Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(11):3222-3229. doi:10.7150/jca.51405 This issue Cite
Research Paper
Targeted Sequencing Analysis of Predominant Histological Subtypes in Resected Stage I Invasive Lung Adenocarcinoma
1. Department of Respiratory Medicine, The Third Affiliated Hospital of Soochow University, 185 Juqian Road, Changzhou, China.
2. Department of Pathology, The Third Affiliated Hospital of Soochow University, 185 Juqian Road, Changzhou, China.
3. Burning Rock Biotech, Guangzhou, Guangdong, 510300, China.
4. Department of Respiratory Medicine, The Affiliated Hospital of Xuzhou Medical University, 99 Huaihai Road, Xuzhou, China.
*These authors contributed equally to this article.
Received 2020-8-2; Accepted 2021-3-15; Published 2021-4-2
Abstract
Objective: Lung adenocarcinoma (LADC) is classified into five main histological subtypes with distinct clinicopathologic characteristics: lepidic-predominant adenocarcinoma (LPA), acinar-predominant adenocarcinoma (APA), papillary-predominant adenocarcinoma (PPA), micropapillary-predominant adenocarcinoma (MPA) and solid-predominant adenocarcinoma (SPA). However, the mutational profiles of predominant histological subtypes have not been well defined. In this study, we aimed to reveal the genomic landscape of 5 main histological subtypes.
Patients and Methods: We performed next-generation sequencing (NGS) in a cohort of 86 stage I invasive adenocarcinoma (IAC) patients, using a customized panel including 168 cancer-associated genes.
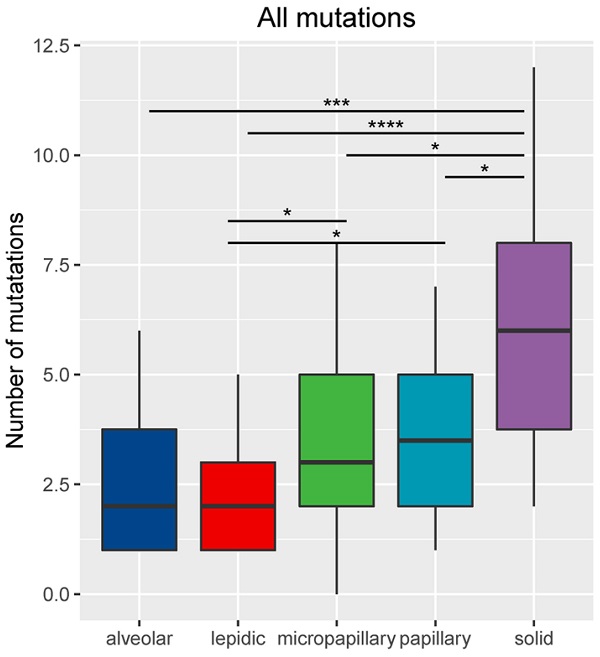
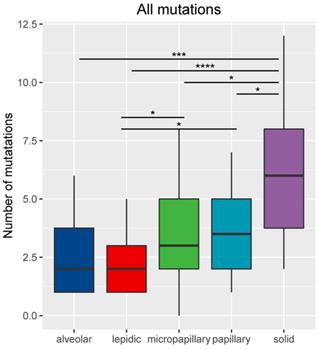
Results: Our analysis identified a total of 302 genomic alterations. Five subtypes showed different mutation profiles with LPA, APA, PPA, MPA and SPA had an average mutation rate of 1.95 (range: 0-5), 2.56 (range: 1-6), 3.5 (range: 1-7), 3.75 (range: 1-8) and 6.05 (range: 2-12), respectively (p=4.17e-06).
Driver mutations occurred in 96.55% (83/86) of all patients. EGFR (73.3%), KRAS (9.3%), ALK (4.7%) and MET (4.7%) are the most commonly mutated lung cancer driver genes, TP53 is the top mutated tumor suppressor gene. SPA patients harbored more driver mutations and higher frequency of TP53 than LPA patients. Interestingly, LRP1B mutations, which has been reported to be associated with high tumor mutation burden and better response to immunotherapy, were only detected from 5 SPA patients (p=0.001). No patients from other four cohorts harbored LRP1B mutations.
Conclusions: We revealed distinctive mutation landscape of the 5 major histological subtypes of LADC, evident by distinctive average mutation rate with SPA and LPA having the highest and lowest average mutation rate, respectively. SPA patients showed higher mutation rate of LRP1B and higher rates for PD-L1 positivity, indicating that SPA patients may have better response to immunotherapy.
Keywords: adenocarcinoma, pathological subtypes, next-generation sequencing, mutational profile, immunotherapy
Introduction
LADC is a heterogeneous tumor, accounting for almost half of all lung cancers. In 2011, the International Association for the Study of Lung Cancer, the American Thoracic Society, and the European Respiratory Society (IASLC/ATS/ERS) proposed a new histologic subtyping system for LADC. IAC was further classified into five major histologic subtypes: LPA, APA, PPA, MPA and SPA [1]. The recently updated 2015 World Health Organization (WHO) classification of lung tumor is generally consistent with the 2011 IALSC/ATS/ERS classification [2]. Studies have well demonstrated that there is an association between IAC subtypes and survival [3-5]. Lepidic-predominant subtype was significantly related to the absence of lymph node metastasis [6] and associated with the most favorable prognostic outcome. Micropapillary and solid predominant subtypes were predictors of increased frequency of lymph node metastasis [7-10] and significantly related to disease recurrence and death [11-17]. Acinar and papillary predominant subtypes had an intermediate prognosis.
Histological subtypes correlate with molecular changes. Numerous studies have evaluated the association between EGFR/KRAS/ALK mutation status and histological subtype. EGFR mutations more commonly occurred in the papillary and micropapillary subtype [18-20], and less commonly occurred in the solid subtype [21]. KRAS mutations and ALK rearrangements were associated with the solid predominant subtype [21, 22].
However, previous studies have focused on several common driver genes, the mutational profiles of predominant histological subtypes have not been well defined. In this study, we aimed to reveal the genomic landscape of the 5 main histological subtypes by targeting 168 cancer-related genes.
Methods
Patients and sample collection
86 patients diagnosed with pathologic stage I (T1-2aN0M0) invasive lung adenocarcinoma were enrolled between January 2015 and December 2018. No patients had received any preoperative chemotherapy or radiotherapy. Tumor staging was performed according to the 8th edition of the TNM classification of the international association for the study of lung cancer (IASLC).
For each patient, surgically resected tumor was obtained immediately post-surgery. Hematoxylin-eosin (H&E) staining was performed on the tissue obtained. Two pathologists estimated and marked the predominant pattern for each tissue sample, according to the 2015 WHO criteria. The predominant pattern was defined as the pattern that occupied most of the tumor, because of small sample size, MPA was defined as an adenocarcinoma with micropapillary component exceeded 10% of the entire tumor area. The number of each subtypes were as follows: LPA (n=20), APA (n=18), PPA (n=16), SPA (n=16) and MPA (n=16). This study was approved by the Review Broad of the Third Affiliated Hospital of Soochow University.
PD-L1 Staining
PD-L1 expression of tumor cells of each sample were determined by IHC using the PD-L1 specific DAKO 22C3 antibody, and was evaluated by a tumor proportion score (TPS). The specimens were considered PD-L1+ (TPS ≥1%) and high PD-L1+ (TPS ≥50%).
Tissue DNA isolation and capture-based targeted DNA sequencing
Tissue DNA was extracted from the predominant pattern marked in each tissue sample using QIAamp DNA FFPE tissue kit (Qiagen) following manufacturer's instructions. A minimum of 50 ng of DNA is required for NGS library construction. Tissue DNA was sheared using Covaris M220 (Covaris, MA, USA), followed by end repair, phosphorylation and adaptor ligation. Fragments between 200-400bp from the sheared tissue DNA were purified (Agencourt AMPure XP Kit, Beckman Coulter, CA, USA), followed by hybridization with capture probes baits, hybrid selection with magnetic beads and PCR amplification. The quality and the size of the fragments were assessed using Qubit 2.0 Fluorimeter with the dsDNA high-sensitivity assay kit (Life Technologies, Carlsbad, CA). Indexed samples were sequenced on Nextseq500 (Illumina, Inc., USA) with paired-end reads and average sequencing depth of 1,000X for tissue samples. A panel of 168 genes including 68 lung cancer-related genes and 100 other genes related to cancer development, spanning 0.273 megabases (Mb) of the human genome, were used for targeted sequencing (Lung Plasma, Burning Rock Biotech, Guangzhou, China).
Sequence data analysis
Sequence data were mapped to the reference human genome (hg19) using Burrows-Wheeler Aligner v.0.7.10 [23]. Local alignment optimization, duplication marking and variant calling were performed using Genome Analysis Tool Kit v.3.2 [24], and VarScan v.2.4.3 [25]. Variants were filtered using the VarScan fpfilter pipeline, loci with depth less than 100 were filtered out. Base-calling in tissue samples required at least 8 supporting reads for single nucleotide variations (SNV) and 5 supporting reads for insertion-deletion variations (INDEL). Variants with population frequency over 0.1% in the ExAC, 1000 Genomes, dbSNP or ESP6500SI-V2 databases were grouped as single nucleotide polymorphisms (SNP) and excluded from further analysis. Remaining variants were annotated with ANNOVAR (2016-02-01 release) [26] and SnpEff v.3.6 [27]. Analysis of DNA translocation was performed using Factera v.1.4.3 [28]. Copy number variations (CNV) were analyzed based on the depth of coverage data of capture intervals. Coverage data were corrected against sequencing bias resulting from GC content and probe design. The average coverage of all captured regions was used to normalize the coverage of different samples to comparable scales. Copy number was calculated based on the ratio between the depth of coverage in tumor samples and average coverage of an adequate number (n>50) of samples without CNV as references per capture interval. CNV is called if the coverage data of the gene region was quantitatively and statistically significant from its reference control. The limit of detection for CNVs is 1.5 and 2.64 for deletions and amplifications, respectively.
Survival analysis
The Kaplan-Meier method was used for survival rate estimation. The log-rank test was used for the comparison of survival curves between three groups. All statistical analysis was performed using R. All tests were two-sided and had a significance level of 0.05.
Results
Characteristics of IAC patients
Clinical characteristics of 86 IAC patients are summarized in Table 1. The median age of all patients was 64 years with a range of 36-78 years. There was no significant difference in age (P=0.511), smoking status (P=0.061) and vascular invasion (P=0.402) between the five groups. SPA group had more male (P=0.031) and stage Ⅰ B patients (P=0.014).
PD-L1 expression status
The positive expression rate of PD-L1 was 5% (1/20), 11.1% (2/18), 43.8% (7/16), 25% (4/16), 62.5% (10/16) in LPA, APA, PPA, MPA and SPA, respectively (P=0.002). In addition, 5.6% (1/18) of APA, 12.5% (2/16) PPA, 6.3% (1/16) of MPA and 18.8% (3/16) of SPA had high PD-L1 TPS. None of LPA had high PD-L1 TPS (P<0.0001) (Table 1).
Survival analysis
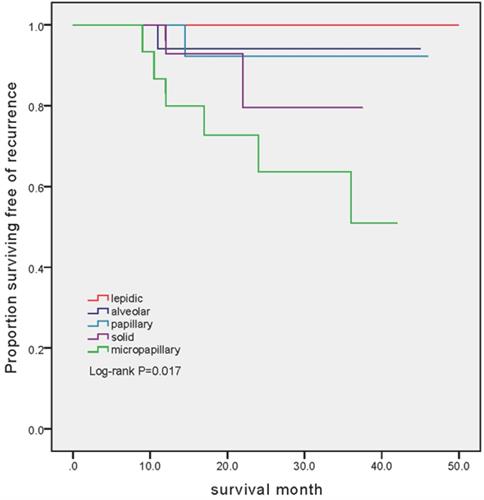
The mean follow-up was 26.3 months (ranged from 8.5 to 50 months). Among 86 patients, 10 (11.6%) recurrences were identified. Disease progressions were markedly different between cohorts (P=0.017). The number of patients developed recurrent disease was 6 (37.5%), 2 (12.5%), 1 (5.56%) and 1 (6.25%) in MPA, SPA, APA and PAA group, respectively. Notably, there was no patient experienced recurrent disease in LPA group (Figure 1).
Patient clinical characteristics
| n=86 | n=20 | n=18 | n=16 | n=16 | n=16 | ||
|---|---|---|---|---|---|---|---|
| Age, years | |||||||
| Range | 36-78 | 40-74 | 48-74 | 45-72 | 43-78 | 36-75 | 0.511 |
| Median | 64 | 59 | 64.5 | 64 | 65 | 64 | |
| Gender | |||||||
| Male | 46 | 8 | 8 | 7 | 12 | 11 | 0.031 |
| Female | 40 | 12 | 10 | 9 | 4 | 5 | |
| Smoking status | |||||||
| Smoker | 23 | 2 | 3 | 6 | 4 | 8 | 0.061 |
| Never-smoker | 63 | 18 | 15 | 10 | 12 | 8 | |
| Clinical stage | |||||||
| Stage IA | 67 | 20 | 13 | 12 | 9 | 13 | 0.014 |
| Stage IB | 19 | 0 | 5 | 4 | 7 | 3 | |
| Vascular invasion | |||||||
| Presence | 2 | 0 | 0 | 1 | 0 | 1 | 0.402 |
| Absence | 84 | 20 | 18 | 15 | 16 | 15 | |
| PD-L1+ | |||||||
| ≥1% | 24 | 1 | 3 | 6 | 10 | 4 | 0.002 |
| <1% | 62 | 19 | 15 | 10 | 6 | 12 | |
| High PD-L1+ | |||||||
| ≥50% | 7 | 0 | 1 | 2 | 3 | 1 | <0.0001 |
| <50% | 79 | 20 | 17 | 14 | 13 | 15 | |
Mutated genes private in LPA, APA, PPA, MPA and SPA group
| Genes private in LPA |
|---|
| MTOR ROS1 JAK1 |
| Genes private in APA |
| INHBA |
| Genes private in PPA |
| KIT PDGFRA MAP3K13 CHEK2 U2AF1 EPHB1 TGFBR2 FGFR2 |
| Genes private in MPA |
| IL7R NTRK1 SETD2 PALB2 VEGFA MEN1 FBXW7 DNMT3A ATR PMS2 |
| Genes private in SPA |
| EPHA5 ERBB4 CDKN1B EPHA7 HIST1H3B NOTCH1 CD274 PAK5 LRP1B MLH1 TERT KDR SOX9 PPP2R1A RET EPHA3 RUNX1 STK11 PIK3CG RAD51C FAT3 MSH2 GRIN2A FGFR1 IKZF1 NTRK3 KEAP1 PTPRD KDM5A ATM ESR1 |
Distinctive genomic profiles of five histological subtypes
For the 86 samples, we identified a total of 302 genomic alterations using a panel consisting of 168 cancer-related genes, including 161 missenses (53%), 50 copy number variances (17%), 33 insert-indels (11%), 19 nonsense (6%), 18 frameshifts (6%), 13 splicing site mutations (4%), 8 fusions (3%). One LPA patient had no detected mutation. Our data revealed that the average mutation rate was 1.95 (range: 0-5), 2.56 (range: 1-6), 3.5 (range: 1-7), 3.75 (range: 1-8) and 6.05 (range: 2-12) in LPA, APA, PPA, MPA and SPA, respectively. The number of total mutations was significantly higher in SPA than in MPA (P <0.05), PPA (P <0.05), APA (P <0.001) and LPA (P<0.0001) (Figure 2). Mutations in EGFR, TP53 and APC were identified in all five histological subtypes. Each subtype also has subtype-specific mutations; LPA, APA, PPA, MPA and SPA have 3, 1, 8, 10 and 31 subtype-specific mutations, respectively. Table 2 shows the specific mutated genes in different groups.
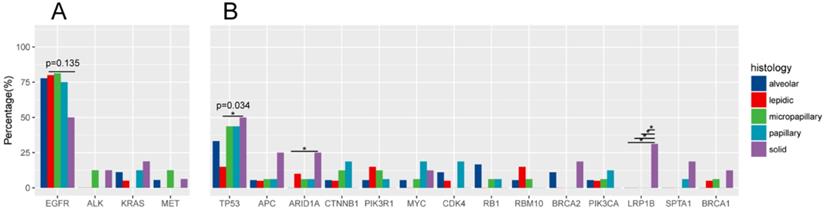
Classic lung cancer driver genes in histological subtypes of IAC
Kaplan-Meier estimates of recurrence-free survival in patients with different subtype. In the LPA group, no patient has developed recurrence (mean follow-up 29.2 months, range 9-50 months). In the APA group, 1 out of 18 (5.56%) experienced recurrent disease (mean follow-up 24.1 months, range 8.5-45 months). In the PPA group, 1 out of 16 (6.25%) experienced recurrent disease (mean follow-up 24.5 months, range 12.5-46 months). In the SPA group, 2 out of 16 (12.5%) experienced recurrent disease (mean follow-up 19.9 months, range 8.5-37.5 months). In the MPA group, 6 out of 16 (37.5%) experienced recurrent disease (mean follow-up 24.3 months, range 8.5-42 months).
Number of mutations (including small nucleotide variations, indels, copy number variations and fusions) based on histological subtype. *Represents p<0.05, **represents P<0.01, ***represents P<0.001, ****represents P<0.0001.
Driver gene mutations was identified in 96.55% (83/86) of all samples (Figure 3A). EGFR was the most frequently mutated common driver (63/86, 73.3%), followed by KRAS (8/86, 9.3%), ALK (4/86, 4.7%), MET (4/86, 4.7%), ERBB2 (3/86, 3.5%), BRAF (3/86, 3.5%), ROS1 (1/86, 1.2%), RET (1/86, 1.2%). EGFR mutations showed a trend of higher prevalence in MPA compared with SPA (13/16, 8/16), but this was not statistically significant (P=0.135) (Figure 4A).
In 20 patients with LPA, EGFR was most frequently mutated (16/20, 80%), followed by KRAS (1/20, 5%), ERBB2 (1/20, 5%), BRAF (1/20, 5%). ALK, MET, RET mutations were not detected. In 18 APA samples, mutant driver genes were EGFR (14/18, 77.8%), KRAS (2/18, 11.1%), MET (1/18, 5.6%). No ALK, ERBB2, BRAF, ROS1 and RET alterations were detected. One patient had no driver gene mutation. Among the 16 PPA patients, driver mutations were only detected in EGFR (12/16, 75%), KRAS (2/16, 12.5%), BRAF (2/16, 12.5%). PPA patients did not have mutations in ALK, ERBB2, BRAF, MET, ROS1 and RET. Among the 16 patients with MPA, driver gene mutations were detected in EGRF (13/16, 81.3%), ALK (2/16, 12.5%), ERBB2 (1/16, 6.3%), MET (2/16, 12.5%). No mutations in KRAS, BRAF, ROS, RET were detected. In 16 patients with SPA, the frequency of mutations - EGFR, KRAS, ALK, ERBB2, MET, RET - was 8/16 (50%), 3/16 (18.9%), 2/16 (12.5%), 1/16 (6.3%), 1/16 (6.3%), 1/16 (6.3%). One ALK missense patient also had concomitant ERBB2 amp. One patient was negative for driver mutations.
TP53 and other mutations
TP53 is the most frequently mutated tumor suppressor gene (Figure.3B), with a frequency of 36.0% (31/86) of all samples and 15% (3/20) of LPA, 33.3% (6/18) of APA, 43.8% (7/16) of PPA, 43.8% (7/16) of MPA, 50% (8/16) of SPA. More TP53-mutant patients were observed in SPA than in LPA. (p=0.034) (Figure 4B). The frequencies of ARIDIA and LRP1B mutation were significantly higher in SPA than in other groups (P<0.05). Interestingly, LRP1B mutations, which has been reported to be associated with high tumor mutation burden and better response to immunotherapy, were only detected from 5 SPA patients (p=0.001).
Mutation landscape of five subtypes. Each column represents a sample. The top bar plot shows the mutation number of each sample. Genetic alterations are presented by various colors. The right column indicated mutated genes. The left column indicated the percent of samples harboring gene variants.
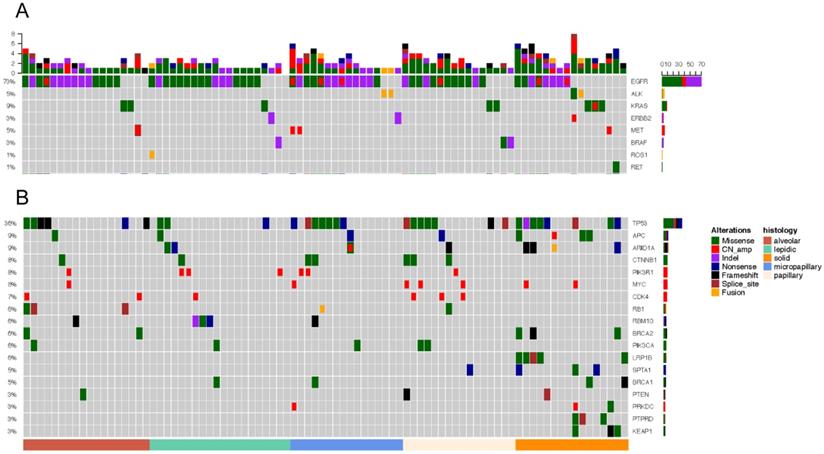
Comparison of gene mutation frequencies in five subtypes.
Discussion
The invasive adenocarcinoma is heterogeneous; however, the molecular features of predominant subtypes are elusive. Numerous literatures have identified the significant associations of driver gene mutations and histological subtypes of invasive adenocarcinoma. The EGFR mutation frequency was found to be higher in micropapillary, papillary, acinar and lepidic predominant component, lower in the solid predominant subtype [19, 20, 22, 29, 30]. Micropapillary component can be used as a predictor of EGFR mutation [19]. Micropapillary component-positive patients with EGFR mutations can benefit from EGFR-TKIs [31, 32]; while solid predominant subtype is a negative response predictor for EGFR-TKI [33]. ALK rearrangements were significantly associated with solid predominant subtype and component of signet-ring cells [34-38]. KRAS mutations were more commonly occurred in invasive mucinous adenocarcinoma [21] and solid predominant tumors [22, 39]. ROS1 fusion were closely associated with solid and acinar patterns [40].
In our study, distinct driver gene mutations were detected in different subtypes. EGFR was the most frequently shared mutated driver gene 73.3% (63/86), which is higher than 40%-50% of whole LADC population. Consistent with previous reports, we found that EGFR mutations were more frequent in MPA, less common in SPA. ALK mutations were only detected in MPA and SPA. In general, EGFR was an important factor in the beginning of lung cancer and then play a decreased role in developed lung cancer and ALK fusions may occur at a later stage in the progression of lung cancer. In addition, we observed that from LPA to SPA, the frequency of TP53 mutations significantly increased. Previous work revealed that TP53 was relatively later molecular event as a key mediator in the invasiveness of lung cancer [41, 42].
Z-Y. Dong et al. found that SPA subtype harbored a notable increase of nonsynonymous mutation and higher rate of transversion/transition based on The Cancer Genome Atlas (TCGA) and Broad database [43]. We also found that even in stageⅠdiseases, SPA had more complex driver gene mutations, higher total number of mutations and more private genes. Notably, LRP1B mutation frequency was 5% (5/100) in the entire cohort, 25% (5/20) were found in SPA, and none in other four subtypes. Li Ding et al. [44] found that mutations in LRP1B was negatively correlated with acinar, papillary and bronchioloalveolar carcinoma (BAC) subtypes and positively correlated with solid subtype. LRP1B mutations, as an important cancer suppressor gene [45], are correlated with higher-TMB and neoantigen burden. Meanwhile, mutation in LRP1B were identified to be associated with better immunotherapy survival outcome in non-small cell lung cancer (NSCLC) and melanoma patients. In LRP1B mutant samples, tumor-infiltrating immune cells were more abundant, which indicated a preferable immune response status [46-48]. Furthermore, PD-L1, as an immunotherapy biomarker, was significantly correlated with higher histologic grades (micropapillary and solid subtypes), compared to mediate histologic grades (acinar and papillary subtypes) and low grades (lepidic subtypes) [49-52]. In our study, the incidences of PD-L1+ and high PD-L1+ lesions were significantly higher in solid subtypes. These findings suggest that SPA patients - especially those harboring LRP1B mutations, may benefit from immunotherapy.
Even with the same TNM stage, our study revealed the distinct mutational profile in different histological subtypes. The intrinsic mutation may determine the malignant grade of various lung adenocarcinoma subtypes. It is estimated that over 50% of patients with early-stage NSCLC will suffered recurrence after surgery [53]. Among patients with lung adenocarcinoma, outcomes after surgical resection vary according to predominant histologic subtype. Many studies have reported that the presence of micropapillary and solid subtypes (predominant or even minor component) has significant prognostic value. Micropapillary and solid subtypes were associated with worse disease-free survival (DFS) and overall survival (OS) [3-5, 11, 54], higher possibility of lymph node metastasis [7-10, 55] and recurrence [13, 15, 31, 56]. The recurrence hazard increased as a function of the percentage of micropapillary and solid pattern [56], while higher percentage of lepidic component was associated with a lower risk for recurrence [57, 58]. Among patients who recurred, solid predominant tumors had earlier, more extra-thoracic, more multisite recurrences and worse postrecurrence survival (PRS) than those with non-solid tumors [12]. In our study, SPA and MPA have higher risk of recurrence compared to those with PPA, APA and LPA (P=0.017).
Recently, the architectural classification of IAC with surgical resection has got more and more attention in clinical practice. Ming-Sound Tsao etc. [59, 60] analysed 575 patients with completely resected lung adenocarcinoma from the LACE-Bio study, and showed the first evidence that micropapillary/solid-predominant histology predict survival benefit from adjuvant chemotherapy in patients with early-stage disease. Shinsuke Sasada [61] revealed that postoperative adjuvant chemotherapy could be considered for non-lepidic predominant tumors even at stage IA. Clinically, stage I IAC patients, which are often underwent uniform treatment and follow-up, might need distinctive therapeutic care based on the histological subtypes with divergent molecular basis.
Our study has several limitations. Firstly, tumor subtypes were extracted from different patients, as they didn't share identical genetic background and relative exposure history, we were unable to detect evolutionary trajectories of the five subtypes. Secondly, the median follow-up time was relatively short, we can't fully investigate the relationship between histologic subtypes, molecular subtypes and clinical efficacy of different treatment protocols. Thirdly, due to the small size of sample with micropapillary occupied most of the area, we selected tumors with micropapillary component exceeded 10% of the entire tumor area as MPA. The genetic mutation profile of MPA may be affected by sample selection.
Conclusions
In summary, in resected stage I IAC, prognoses of solid or micropapillary predominant subtypes were apparently worse than that of other subtypes, different histological subtypes had distinct mutational profiles. SPA harbored more complex mutation profile even at stage I and higher mutation rate of LRP1B. The rates for PD-L1 positivity and high TPS were significantly higher in SPA. Clinically, SPA patients may benefit from immunotherapy. Stage I IAC patients, which are often underwent undifferentiated treatment and follow-up, might need distinctive therapeutic care based on the histological subtypes with unique genetic profiles.
Abbreviations
LADC: lung adenocarcinoma; LPA: lepidic-predominant adenocarcinoma; APA: acinar-predominant adenocarcinoma; PPA: papillary-predominant adenocarcinoma; MPA: micropapillary-predominant adenocarcinoma; SPA: solid-predominant adenocarcinoma; NGS: next-generation sequencing; IAC: Invasive adenocarcinoma; IASLC/ATS/ERS: International Association for the Study of Lung Cancer, the American Thoracic Society, and the European Respiratory Society; WHO: World Health Organization; SNV: single nucleotide variations; INDEL: insertion-deletion variations; SNP: single nucleotide polymorphisms; CNV: copy number variations; TCGA: The Cancer Genome Atlas; BAC: bronchioloalveolar carcinoma; NSCLC: non-small cell lung cancer; DFS: disease-free survival; OS: overall survival; PRS: postrecurrence survival.
Acknowledgements
This work was funded by grants from the International Science and Technology Cooperation Project of the Changzhou Science and Technology Bureau (No. CZ20140016), the General Program of National Natural Science Foundation of China (No. 81970080).
Compliance with Ethical Standards
This study was approved by the Review Broad of the Third Affiliated Hospital of Soochow University. Our research involved human participants.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y. et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. Journal of thoracic oncology. 2011;6:244-85
2. Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JH, Beasley MB. et al. The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. Journal of thoracic oncology. 2015;10:1243-60
3. Warth A, Muley T, Meister M, Stenzinger A, Thomas M, Schirmacher P. et al. The novel histologic International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification system of lung adenocarcinoma is a stage-independent predictor of survival. J Clin Oncol. 2012;30:1438-46
4. Yoshizawa A, Motoi N, Riely GJ, Sima CS, Gerald WL, Kris MG. et al. Impact of proposed IASLC/ATS/ERS classification of lung adenocarcinoma: prognostic subgroups and implications for further revision of staging based on analysis of 514 stage I cases. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2011;24:653-64
5. Russell PA, Wainer Z, Wright GM, Daniels M, Conron M, Williams RA. Does lung adenocarcinoma subtype predict patient survival?: A clinicopathologic study based on the new International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary lung adenocarcinoma classification. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2011;6:1496-504
6. Xu L, Tavora F, Burke A. Histologic features associated with metastatic potential in invasive adenocarcinomas of the lung. The American journal of surgical pathology. 2013;37:1100-8
7. Zhao Y, Wang R, Shen X, Pan Y, Cheng C, Li Y. et al. Minor Components of Micropapillary and Solid Subtypes in Lung Adenocarcinoma are Predictors of Lymph Node Metastasis and Poor Prognosis. Annals of surgical oncology. 2016;23:2099-105
8. Hung JJ. Histologic subtype component predicts lymph node micrometastasis and prognosis in patients with stage I lung adenocarcinoma. Journal of thoracic disease. 2017;9:3623-5
9. Dai C, Xie H, Kadeer X, Su H, Xie D, Ren Y. et al. Relationship of Lymph Node Micrometastasis and Micropapillary Component and Their Joint Influence on Prognosis of Patients With Stage I Lung Adenocarcinoma. The American journal of surgical pathology. 2017;41:1212-20
10. Yeh YC, Kadota K, Nitadori J, Sima CS, Rizk NP, Jones DR. et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification predicts occult lymph node metastasis in clinically mediastinal node-negative lung adenocarcinoma. Eur J Cardiothorac Surg. 2016;49:e9-e15
11. Miyahara N, Nii K, Benazzo A, Hoda MA, Iwasaki A, Klepetko W. et al. Solid predominant subtype in lung adenocarcinoma is related to poor prognosis after surgical resection: A systematic review and meta-analysis. Eur J Surg Oncol. 2019;45:1156-62
12. Ujiie H, Kadota K, Chaft JE, Buitrago D, Sima CS, Lee MC. et al. Solid Predominant Histologic Subtype in Resected Stage I Lung Adenocarcinoma Is an Independent Predictor of Early, Extrathoracic, Multisite Recurrence and of Poor Postrecurrence Survival. J Clin Oncol. 2015;33:2877-84
13. Hung JJ, Yeh YC, Wu YC, Chou TY, Hsu WH. Prognostic Factors in Completely Resected Node-Negative Lung Adenocarcinoma of 3 cm or Smaller. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2017;12:1824-33
14. Cha MJ, Lee HY, Lee KS, Jeong JY, Han J, Shim YM. et al. Micropapillary and solid subtypes of invasive lung adenocarcinoma: clinical predictors of histopathology and outcome. J Thorac Cardiovasc Surg. 2014;147:921-8 e2
15. Yoshida Y, Nitadori JI, Shinozaki-Ushiku A, Sato J, Miyaji T, Yamaguchi T. et al. Micropapillary histological subtype in lung adenocarcinoma of 2 cm or less: impact on recurrence and clinical predictors. General thoracic and cardiovascular surgery. 2017;65:273-9
16. Zombori T, Nyari T, Tiszlavicz L, Palfoldi R, Csada E, Geczi T. et al. The more the micropapillary pattern in stage I lung adenocarcinoma, the worse the prognosis-a retrospective study on digitalized slides. Virchows Arch. 2018;472:949-58
17. Tsubokawa N, Mimae T, Sasada S, Yoshiya T, Mimura T, Murakami S. et al. Negative prognostic influence of micropapillary pattern in stage IA lung adenocarcinoma. Eur J Cardiothorac Surg. 2016;49:293-9
18. Shim HS, Lee DH, Park EJ, Kim SH. Histopathologic characteristics of lung adenocarcinomas with epidermal growth factor receptor mutations in the International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society lung adenocarcinoma classification. Archives of pathology & laboratory medicine. 2011;135:1329-34
19. Chao L, Yi-Sheng H, Yu C, Li-Xu Y, Xin-Lan L, Dong-Lan L. et al. Relevance of EGFR mutation with micropapillary pattern according to the novel IASLC/ATS/ERS lung adenocarcinoma classification and correlation with prognosis in Chinese patients. Lung cancer. 2014;86:164-9
20. Song Z, Zhu H, Guo Z, Wu W, Sun W, Zhang Y. Correlation of EGFR mutation and predominant histologic subtype according to the new lung adenocarcinoma classification in Chinese patients. Medical oncology. 2013;30:645
21. Jiang L, Mino-Kenudson M, Roden AC, Rosell R, Molina MA, Flores RM. et al. Association between the novel classification of lung adenocarcinoma subtypes and EGFR/KRAS mutation status: A systematic literature review and pooled-data analysis. Eur J Surg Oncol. 2019;45:870-6
22. Dong Y-J, Cai Y-R, Zhou L-J, Su DAN, Mu J, Chen X-J. et al. Association between the histological subtype of lung adenocarcinoma, EGFR/KRAS mutation status and the ALK rearrangement according to the novel IASLC/ATS/ERS classification. Oncology letters. 2016;11:2552-8
23. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England). 2009;25:1754-60
24. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20:1297-303
25. Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L. et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome research. 2012;22:568-76
26. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164-e
27. Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80-92
28. Newman AM, Bratman SV, Stehr H, Lee LJ, Liu CL, Diehn M. et al. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics (Oxford, England). 2014;30:3390-3
29. Yanagawa N, Shiono S, Abiko M, Ogata SY, Sato T, Tamura G. The correlation of the International Association for the Study of Lung Cancer (IASLC)/American Thoracic Society (ATS)/European Respiratory Society (ERS) classification with prognosis and EGFR mutation in lung adenocarcinoma. The Annals of thoracic surgery. 2014;98:453-8
30. Motoi N, Szoke J, Riely G, Seshan V, Kris M, Rusch V. et al. Lung adenocarcinoma: modification of the 2004 WHO mixed subtype to include the major histologic subtype suggests correlations between papillary and micropapillary adenocarcinoma subtypes, EGFR mutations and gene expression analysis. The American journal of surgical pathology. 2008;32:810-27
31. Sumiyoshi S, Yoshizawa A, Sonobe M, Kobayashi M, Fujimoto M, Tsuruyama T. et al. Pulmonary adenocarcinomas with micropapillary component significantly correlate with recurrence, but can be well controlled with EGFR tyrosine kinase inhibitors in the early stages. Lung cancer. 2013;81:53-9
32. Zhang Y, Wang R, Cai D, Li Y, Pan Y, Hu H. et al. A comprehensive investigation of molecular features and prognosis of lung adenocarcinoma with micropapillary component. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2014;9:1772-8
33. Yoshida T, Ishii G, Goto K, Yoh K, Niho S, Umemura S. et al. Solid predominant histology predicts EGFR tyrosine kinase inhibitor response in patients with EGFR mutation-positive lung adenocarcinoma. Journal of cancer research and clinical oncology. 2013;139:1691-700
34. Zhao R, Zhang J, Han Y, Shao J, Zhu L, Xiang C. et al. Clinicopathological Features of ALK Expression in 9889 Cases of Non-small-Cell Lung Cancer and Genomic Rearrangements Identified by Capture-Based Next-Generation Sequencing: A Chinese Retrospective Analysis. Molecular diagnosis & therapy. 2019;23:395-405
35. Li P, Gao Q, Jiang X, Zhan Z, Yan Q, Li Z. et al. Comparison of Clinicopathological Features and Prognosis between ALK Rearrangements and EGFR Mutations in Surgically Resected Early-stage Lung Adenocarcinoma. J Cancer. 2019;10:61-71
36. Yu Y, Ding Z, Zhu L, Teng H, Lu S. Frequencies of ALK rearrangements in lung adenocarcinoma subtypes: a study of 2299 Chinese cases. SpringerPlus. 2016;5:894
37. Wang H, Zhang W, Wang K, Li X. Correlation between EML4-ALK, EGFR and clinicopathological features based on IASLC/ATS/ERS classification of lung adenocarcinoma. Medicine. 2018;97:e11116
38. Possidente L, Landriscina M, Patitucci G, Borgia L, Lalinga V, Vita G. ALK rearrangement in specific subtypes of lung adenocarcinoma: immunophenotypic and morphological features. Medical oncology. 2017;34:76
39. Kadota K, Sima CS, Arcila ME, Hedvat C, Kris MG, Jones DR. et al. KRAS Mutation Is a Significant Prognostic Factor in Early-stage Lung Adenocarcinoma. The American journal of surgical pathology. 2016;40:1579-90
40. Chen YF, Hsieh MS, Wu SG, Chang YL, Shih JY, Liu YN. et al. Clinical and the prognostic characteristics of lung adenocarcinoma patients with ROS1 fusion in comparison with other driver mutations in East Asian populations. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2014;9:1171-9
41. Chen H, Carrot-Zhang J, Zhao Y, Hu H, Freeman SS, Yu S. et al. Genomic and immune profiling of pre-invasive lung adenocarcinoma. Nat Commun. 2019;10:5472
42. Hu X, Fujimoto J, Ying L, Fukuoka J, Ashizawa K, Sun W. et al. Multi-region exome sequencing reveals genomic evolution from preneoplasia to lung adenocarcinoma. Nat Commun. 2019;10:2978
43. Dong Z, Zhong W, Liu S, Xie Z, Wu S, Wu Y. Potential predictive value for adjuvant PD-1 blockade based on histologic subtype in resected lung adenocarcinoma. Annals of Oncology. 2016 27
44. Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K. et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069-75
45. Liu C-X, Musco S, Lisitsina NM, Yaklichkin SY, Lisitsyn NA. Genomic organization of a new candidate tumor suppressor gene, LRP1B. Genomics. 2000;69:271-4
46. Lan S, Li H, Liu Y, Ma L, Liu X, Liu Y. et al. Somatic mutation of LRP1B is associated with tumor mutational burden in patients with lung cancer. Lung cancer. 2019;132:154-6
47. Chen H, Chong W, Wu Q, Yao Y, Mao M, Wang X. Association of LRP1B Mutation With Tumor Mutation Burden and Outcomes in Melanoma and Non-small Cell Lung Cancer Patients Treated With Immune Check-Point Blockades. Frontiers in immunology. 2019;10:1113
48. Zhu J, Tucker MD, Kao C, Labriola M, Cheris S, Datto MB. et al. Immune checkpoint inhibitor response in tumors with LRP1B variants. American Society of Clinical Oncology. 2019 p. e14291
49. Majithia N, Aubry M, Murphy S, Mansfield A. Heterogeneity of PD-L1 expression between invasive and lepidic components of lung adenocarcinomas. International Journal of Radiation Oncology• Biology• Physics. 2019;104:237-8
50. Zhang Y, Wang L, Li Y, Pan Y, Wang R, Hu H. et al. Protein expression of programmed death 1 ligand 1 and ligand 2 independently predict poor prognosis in surgically resected lung adenocarcinoma. OncoTargets and therapy. 2014;7:567
51. Ng Kee Kwong F, Laggner U, McKinney O, Croud J, Rice A, Nicholson AG. Expression of PD-L1 correlates with pleomorphic morphology and histological patterns of non-small-cell lung carcinomas. Histopathology. 2018;72:1024-32
52. Ahmad A, Kim H, Kwon HJ, Park SY, Park Y, Park E. et al. Clinicopathological analysis and prognostic significance of programmed cell death-ligand 1 protein and mRNA expression in non-small cell lung cancer. PloS one. 2018;13:e0198634
53. Chansky K, Sculier JP, Crowley JJ, Giroux D, Van Meerbeeck J, Goldstraw P. The International Association for the Study of Lung Cancer Staging Project: prognostic factors and pathologic TNM stage in surgically managed non-small cell lung cancer. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2009;4:792-801
54. Xu S, Xi J, Jiang W, Lu S, Wang Q. Solid component and tumor size correlate with prognosis of stage IB lung adenocarcinoma. The Annals of thoracic surgery. 2015;99:961-7
55. Cheng X, Zheng D, Li Y, Li H, Sun Y, Xiang J. et al. Tumor histology predicts mediastinal nodal status and may be used to guide limited lymphadenectomy in patients with clinical stage I non-small cell lung cancer. J Thorac Cardiovasc Surg. 2018;155:2648-56 e2
56. Takahashi Y, Eguchi T, Tan KS, Tano Z, Travis W, Jones D. et al. Recurrence Dynamics in Resected Pathological Stage I Lung Adenocarcinoma Depend on the IASLC/ATS/ERS Histological Subtype. Journal of Thoracic Oncology. 2017;12:S2032
57. Strand TE, Rostad H, Strom EH, Hasleton P. The percentage of lepidic growth is an independent prognostic factor in invasive adenocarcinoma of the lung. Diagnostic pathology. 2015;10:94
58. Kadota K, Villena-Vargas J, Yoshizawa A, Motoi N, Sima CS, Riely GJ. et al. Prognostic significance of adenocarcinoma in situ, minimally invasive adenocarcinoma, and nonmucinous lepidic predominant invasive adenocarcinoma of the lung in patients with stage I disease. The American journal of surgical pathology. 2014;38:448
59. Tsao MS, Marguet S, Le Teuff G, Lantuejoul S, Shepherd FA, Seymour L. et al. Subtype Classification of Lung Adenocarcinoma Predicts Benefit From Adjuvant Chemotherapy in Patients Undergoing Complete Resection. J Clin Oncol. 2015;33:3439-46
60. Russell PA, Wright GM. Predominant histologic subtype in lung adenocarcinoma predicts benefit from adjuvant chemotherapy in completely resected patients: discovery of a holy grail? Ann Transl Med. 2016;4:16
61. Sasada S, Miyata Y, Mimae T, Mimura T, Okada M. Impact of Lepidic Component Occupancy on Effects of Adjuvant Chemotherapy for Lung Adenocarcinoma. The Annals of thoracic surgery. 2015;100:2079-86
Author contact
![]() Corresponding authors: Chong Li, MD, Department of Respiratory Medicine, The Third Affiliated Hospital of Soochow University, 185 Juqian Road, Changzhou, 213003, China, E-mail: zeyou06com. Qing Li, MD, Department of Pathology, The Third Affiliated Hospital of Soochow University, 185 Juqian Road, Changzhou, 213003, China, E-mail: liqblkcom.
Corresponding authors: Chong Li, MD, Department of Respiratory Medicine, The Third Affiliated Hospital of Soochow University, 185 Juqian Road, Changzhou, 213003, China, E-mail: zeyou06com. Qing Li, MD, Department of Pathology, The Third Affiliated Hospital of Soochow University, 185 Juqian Road, Changzhou, 213003, China, E-mail: liqblkcom.

 Global reach, higher impact
Global reach, higher impact