3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(8):2199-2205. doi:10.7150/jca.50630 This issue Cite
Research Paper
Proteomic profiling reveals a signature for optimizing prognostic prediction in Colon Cancer
1. Department of Colorectal Surgery, Fudan University Shanghai Cancer Center, Shanghai 200032, China.
2. Department of Oncology, Shanghai Medical College, Fudan University, Shanghai 200032, China.
*These authors contributed equally to this work.
Received 2020-7-13; Accepted 2020-12-26; Published 2021-2-22
Abstract
Previous studies developed prognostic signatures largely depended on transcriptome profiles. The purpose of our present study was to develop a proteomic signature to optimize the evaluation of prognosis of colon cancer patients. The proteomic data of colon cancer patient cohorts were downloaded from The Cancer Proteome Atlas (TCPA). Patients were randomized 3:2 to train set and internal validation set. Univariate Cox regression and lasso Cox regression analysis were performed to identify the prognostic proteins. A four-protein signature was developed to divide patients into a high-risk group and low-risk group with significantly different survival outcomes in both train set and internal validation set. Time-dependent receiver-operating characteristic at 1 year demonstrated that the proteomic signature presented more prognostic accuracy [area under curve (AUC = 0.704)] than the American Joint Commission on Cancer tumor-node-metastasis (AJCC-TNM) staging system (AUC = 0.681) in entire set. In conclusion, we developed a proteomic signature which can improve prognostic accuracy of patients with colon cancer and optimize the therapeutic and follow-up strategies.
Keywords: proteomic profiling, colon cancer, prognosis
Introduction
Colon cancer is one of the most common malignancies worldwide [1]. Radical surgery alone or combined with adjuvant chemotherapy is the standard regimen for management of colon cancer without distant metastasis. However, about 25%-40% patients will suffer recurrence and metastasis after receiving standardized treatment [2]. The early detection and management of relapse contribute to improve prognosis. To date, prognostic prediction is largely depended on the tumor, lymph node, metastasis (TNM) staging system [3]. On this basis, novel prognostic models have been developed to improve prognostic prediction and optimize the therapeutic and follow-up strategies using clinicopathologic and genetic factors [4, 5]. Notably, with the advance of genome-sequencing technologies, gene signatures at mRNA level presented an excellent prediction of colon cancer prognosis [6-8]. However, only limited studies developed signatures at the level of protein to guide patients' prognostic stratification [9, 10].
Mass spectrometry-based proteomics can detect global protein abundance and post-translational modifications and provide comprehensive biological perspectives, which could not be replaced by genomic analysis alone [11, 12]. In the present study, we identified robust prognostic proteins and constructed a proteomic classifier in colon cancer using The Cancer Proteome Atlas (TCPA) database. To our best knowledge, the proteomic signatures have not been reported in colon cancer previously.
Materials and Methods
The proteomic data (Level 4) of colon cancer patient cohorts (COAD) were downloaded from The Cancer Proteome Atlas (TCPA) (https://www.tcpaportal.org). Package impute (Bioconductor) was applied to impute the missing values. The clinical data were downloaded from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov).
Development and validation of a proteomic signature
Firstly, patients were randomized 3:2 to train series and internal validation series. Univariate Cox regression analysis was performed to identify the biomarkers for prognosis in train set. Proteins with significant differences (p<0.05) were selected for LASSO Cox regression model. Finally, a multivariate Cox regression analysis was conducted to construct a multi-protein-based classifier for prognostic prediction of colon cancer patients. According to specific risk score formula, patients were divided into high-risk and low-risk groups with significantly different survival outcomes using the median value of the train series as the cutoff point.
Statistical analysis
Kaplan-Meier curve was depicted to compare survival differences between high-risk group and low-risk group. Multivariate Cox regression analysis was conducted to identify independent prognostic factors. Receiver-operating characteristic (ROC) curve was plotted to evaluate the prognostic or predictive accuracy of the proteomic signature and clinicopathological factors. All statistical analyses were performed with R (version 3.6.1, https://www.r-project.org/).
Results
Development of a proteomic signature from the train set
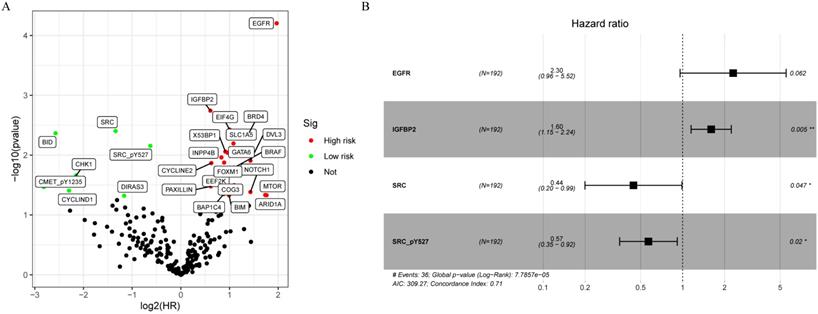
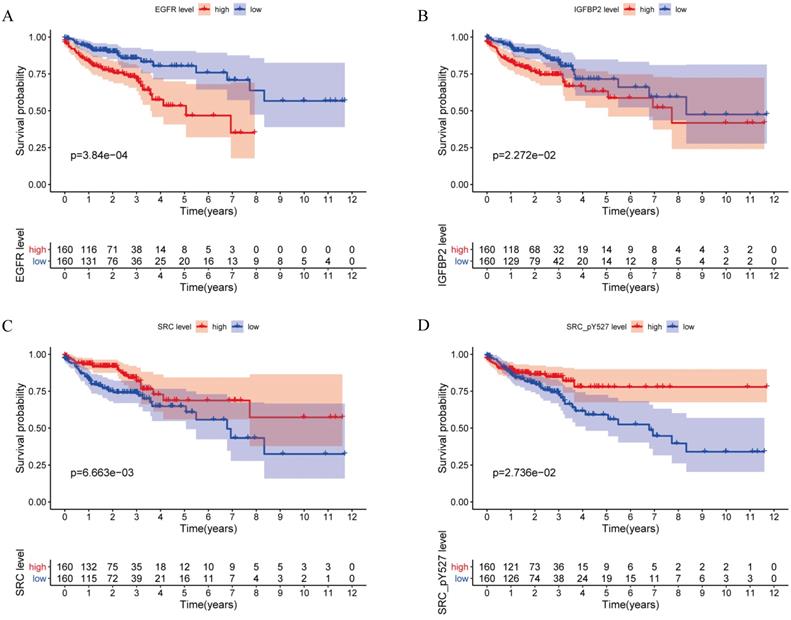
A total of 315 colon cancer patients with complete proteomic profiling and survival data were included in our study. Patients were randomized 3:2 to train set and internal validation set. Twenty-five robust prognostic proteins were identified using univariate Cox regression analysis (Figure 1A). Lasso Cox regression and stepwise multivariate Cox regression were performed to construct a proteomic signature. Finally, a four-protein signature was developed and forest plot was presented (Figure 1B). The risk score = (0.834071775625132 × expression level of EGFR) + (0.471960975428313 × expression level of IGFBP2) + (-0.810781951083818 × expression level of SRC) + (-0.563796255046605 × expression level of SRC_pY527). Kaplan-Meier curves were plotted for each protein using the median value of the protein as the cutoff point. High expression of EGFR or IGFBP2 was associated with poor prognosis while high expression of SRC or SRC_pY527 predicted superior survival in colon cancer (Figure 2).
The prognostic value of the four-protein signature in train set and internal validation set
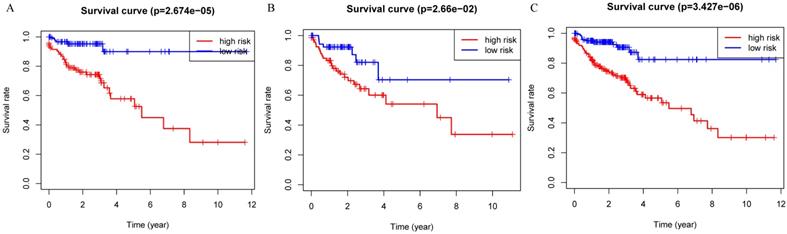
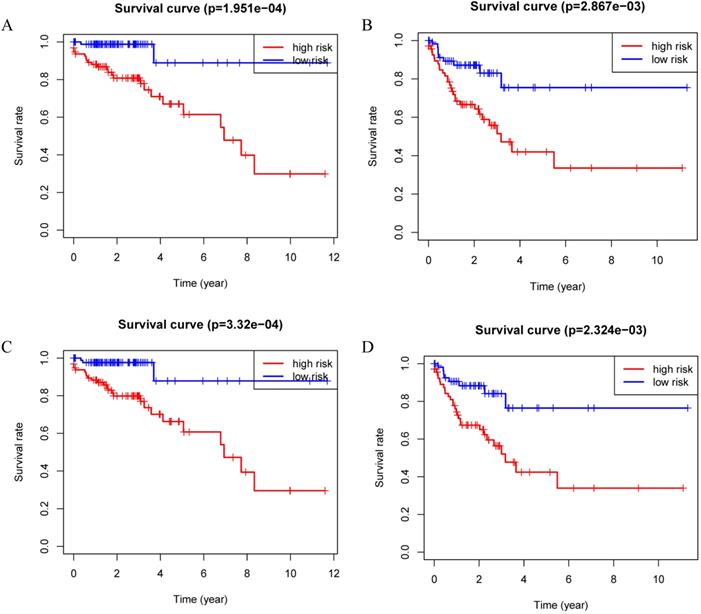
Patients were divided into a low-risk group and high-risk group using the median risk score as the cutoff value. Patients with higher risk scores had a worse prognosis as compared to those with lower risk scores in train set, internal validation set and entire set (Figure 3). Stratified analysis revealed that the four-protein signature still had prognostic values in stage I+II, stage III+IV, lymph node positive and lymph node negative subgroups (Figure 4). The distribution of the proteomic risk score, the survival status of patients and heatmap of the proteomic expression profiles were also presented (Figure S1, S2 and S3).
Development of a proteomic signature. A. Volcano plot of univariate Cox regression analysis. B. Forest plot of the multivariate Cox regression analysis.
Kaplan-Meier curves for EGFR, IGFBP2, SRC and SRC_pY527.
Kaplan-Meier curves for the proteomic signature. A. Train set; B. Internal validation set; C. Entire set.
Independence and accuracy of the proteomic signature in predicting prognosis
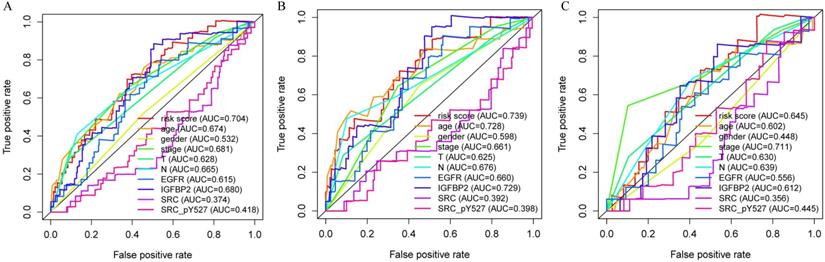
Multivariate analysis showed that our proteomic signature remained an independent prognostic factor in entire set (Table 1). Clinicopathological characteristics of colon cancer patients in TCPA database were detailed in Table S1. Additionally, we performed ROC analysis to compare the sensitivity and specificity of prognostic prediction among proteomic signature, single protein, age, gender, T stage, N stage and AJCC stage. Time-dependent receiver-operating characteristic at 1 year demonstrated that the proteomic signature presented more prognostic accuracy [area under curve (AUC = 0.704)] than the American Joint Commission on Cancer tumor-node-metastasis (AJCC-TNM) staging system (AUC = 0.681) in entire set (Figure 5A). The AUC values in train and internal validation set were also presented and compared (Figure 5B, 5C).
Kaplan-Meier curves for the proteomic signature in subgroups. A. Stage I and stage II; B. Stage III and stage IV; C. Lymph node negative; D. Lymph node positive.
Receiver operating characteristic (ROC) analysis of the sensitivity and specificity of the proteomic signature, each protein and clinicopathological features. A. Entire set; B. Train set; C. Internal validation set.
Constructing protein co-expression networks
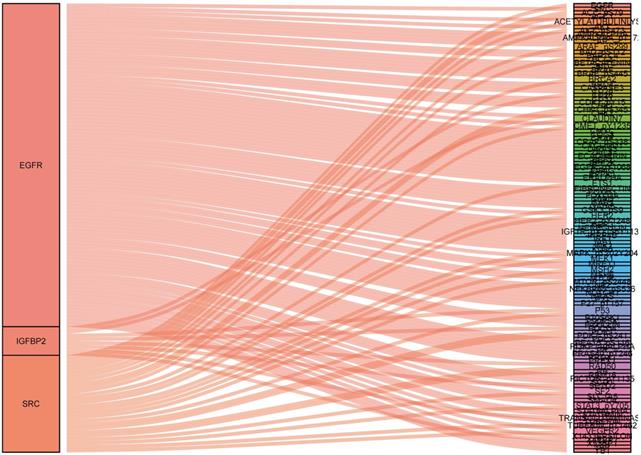
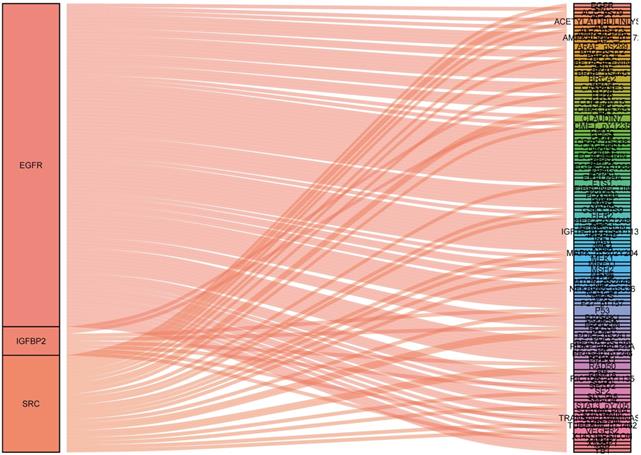
To identify what proteins were significantly associated with the expression of EGFR, IGFBP2, SRC and SRC_pY527 (R>0.2 or R<-0.2, p<0.05), sankey diagram was plotted (Figure 6). EGFR had more co-expressive proteins than the others.
Univariable and multivariable Cox regression analysis in colon cancer
| Variable | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| Proteomic signature | 1.207 (1.122-1.298) | <0.001 | 1.158 (1.070-1.254) | <0.001 |
| Age | 1.053 (1.025-1.081) | <0.001 | 1.053 (1.026-1.080) | <0.001 |
| Gender | 0.913 (0.558-1.493) | 0.717 | 1.095 (0.658-1.820) | 0.727 |
| Stage | 1.957 (1.474-2.598) | <0.001 | 2.201 (1.629-2.975) | <0.001 |
Sankey diagram of the correlations between proteins.
Discussion
Integrated transcriptome profiling of colon cancer has increased our knowledge of molecular features relevant to carcinogenesis. A number of studies developed and validated multigene signatures to predict prognosis based on global mRNAs profiling. Recently, global proteomic data which provided novel insights into the comprehensive understanding of cancers, have become focus of attention [13]. Compared with a single proteomic biomarker, the combination of the prognostic proteins may have better predictive efficacy.
In this study, we established a novel proteomic signature (including EGFR, IGFBP2, SRC and SRC_pY527) for prognostic prediction of colon cancer using TCPA database. The survival curves revealed a significant separation between low-risk and high-risk patients in both training set and internal validation set. Stratified by AJCC stage and lymph node status, the proteomic signature remained an excellent prognostic model. Time-dependent ROC at 1 year demonstrated that our signature had the most significant accuracy in predicting prognosis as compared to other indicators, indicating that the risk model developed from the four proteins could be a useful tool for colon cancer survival prediction.
Epidermal growth factor receptor (EGFR), a member of the subclass I of the receptor tyrosine kinase super-family, is overexpressed in 49% to 82% of colorectal cancer [14-16]. EGFR is one of the most promising targets for the management of metastatic colorectal cancer. However, EGFR testing for colorectal cancer patients has no predictive value of response to EGFR inhibitors [17, 18]. The RAS/RAF/MAPK pathway is downstream of EGFR. Evidence indicated that RAS and BRAF status had predictive value of response to cetuximab or panitumumab therapy [19-21].
Insulin-like growth factor-binding protein 2 (IGFBP2) is a member of the IGFBP family that bind IGFs with high affinity [22]. Previous studies suggested that IGFBP2 expression was upregulated in multiple tumors [23-25]. Recently, Liu et al reported that IGFBP2 promoted vasculogenic mimicry formation via targeting CD144 and MMP2 expression in glioma [26]. Gao et al indicated that IGFBP2 could drive epithelial-mesenchymal transition and invasive character by activating the NF-κB pathway in pancreatic ductal adenocarcinoma [27]. Nevertheless, the function and mechanisms of IGFBP2 in colon cancer remains unclear. Our study revealed that high expression IGFBP2 indicated poor prognosis.
Proto-oncogene tyrosine-protein kinase Src contains an SH3 domain, an SH2 domain, a protein-tyrosine kinase domain, and a regulatory tail and participates in multiple biological processes [28]. Src could be activated by upstream signaling pathways to form phospho-Src (p-Src), and p-Src could activate downstream signaling pathways by phosphorylating the target proteins [29, 30]. Recent studies suggested that Src family kinases were involved in carcinogenesis. Hu et al demonstrated that the expression of Src and p-Src was significantly upregulated in osteochondroma and could be used as robust indicators to predict prognosis [31]. Singh et al found that Src and p-Src could promote colon cancer invasion and metastasis [32]. Intriguingly, our study indicated that high expression of SRC and SRC_pY527 was associated with superior prognosis in colon cancer patients. Further precise regulation mechanisms of SRC and SRC_pY527 in colon cancer are needed to be explored.
Finally, we also identified several proteins which significantly associated with the expression of EGFR, IGFBP2, SRC and SRC_pY527. These proteins should also be further explored. Several limitations are needed to address in our present study. Firstly, only over two hundred proteins were identified in TCPA database, the information about a lot of critical proteins were missing. Secondly, the lack of external validation resulted in limited clinical value of our signature. Further research regarding its external validation and clinical utility are needed. Lastly, molecular biology experiments are necessary for clarifying the underlying molecular mechanism of our proteomic signature.
In conclusion, our study established a novel proteomic signature for improving prognostic prediction in colon cancer, which may assist to develop individual therapeutic and follow-up strategies.
Supplementary Material
Supplementary figures.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant NO. 81972260; NO. 81772599; NO. 81702353) and Shanghai Municipal Natural Science Foundation (17ZR1406400). The funders had no role in the study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Ethics approval and consent to participate
This study was approved by the Ethical Committee and Institutional Review Board of the FUSCC. This study was performed based on the publicly available TCPA/TCGA database. We did not use personal identifying information. The informed consent was not necessary for this study.
Author Contribution Statements
XL, YM and RW contributed to conception and design. ZS and DL improved the study design and contributed to the interpretation of results. YM collected the data. SC performed data processing and statistical analysis. DL and ZS wrote the manuscript. YM revised the manuscript. All authors approved the final version.
Data Statement
The datasets included in our current study are available in the TCGA-COAD (https://portal.gdc.cancer.gov) and The Cancer Proteome Atlas (https://tcpaportal.org/).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians. 2018;68:394-424
2. Tjandra JJ, Chan MK. Follow-up after curative resection of colorectal cancer: a meta-analysis. Dis Colon Rectum. 2007;50:1783-99
3. Weiser MR, Gonen M, Chou JF, Kattan MW, Schrag D. Predicting survival after curative colectomy for cancer: individualizing colon cancer staging. J Clin Oncol. 2011;29:4796-802
4. Tong D, Tian Y, Zhou T, Ye Q, Li J, Ding K. et al. Improving prediction performance of colon cancer prognosis based on the integration of clinical and multi-omics data. BMC Med Inform Decis Mak. 2020;20:22
5. Wang Z, Wang Y, Yang Y, Luo Y, Liu J, Xu Y. et al. A competing-risk nomogram to predict cause-specific death in elderly patients with colorectal cancer after surgery (especially for colon cancer). World J Surg Oncol. 2020;18:30
6. Yang H, Lin HC, Liu H, Gan D, Jin W, Cui C. et al. A 6 lncRNA-Based Risk Score System for Predicting the Recurrence of Colon Adenocarcinoma Patients. Front Oncol. 2020;10:81
7. Ma R, Zhao Y, He M, Zhao H, Zhang Y, Zhou S. et al. Identifying a ten-microRNA signature as a superior prognosis biomarker in colon adenocarcinoma. Cancer Cell Int. 2019;19:360
8. Dai W, Feng Y, Mo S, Xiang W, Li Q, Wang R. et al. Transcriptome profiling reveals an integrated mRNA-lncRNA signature with predictive value of early relapse in colon cancer. Carcinogenesis. 2018;39:1235-44
9. Kwon OK, Ha YS, Na AY, Chun SY, Kwon TG, Lee JN. et al. Identification of Novel Prognosis and Prediction Markers in Advanced Prostate Cancer Tissues Based on Quantitative Proteomics. Cancer Genomics Proteomics. 2020;17:195-208
10. Ku X, Xu Y, Cai C, Yang Y, Cui L, Yan W. In-Depth Characterization of Mass Spectrometry-Based Proteomic Profiles Revealed Novel Signature Proteins Associated with Liver Metastatic Colorectal Cancers. Anal Cell Pathol (Amst). 2019;2019:7653230
11. Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi Z. et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513:382-7
12. Gao Q, Zhu H, Dong L, Shi W, Chen R, Song Z. et al. Integrated Proteogenomic Characterization of HBV-Related Hepatocellular Carcinoma. Cell. 2019;179:561-77 e22
13. Ellis MJ, Gillette M, Carr SA, Paulovich AG, Smith RD, Rodland KK. et al. Connecting genomic alterations to cancer biology with proteomics: the NCI Clinical Proteomic Tumor Analysis Consortium. Cancer Discov. 2013;3:1108-12
14. McKay JA, Murray LJ, Curran S, Ross VG, Clark C, Murray GI. et al. Evaluation of the epidermal growth factor receptor (EGFR) in colorectal tumours and lymph node metastases. Eur J Cancer. 2002;38:2258-64
15. Yen LC, Uen YH, Wu DC, Lu CY, Yu FJ, Wu IC. et al. Activating KRAS mutations and overexpression of epidermal growth factor receptor as independent predictors in metastatic colorectal cancer patients treated with cetuximab. Ann Surg. 2010;251:254-60
16. Spano JP, Lagorce C, Atlan D, Milano G, Domont J, Benamouzig R. et al. Impact of EGFR expression on colorectal cancer patient prognosis and survival. Ann Oncol. 2005;16:102-8
17. Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A. et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337-45
18. Saltz LB, Meropol NJ, Loehrer PJ Sr, Needle MN, Kopit J, Mayer RJ. Phase II trial of cetuximab in patients with refractory colorectal cancer that expresses the epidermal growth factor receptor. J Clin Oncol. 2004;22:1201-8
19. Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ. et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626-34
20. De Roock W, Piessevaux H, De Schutter J, Janssens M, De Hertogh G, Personeni N. et al. KRAS wild-type state predicts survival and is associated to early radiological response in metastatic colorectal cancer treated with cetuximab. Ann Oncol. 2008;19:508-15
21. Sorich MJ, Wiese MD, Rowland A, Kichenadasse G, McKinnon RA, Karapetis CS. Extended RAS mutations and anti-EGFR monoclonal antibody survival benefit in metastatic colorectal cancer: a meta-analysis of randomized, controlled trials. Ann Oncol. 2015;26:13-21
22. Chua CY, Liu Y, Granberg KJ, Hu L, Haapasalo H, Annala MJ. et al. IGFBP2 potentiates nuclear EGFR-STAT3 signaling. Oncogene. 2016;35:738-47
23. Du Y, Wang P. Upregulation of MIIP regulates human breast cancer proliferation, invasion and migration by mediated by IGFBP2. Pathol Res Pract. 2019;215:152440
24. Hu X, Chen M, Liu W, Li Y, Fu J. Preoperative plasma IGFBP2 is associated with nodal metastasis in patients with penile squamous cell carcinoma. Urol Oncol. 2019;37:452-61
25. Shen F, Song C, Liu Y, Zhang J, Wei Song S. IGFBP2 promotes neural stem cell maintenance and proliferation differentially associated with glioblastoma subtypes. Brain Res. 2019;1704:174-86
26. Liu Y, Li F, Yang YT, Xu XD, Chen JS, Chen TL. et al. IGFBP2 promotes vasculogenic mimicry formation via regulating CD144 and MMP2 expression in glioma. Oncogene. 2019;38:1815-31
27. Gao S, Sun Y, Zhang X, Hu L, Liu Y, Chua CY. et al. IGFBP2 Activates the NF-kappaB Pathway to Drive Epithelial-Mesenchymal Transition and Invasive Character in Pancreatic Ductal Adenocarcinoma. Cancer Res. 2016;76:6543-54
28. Roskoski R Jr. Src protein-tyrosine kinase structure, mechanism, and small molecule inhibitors. Pharmacol Res. 2015;94:9-25
29. Zhang XT, Ding L, Kang LG, Wang ZY. Involvement of ER-alpha36, Src, EGFR and STAT5 in the biphasic estrogen signaling of ER-negative breast cancer cells. Oncol Rep. 2012;27:2057-65
30. Fan P, McDaniel RE, Kim HR, Clagett D, Haddad B, Jordan VC. Modulating therapeutic effects of the c-Src inhibitor via oestrogen receptor and human epidermal growth factor receptor 2 in breast cancer cell lines. Eur J Cancer. 2012;48:3488-98
31. Hu C, Deng Z, Zhang Y, Yan L, Cai L, Lei J. et al. The prognostic significance of Src and p-Src expression in patients with osteosarcoma. Med Sci Monit. 2015;21:638-45
32. Singh AB, Sharma A, Dhawan P. Claudin-1 expression confers resistance to anoikis in colon cancer cells in a Src-dependent manner. Carcinogenesis. 2012;33:2538-47
Author contact
![]() Corresponding author: Xinxiang Li, E-mail: 1149lxxcom; Yanlei Ma, E-mail: yanleimaedu.cn and Renjie Wang, E-mail: wangbladejaycom.
Corresponding author: Xinxiang Li, E-mail: 1149lxxcom; Yanlei Ma, E-mail: yanleimaedu.cn and Renjie Wang, E-mail: wangbladejaycom.

 Global reach, higher impact
Global reach, higher impact