004;6(2):106-112
23. Ha CH, Wang W, Jhun BS, Wong C, Hausser A, Pfizenmaier K, McKinsey TA, Olson EN, Jin ZG. Protein kinase D-dependent phosphorylation and nuclear export of histone deacetylase 5 mediates vascular endothelial growth factor-induced gene expression and angiogenesis. J Biol Chem. 2008;283(21):14590-14599
24. Bastea LI, Doppler H, Balogun B, Storz P. Protein kinase D1 maintains the epithelial phenotype by inducing a DNA-bound, inactive SNAI1 transcriptional repressor complex. PLoS One. 2012;7(1):e30459
25. Du C, Zhang C, Hassan S, Biswas MHU, Balaji KC. Protein kinase D1 suppresses epithelial-to-mesenchymal transition through phosphorylation of Snail (vol 70, pg 7810, 2010). Cancer Research. 2010;70(23):10015-10015
26. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7-30
27. Borges S, Doppler H, Perez EA, Andorfer CA, Sun Z, Anastasiadis PZ, Thompson E, Geiger XJ, Storz P. Pharmacologic reversion of epigenetic silencing of the PRKD1 promoter blocks breast tumor cell invasion and metastasis. Breast Cancer Res. 2013;15(2):R66
28. Borges S, Perez EA, Thompson EA, Radisky DC, Geiger XJ, Storz P. Effective Targeting of Estrogen Receptor-Negative Breast Cancers with the Protein Kinase D Inhibitor CRT0066101. Mol Cancer Ther. 2015;14(6):1306-1316
29. Huck B, Duss S, Hausser A, Olayioye MA. Elevated protein kinase D3 (PKD3) expression supports proliferation of triple-negative breast cancer cells and contributes to mTORC1-S6K1 pathway activation. J Biol Chem. 2014;289(6):3138-3147
30. Hao Q, McKenzie R, Gan H, Tang H. Protein kinases D2 and D3 are novel growth regulators in HCC1806 triple-negative breast cancer cells. Anticancer Res. 2013;33(2):393-399
31. Liu Y, Li J, Zhang J, Yu Z, Yu S, Wu L, Wang Y, Gong X, Wu C, Cai X. et al. Oncogenic Protein Kinase D3 Regulating Networks in Invasive Breast Cancer. Int J Biol Sci. 2017;13(6):748-758
32. Liu Y, Song H, Yu S, Huang KH, Ma X, Zhou Y, Yu S, Zhang J, Chen L. Protein Kinase D3 promotes the cell proliferation by activating the ERK1/c-MYC axis in breast cancer. J Cell Mol Med. 2020;24(3):2135-2144
33. Liu Y, Li J, Ma Z, Zhang J, Wang Y, Yu Z, Lin X, Xu Z, Su Q, An L. et al. Oncogenic functions of protein kinase D2 and D3 in regulating multiple cancer-related pathways in breast cancer. Cancer Med. 2019;8(2):729-741
34. Lieb WS, Lungu C, Tamas R, Berreth H, Rathert P, Storz P, Olayioye MA, Hausser A. The GEF-H1/PKD3 signaling pathway promotes the maintenance of triple-negative breast cancer stem cells. Int J Cancer. 2020;146(12):3423-3434
35. Doppler H, Bastea LI, Borges S, Spratley SJ, Pearce SE, Storz P. Protein kinase d isoforms differentially modulate cofilin-driven directed cell migration. PLoS One. 2014;9(5):e98090
36. Huck B, Kemkemer R, Franz-Wachtel M, Macek B, Hausser A, Olayioye MA. GIT1 phosphorylation on serine 46 by PKD3 regulates paxillin trafficking and cellular protrusive activity. J Biol Chem. 2012;287(41):34604-34613
37. Chen J, Deng F, Singh SV, Wang QJ. Protein kinase D3 (PKD3) contributes to prostate cancer cell growth and survival through a PKCepsilon/PKD3 pathway downstream of Akt and ERK 1/2. Cancer Res. 2008;68(10):3844-3853
38. Li L, Hua L, Fan H, He Y, Xu W, Zhang L, Yang J, Deng F, Zeng F. Interplay of PKD3 with SREBP1 Promotes Cell Growth via Upregulating Lipogenesis in Prostate Cancer Cells. J Cancer. 2019;10(25):6395-6404
39. LaValle CR, Zhang L, Xu S, Eiseman JL, Wang QJ. Inducible silencing of protein kinase D3 inhibits secretion of tumor-promoting factors in prostate cancer. Mol Cancer Ther. 2012;11(7):1389-1399
40. He JH, Li BX, Han ZP, Zou MX, Wang L, Lv YB, Zhou JB, Cao MR, Li YG, Zhang JZ. Snail-activated long non-coding RNA PCA3 up-regulates PRKD3 expression by miR-1261 sponging, thereby promotes invasion and migration of prostate cancer cells. Tumour Biol. 2016
41. Zou Z, Zeng F, Xu W, Wang C, Ke Z, Wang QJ, Deng F. PKD2 and PKD3 promote prostate cancer cell invasion by modulating NF-kappaB- and HDAC1-mediated expression and activation of uPA. J Cell Sci. 2012;125(Pt 20):4800-4811
42. Zhang JW, Zhang YJ, Wang JQ, Zhang SS, Zhao YJ, Ren H, Chu YM, Feng L, Wang CF. Protein kinase D3 promotes gastric cancer development through p65/6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 activation of glycolysis. Exp Cell Res. 2019;380(2):188-197
43. Chen J, Shen Q, Labow M, Gaither LA. Protein kinase D3 sensitizes RAF inhibitor RAF265 in melanoma cells by preventing reactivation of MAPK signaling. Cancer Res. 2011;71(12):4280-4291
44. Yang H, Xu M, Chi X, Yan Q, Wang Y, Xu W, Zhuang K, Li A, Liu S. Higher PKD3 expression in hepatocellular carcinoma (HCC) tissues predicts poorer prognosis for HCC patients. Clin Res Hepatol Gastroenterol. 2017;41(5):554-563
45. Monovich L, Vega RB, Meredith E, Miranda K, Rao C, Capparelli M, Lemon DD, Phan D, Koch KA, Chapo JA. et al. A novel kinase inhibitor establishes a predominant role for protein kinase D as a cardiac class IIa histone deacetylase kinase. FEBS Lett. 2010;584(3):631-637
46. Meredith EL, Beattie K, Burgis R, Capparelli M, Chapo J, Dipietro L, Gamber G, Enyedy I, Hood DB, Hosagrahara V. et al. Identification of potent and selective amidobipyridyl inhibitors of protein kinase D. J Med Chem. 2010;53(15):5422-5438
47. Meredith EL, Ardayfio O, Beattie K, Dobler MR, Enyedy I, Gaul C, Hosagrahara V, Jewell C, Koch K, Lee W. et al. Identification of orally available naphthyridine protein kinase D inhibitors. J Med Chem. 2010;53(15):5400-5421
48. George KM, Frantz MC, Bravo-Altamirano K, Lavalle CR, Tandon M, Leimgruber S, Sharlow ER, Lazo JS, Wang QJ, Wipf P. Design, Synthesis, and Biological Evaluation of PKD Inhibitors. Pharmaceutics. 2011;3(2):186-228
49. Gamber GG, Meredith E, Zhu QM, Yan WL, Rao C, Capparelli M, Burgis R, Enyedy I, Zhang JH, Soldermann N. et al. 3,5-Diarylazoles as novel and selective inhibitors of protein kinase D. Bioorg Med Chem Lett. 2011;21(5):1447-1451
50. Tandon M, Salamoun JM, Carder EJ, Farber E, Xu S, Deng F, Tang H, Wipf P, Wang QJ. SD-208, a novel protein kinase D inhibitor, blocks prostate cancer cell proliferation and tumor growth in vivo by inducing G2/M cell cycle arrest. PLoS One. 2015;10(3):e0119346
51. Evans IM, Bagherzadeh A, Charles M, Raynham T, Ireson C, Boakes A, Kelland L, Zachary IC. Characterization of the biological effects of a novel protein kinase D inhibitor in endothelial cells. Biochem J. 2010;429(3):565-572
52. Wei N, Chu E, Wipf P, Schmitz JC. Protein kinase d as a potential chemotherapeutic target for colorectal cancer. Mol Cancer Ther. 2014;13(5):1130-1141
53. Lavalle CR, Bravo-Altamirano K, Giridhar KV, Chen J, Sharlow E, Lazo JS, Wipf P, Wang QJ. Novel protein kinase D inhibitors cause potent arrest in prostate cancer cell growth and motility. BMC Chem Biol. 2010;10:5
54. Harikumar KB, Kunnumakkara AB, Ochi N, Tong Z, Deorukhkar A, Sung B, Kelland L, Jamieson S, Sutherland R, Raynham T. et al. A novel small-molecule inhibitor of protein kinase D blocks pancreatic cancer growth in vitro and in vivo. Mol Cancer Ther. 2010;9(5):1136-1146
55. Liu Y, Wang Y, Yu S, Zhou Y, Ma X, Su Q, An L, Wang F, Shi A, Zhang J. et al. The Role and Mechanism of CRT0066101 as an Effective Drug for Treatment of Triple-Negative Breast Cancer. Cell Physiol Biochem. 2019;52(3):382-396
56. Rozengurt E. Protein kinase D signaling: multiple biological functions in health and disease. Physiology (Bethesda). 2011;26(1):23-33
57. Chen JA, Lu LB, Feng Y, Wang H, Dai LL, Li Y, Zhang P. PKD2 mediates multi-drug resistance in breast cancer cells through modulation of P-glycoprotein expression. Cancer Lett. 2011;300(1):48-56
58. Bertoli G, Cava C, Castiglioni I. MicroRNAs: New Biomarkers for Diagnosis, Prognosis, Therapy Prediction and Therapeutic Tools for Breast Cancer. Theranostics. 2015;5(10):1122-1143
59. Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M. et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441(7089):111-114
60. Yan LX, Wu QN, Zhang Y, Li YY, Liao DZ, Hou JH, Fu J, Zeng MS, Yun JP, Wu QL. et al. Knockdown of miR-21 in human breast cancer cell lines inhibits proliferation, in vitro migration and in vivo tumor growth. Breast Cancer Res. 2011;13(1):R2
Author contact
![]() Corresponding authors: Liming Chen, E-mail:chenliming1981edu.cn; Zhenghong Yu, E-mail:13327800182cn.
Corresponding authors: Liming Chen, E-mail:chenliming1981edu.cn; Zhenghong Yu, E-mail:13327800182cn.

 Global reach, higher impact
Global reach, higher impact