Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(11):3249-3256. doi:10.7150/jca.55558 This issue Cite
Research Paper
Hsa_circ_0091581 promotes glioma progression by regulating RMI1 via sponging miR-1243-5p
1. Department of Neurosurgery, People's Hospital of Xuancheng City, Xuancheng, Anhui, China.
2. Department of Neurosurgery, The Affiliated Brain Hospital of Nanjing Medical University, Nanjing, Jiangsu, China.
Received 2020-11-6; Accepted 2021-3-4; Published 2021-4-2
Abstract
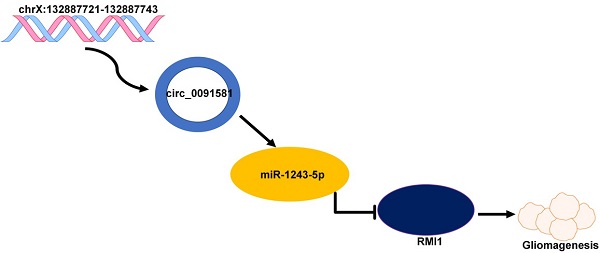
Glioma is a pervasive malignancy and the main cause of cancer-related deaths worldwide. Circular RNA is an important subject of cancer research, and its role and function in glioma are poorly understood. This study demonstrated that hsa_circ_0091581 is upregulated in glioma tissues and cells. The results of the CCK-8, EdU, and transwell assays indicated that hsa_circ_0091581 promotes proliferation, migration, and invasion of glioma cells. The results of the luciferase reporter and RNA immunoprecipitation assays indicated that the mechanism of the effects of hsa_circ_0091581 on glioma cells involves sponging miR-1243-5p to regulate RMI1. The results of the rescue experiments indicated that hsa_circ_0091581 regulates proliferation, migration, and invasion of glioma cells by targeting RMI1 in a miR-1243-5p dependent manner. The results of the nude mice xenograft assays showed that knockdown of hsa_circ_0091581 inhibits glioma growth in vivo. Thus, our study determined the role of hsa_circ_0091581/miR-1243-5p/RMI1 in glioma and suggests that this axis may be a novel therapeutic target in glioma.
Keywords: circ_0091581, miR-1243-5p, RMI1, glioma.
Introduction
Glioma is a pervasive malignancy and the main cause of cancer-related deaths worldwide [1]. Glioblastoma multiforme (GBM), WHO Ⅳ grade glioma, is characterized by the presence of poorly differentiated stromal cells around focal necrosis and capillary proliferation [2, 3]. The latest statistics shows that GBM accounts for 12%~15% of all intracranial malignant tumors and 50%~60% of astrocytic tumors [4]. Current systematic treatment strategies for glioma include surgery, chemotherapy, radiotherapy and tumor treating fields (TTF) and have introduced certain innovations; however, GBM grows rapidly, has a poor prognosis, and usually results in death within one year after diagnosis [5]. Therefore, investigation of the molecular mechanisms of glioma occurrence and progression is very important.
Circular RNA (circRNA) is a specific type of RNA molecule and has been extensively investigated in the fields of RNA and disease research [6]. CircRNA molecules differ from linear RNA (with 5' and 3' ends) and have a closed circular structure, which is not affected by RNA enzymes [7, 8]. From the cell biology point of view, circRNAs are closely related to the biological functions of cells, such as proliferation, differentiation, and apoptosis [9, 10]. Recent studies have shown that circRNAs play an important regulatory role in the growth and development of organisms and occurrence and development of diseases [11, 12]. CircRNAs has attracted attention of the scientists in the field of cancer research. Numerous recent studies have suggested that circRNAs play an important role in tumor progression, and certain circRNAs can even be used as targets for early diagnosis, prognosis assessment and targeted therapy of tumors. For example, circ-ZKSCAN1 has been reported as a prognostic factor of bladder cancer recurrence [13]; circ-UMAD1 in peripheral circulation may be a biomarker of lymph node metastasis in thyroid cancer [14]; hsa_circ_0003829 may act as a diagnostic predictor in oral squamous cell carcinoma [15]. In glioma, accumulating evidence indicates that circRNAs are closely associated with glioma progression [1, 16, 17]; however, progress of the sequencing technology resulted in identification of additional circRNAs, and the relationships between circRNAs and glioma progression require further investigation.
In this study, we report that hsa_circ_0091581 is upregulated in glioma tissues and glioma cells. Function assays showed that hsa_circ_0091581 promotes proliferation, migration, and invasion of glioma cells. Using online public databases and RNA pull-down, RNA immunoprecipitation, and luciferase reporter assays, we determined the mechanism of the effect of hsa_circ_0091581 on glioma progression. Overall, our study is the first to report that hsa_circ_0091581 promotes glioma proliferation, migration, and invasion by targeting the miR-1243-5p/RMI1 axis that may be a potential target for glioma treatment.
Methods
Clinical samples
Glioma and corresponding adjacent nonneoplastic tissue samples (n=20) were obtained from patients diagnosed with glioma and admitted to the People's Hospital of Xuancheng City from June 2014 to September 2018. The samples were stored in liquid nitrogen immediately after resection. The pathological grade of the tumors was independently determined by two senior pathologists. Written informed consent was obtained from all patients. This study was approved by the Ethics Committee of People's Hospital of Xuancheng City.
Cell culture and transfection
Glioma cell lines (U138, U87, U251, A172, and T98G) were obtained from the American Type Culture Collection (ATCC, MD, USA). Normal astrocytes were purchased from Procell (Wuhan, China). All cell lines were grown in Dulbecco's modified Eagle's medium (DMEM, Gibco, NY, USA) containing 10% fetal bovine serum (FBS, HyClone, UT, USA) and maintained at 5%CO2 and 37°C. The synthetic nucleotides and constructs used in this study were provided by GenePharma (Shanghai, China), and the transfection was performed using Lipofectamine 3000 (Invitrogen, CA, USA).
Quantitative real-time PCR (qRT-PCR)
Total RNA was isolated from the cells or tissues by using TRIzol reagent (Invitrogen, CA, USA). Qualified RNA was reverse transcribed into complementary deoxyribonucleic acid (cDNA) using a PrimeScript RT kit (Invitrogen, CA, USA); qRT-PCR was performed using SYBR® Premix Ex Taq™ (TaKaRa, Tokyo, Japan) at 92°C for 10 min and 40 cycles at 92°C for 10 s and 60°C for 1 min. GAPDH was used as an internal control. The relative level of circ_0091581, miR-1243-5p, and RMI1 was calculated by the 2-ΔΔCt method. The primers used in this study were shown in Table 1.
The primers used in this study.
| Gene | Forward | Reverse |
|---|---|---|
| Circ_0091581 | GGAGAACGTACTGCTTGGTC | TGGAGTCAGGCTTGGGTAGT |
| miR-1243-5p | GTCAACTGGATCAATTATAGG | GTGCAGGGTCCGAGGT |
| RMI1 | GTGCGATCCTCAAGAGCGTA | CAGACATCCATCAGCCGGAC |
| GAPDH | TCGACAGTCAGCCGCATCTTCTTT | ACCAAATCCGTTGACTCCGACCTT |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
RNase R digestion assay
Isolated RNA (2 µg) was incubated with RNase R (3 U/µg) or digestion buffer at 37°C for 30 min. After the solution was purified, qPCR was performed to determine the RNA levels.
Actinomycin D assay
Glioma cells were incubated with actinomycin D (3 µg/ml, Sigma, CA, USA) to block the transcription of mRNAs for 0 h, 8 h, 16 h, and 24 h. After cells were harvested, circular GPC3 (hsa_circ_0091581) and linear GPC3 RNAs were quantified by qRT-PCR to determine the half-life of RNA.
Western bolt
Total protein was isolated by using RIPA lysis buffer (Keygene, Shanghai, China). After separation by SDS-PAGE, the proteins were transferred to PVDF membranes (Millipore, MA, USA). Then, the membrane was incubated with primary and secondary antibodies (Proteintech, IL, USA). Finally, the signals were detected by an Image Quant LAS 4000 system (GE, USA).
Luciferase reporter assay
The synthetic nucleotide and constructs used in this assay were provided by GenePharma (Shanghai, China). The pmirGLO-circ_0091581-WT or pmirGLO-circ_0091581-MUT vectors were cotransfected with NC mimics or miR-1243-5p mimics into U87 and U251 cells. The pmirGLO-RMI1-WT or pmirGLO-RMI1-MUT vectors were cotransfected with NC mimics or miR-1243-5p mimics into U87 and U251 cells. Transfection was carried out by using Lipofectamine 3000 (Invitrogen, CA, USA). After 48 h, final luciferase activity was assessed using a luciferase reporter assay system (Promega, WI, USA).
RNA immunoprecipitation (RIP)
RIP assay was performed using a Magna RIP RNA-binding protein immunoprecipitation kit (Millipore, MA, USA). The treated U87 and U251 cells were lysed in a lysis buffer containing protease and RNase inhibitors. Then, the cell lysates were incubated in a RIP buffer with magnetic beads conjugated with an anti-human Ago2 antibody (Millipore), and normal IgG (Millipore) were used as a negative control. Finally, the coprecipitated RNAs were eluted from the beads and assayed by qRT-PCR.
Cell counting kit-8 (CCK-8) assay
Cells (2,000/well) were inoculated in a 96-well plate. At 12 h, 24 h, 48 h, and 72 h, the optical density at 450 nm at the indicated time was recorded using a CCK-8 kit (Beyotime, Shanghai, China), and viability curves were constructed.
5-Ethynyl-2-deoxyuridine (EdU) assay
Cells (20,000/well) were plated in a 96-well plate overnight. Then, the cells were incubated in 4% methanol for 30 min followed by permeabilization in 0.5% Triton X-100 (Keygene, Shanghai, China) for 10 min. Then, the cells were incubated with 1×ApollorR (RiboBio, Guangzhou, China) for 30 min. Cells were stained by 4',6-diamidino-2-phenylindole (DAPI) for another 30 min in the dark. Finally, EdU-positive cells were counted.
Transwell assay
Cells (40,000/well) were suspended in DMEM without FBS and added to the upper section of a transwell chamber (8 µm; Millipore, MA, USA). DMEM containing FBS (10%) (600 µl) was added to the bottom of a 24-well plate with inserts. After culture for 48 h, the cells migrated or invaded to the bottom chamber were fixed and stained. In invasion assay, the chambers were precoated with Matrigel (200 mg/ml, BD, NJ, USA), and in migration assay, the precoating was not performed.
Tumor xenograft assay
Eight female nude mice aged 5-6 weeks were purchased from Beijing Laboratory Animal Center (Beijing, China). U87 cells (1,000,000) transfected with sh-circ_0091581 or a negative control were injected subcutaneously into mice. Three weeks later, the tumors formed in mice were evaluated (equation: Volume=length×width2×0.5).
Statistical analysis
The data are presented as the mean ± standard error and were analyzed by SPSS (Statistical Product and Service Solutions, Version 17.0, IL, USA). Figures were edited using GraphPad Prism (Version X; CA, USA). Two-paired independent t-test was performed to assess the differences between the groups. Differences were considered significant at P< 0.05.
Results
The expression and characteristics of hsa_circ_0091581 in glioma
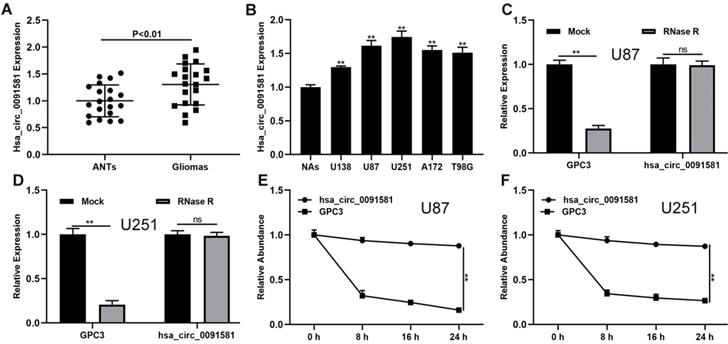
The expression of hsa_circ_0091581 in glioma and the corresponding adjacent nonneoplastic tissue (ANTs) was assayed by qRT-PCR. Compared with ANTs, hsa_circ_0091581 was significantly upregulated in glioma tissues (Fig. 1A). Moreover, compared with normal astrocytes (NAs), hsa_circ_0091581 was considerably upregulated in glioma cells (Fig. 1B). To validate the circular characteristics of hsa_circ_0091581, U87 and U251 cells were treated with RNase R. The results showed that RNase R did not digest hsa_circ_0091581 (Fig. 1C, D). Furthermore, the results of the actinomycin D assay indicated that the circular transcript (hsa_circ_0091581) is more stable than the linear transcript (GPC3) in U87 and U251 cells (Fig. 1E, F). There results suggest that hsa_circ_0091581 may have important functions in glioma.
Hsa_ circ_0091581 promotes the proliferation, migration, and invasion of glioma cells in vitro
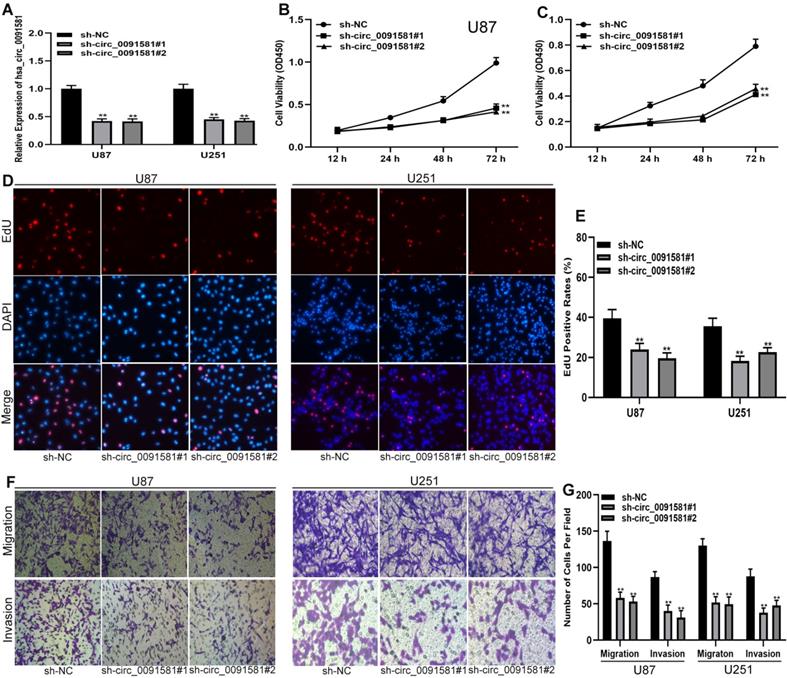
To test the functions of hsa_circ_0091581 in glioma cells, short hairpin RNAs (shRNAs) and corresponding negative control were used to construct three cell models with various expression levels of hsa_circ_0091581 (Fig. 2A). CCK-8 assay showed that hsa_circ_0091581 knockdown inhibits the proliferation of U87 and U251 cells (Fig. 2B, C). The results of the EdU assay are similar to that of the CCK-8 assay (Fig. 2D, E). The results of the transwell assay indicated that hsa_circ_0091581 downregulation suppresses the migration and invasion of U87 and U251 cells (Fig. 2F-G).
Hsa_ circ_0091581 functions as a sponge of miR-1243-5p
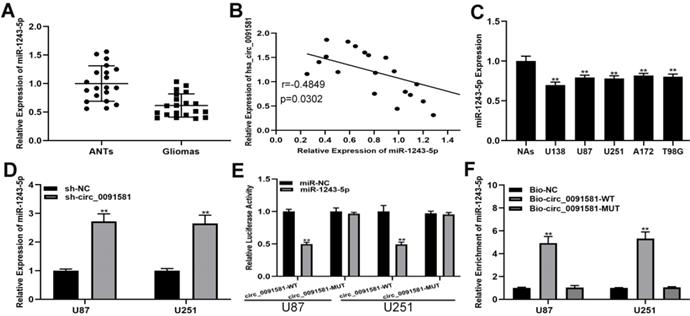
A number of studies have shown that circRNAs can regulate the progress of glioma by adsorbing miRNAs, which called competing endogenous RNAs (ceRNA) mechanism [18, 19]. An online database (circInteractome, https://circinteractome.nia.nih.gov/) was searched to selected 28 miRNAs as the sponge targets of hsa_circ_0091581. miRNAs with the top ten scores were selected for further screening, and miR-1243-5p was of interest. Initially, we detected the expression of miR-1243-5p in the clinical samples and found that miR-1243-5p was downregulated in glioma versus ANTs (Fig. 3A). Pearson correlation analysis indicated an inverse correlation between hsa_circ_0091581 and miR-1243-5p (Fig. 3B). Predictably, comparison with NAs indicated that miR-1243-5p is downregulated in glioma cells (Fig. 3C). Additionally, qRT-PCR showed that knockdown of hsa_circ_0091581 can upregulate miR-1243-5p in U87 and U251 cells (Fig. 3D). Moreover, the results of the luciferase reporter assays indicated that miR-1243-5p can decrease the luciferase activity in the case of hsa_circ_0091581-WT and has no effect on the expression of hsa_circ_0091581-MUT (Fig. 3E). Finally, the results of the RIP assay indicated that miR-1243-5p is enriched in the Bio-hsa_circ_0091581 group (Fig. 3F). These results demonstrated that hsa_circ_0091581 can function as a sponge of miR-1243-5p.
Hsa-circ_0091581 is upregulated in glioma tissues and cells. (A) The expression of hsa_circ_0091581 in glioma and adjacent nonneoplastic tissue (n=20). (B) The expression of hsa_circ_0091581 in glioma cells. (C-D) The RNase R assay confirmed the circular structure of hsa_circ_0091581 in U87 and LN229 cells. (E-F) Circular transcript of GPC3 (hsa_circ_0091581) was more stable than linear transcript according to the results of the actinomycin D treatment assays in U87 and LN229 cells.
RMI1 is the functional target of miR-1243-5p
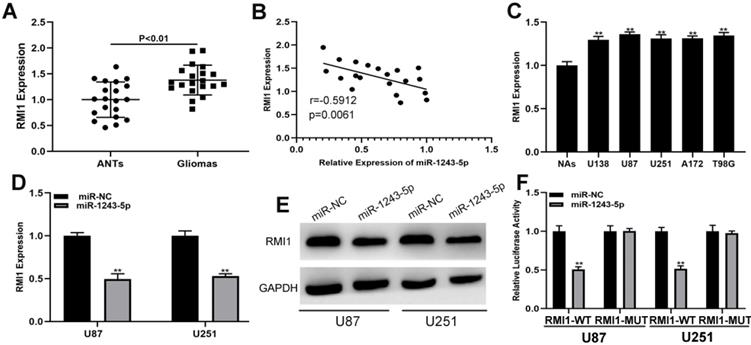
The targets of miR-1243-5p were identified by screening an online database (targetScan, http://www.targetscan.org/vert_72/), and RMI1 was selected for further investigations because of its high score. The expression of RMI1 in clinical samples indicated that RMI1 was upregulated in glioma tissues compared with the level in ANTs (Fig. 4A). The result of Pearson correlation analysis showed that RMI1 is negatively associated with miR-1243-5p (Fig. 4B). Additionally, the expression of RMI1 was higher in glioma cells than that in NAs (Fig. 4C). Furthermore, the results of the qRT-PCR and western blot assays indicated that miR-1243-5p can repress RMI1 expression in U87 and U251 cells (Fig. 4D, E). Finally, the results of the luciferase reporter assay indicated that miR-1243-5p can decrease the luciferase activity in the case of RMI1-WT and has no effect on the expression of RMI1-MUT (Fig. 4F). Overall, these results indicated that RMI1 is the function target of miR-1243-5p.
Hsa_ circ_0091581 regulates the proliferation, migration, and invasion of glioma cells in a miR-1243-5p-dependent manner
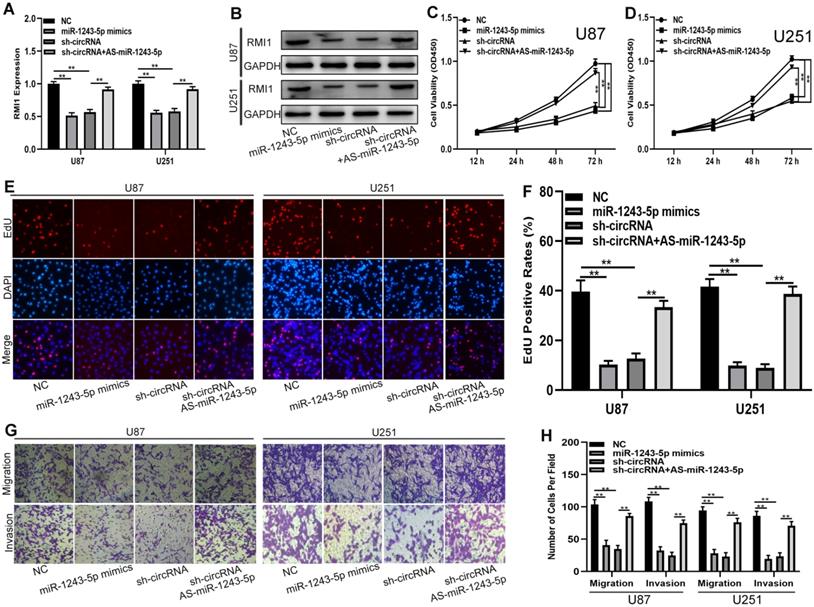
To determine whether the function of hsa_circ_0091581 in glioma cells is dependent on the miR-1243-5p/RMI1axis, shRNAs, miR-1243-5p mimics, and miR-1243-5p inhibitors were used to construct four cell models. The expression of RMI1 in the four cell models was tested by qRT-PCR and western blot (Fig. 5A, B). The results of the CCK-8 and EdU assays showed that miR-1243-5p overexpression and hsa_circ_0091581 downregulation can suppress the proliferation of U87 and U251 cells, and downregulation of miR-1243-5p can restore the inhibitory effect of hsa_circ_0091581 downregulation on the proliferation (Fig. 5C-F). Moreover, the results of the transwell assay indicated that upregulation of miR-1243-5p and knockdown of hsa_circ_0091581 can inhibit the migration and invasion of U87 and U251 cells, and the inhibitory effect of hsa_circ_0091581 can be rescued by miR-1243-5p inhibitors (Fig. 5G, H). These findings suggested that hsa_circ_0091581 regulates the proliferation, migration, and invasion of glioma cells by targeting RMI1 in a miR-1243-5p dependent manner.
Downregulation of hsa-circ_0091581 inhibits the proliferation, migration, and invasion of glioma cells. (A) qRT-PCR assay of the expression of hsa_circ_0091581 and GPC3 in U87 and U251 cells transfected with sh-NC or sh-circ_0091581. (B-C) Cell proliferation was assessed by CCK-8 assay after hsa_circ_0091581 downregulation in U87 and U251 cells. (D-E) Cell proliferation was assessed by EdU assay after hsa_circ_0091581 downregulation in U87 and U251 cells. (F-G) Cell migration and invasion were evaluated by transwell assay after hsa_circ_0091581 downregulation in U87 and U251 cells.
Hsa_circ_0091581 sponges miR-1243-5p in U87 and U251 cells. (A) The expression of miR-1243-5p in glioma and adjacent nonneoplastic tissue (n=20). (B) Pearson correlation analysis indicated a reverse correlation between hsa_circ_0091581 and miR-1243-5p. (C) The expression of miR-1243-5p in glioma cells. (D) qRT-PCR assay of the expression of miR-1243-5p in U87 and U251 cells transfected with sh-NC or sh-circ_0091581. (E) The relative luciferase activity was assayed in U87 and U251 cells cotransfected with miR-1243-5p mimics or miR-NC and hsa_circ_0091581 wild type or mutant luciferase reporter vectors. (F) Biotinylated RIP assay of the cells transfected with Bio-circ_0091581, Bio-circ_0091581-MT, or a control vector.
RMI1 is the functional target of miR-1243-5p in U87 and U251 cells. (A) The expression of RMI1 in glioma and adjacent nonneoplastic tissue (n=20). (B) Pearson correlation analysis indicated a reverse correlation between RMI1 and miR-1243-5p. (C) The expression of RMI1 in glioma cells. (D-E) qRT-PCR and western blot assays of the expression of RMI1 in U87 and U251 cells transfected with miR-NC or miR-1243-5p mimics. (F) The relative luciferase activity was assayed in U87 and U251 cells cotransfected with miR-1243-5p mimics or miR-NC and RMI1 wild type or mutant luciferase reporter vectors.
Hsa_circ_0091581 promotes the proliferation, migration, and invasion of glioma cells by targeting RMI1 in a miR-1243-5p dependent manner. (A-B) qRT-PCR and western blot assay of the expression of RMI1 in U87 and U251 cells transfected with sh-NC+miR-NC, sh-NC+miR-1243-5p mimics, miR-NC+sh-circ_0091581, or sh-circ_0091581+miR-1243-5p inhibitors. (C-D) CCK-8 assay results indicate that miR-1243-5p mimics and sh-circ_0091581 inhibit the proliferation of U87 and U251 cells, and the inhibitory effect of sh-circ_0091581 is restored by miR-1243-5p inhibitors. (E-F) EdU assay results indicate that miR-1243-5p mimics and sh-circ_0091581 inhibit the proliferation of U87 and U251 cells, and the inhibitory effect of sh-circ_0091581 is restored by miR-1243-5p inhibitors. (G-H) Transwell assay results indicate that miR-1243-5p mimics and sh-circ_0091581 inhibit the migration and invasion of U87 and U251 cells, and the inhibitory effect of sh-circ_0091581 is restored by miR-1243-5p inhibitors.
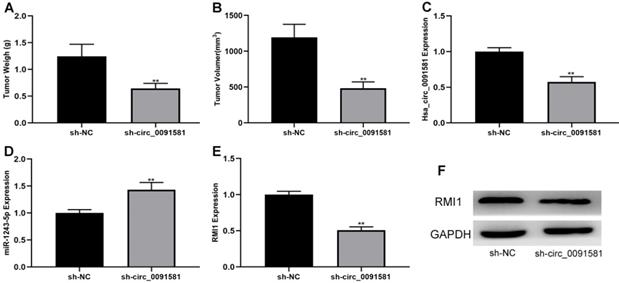
Hsa_circ_0091581 promotes glioma growth in vivo. (A-B) Downregulation of hsa_circ_0091581 decreases tumor growth in vivo. (C) The expression of hsa_circ_0091581 in the tumors formed in the two groups of mice. (D) Downregulation of hsa_circ_0091581 increases miR-1243-5p expression in the tumor. (E-F) Downregulation of hsa_circ_0091581 reduces RMI1 expression in the tumor.
Knockdown of hsa_circ_0091581 inhibits glioma growth in a nude mice xenograft model
To determine whether hsa_circ_0091581 can suppress glioma growth in vivo, a nude mice xenograft model was generated. U87 cells (1,000,000) transfected with sh-circ_0091581 or negative control were injected into the right shoulder of mice. After 21 days, the tumors formed in mice were evaluated. The results showed that tumors formation in the sh-circ_0091581 group had lower weight and volume compared with those in the negative control group (Fig. 6A, B). Moreover, the results of qRT-PCR and/or western blot indicated that hsa_circ_0091581 and RMI1 were downregulated and miR-1243-5p was upregulated in sh-circ_0091581 group compared with those in the negative control group (Fig. 6C-F). Thus, knockdown of hsa_circ_0091581 inhibits glioma growth in vivo, and this effect is associated with miR-1243-5p and RMI1.
Discussion
Glioma has always been an important subject of research and a challenge for neurosurgery [20, 21]. At present, the treatment of glioma mainly includes surgical resection followed by adjuvant radiotherapy and chemotherapy [22]. Recently, a variety of new treatment methods of glioma have emerged and are used in clinic to improve the prognosis of patients to an extent; however, the median survival time of glioma patients has not improved significantly[23]. Thus, investigation of the molecular mechanisms of glioma pathogenesis and progression is important. Rapid development of next generation sequencing (NGS) technology results in detection of an increasing number of noncoding RNAs (ncRNAs) and especially circRNAs [24]. circRNAs play an important role in the development of human diseases. For example, hsa_circ_0026416 acts as an oncogene in colorectal cancer [25]; circCFL1 can promote the progression of diffuse large B-cell lymphoma [26]; and hsa_circ_0001068 can be a novel biomarker of ovarian cancer [27]. Hsa_circ_0091581 is a novel circRNA transcribed from the glypican-3 gene (GPC3). hsa_circ_0091581 plays an oncogenic role in hepatocellular carcinoma[28, 29]; however, its role in glioma was not investigated previously. In present study, we show that hsa_circ_0091581 is upregulated in glioma tissues and cell lines and can promote the proliferation, migration, and invasion of glioma cells in vitro.
To determine the mechanism of action of hsa_circ_0091581 on glioma proliferation, migration, and invasion, we used the results described in the literature that suggested that circRNAs can play a regulatory role by adsorbing miRNAs [30-32]. Hsa_circ_0001785 sponges miR-942 to regulate breast cancer progression [33]; hsa_circ_0005556 sponges miR-4270 to facilitate gastric cancer progression [34]; and hsa_circ_0004913 act as a sponge for miR-184 and is thus involved in the progress of hepatocellular carcinoma [35]. Ji and Wei et al. reported that hsa_circ_0091581 is involved in hepatocellular carcinoma progression by sponging miR-591 and miR-526b [28, 29]. In the present study, hsa_circ_0091581 was able to sponge miR-1243-5p involved in the progression of pancreatic cancer as a target of hsa_circ_0002570 [36]. Then, a search of an online database and luciferase reporter assays were used to select RecQ-mediated genome instability protein 1 (RMI1) as the target of miR-1243-5p. RMI1 plays an important role in DNA repair and is closely associated with many human diseases, including tumors, obesity, and Bloom syndrome [37-39]. However, the role of RMI1 in glioma has not been reported previously. Our data indicate that RMI1 is upregulated in glioma tissues and cell lines, and qRT-PCR, western blot, and functional assays demonstrated that RMI1 is a functional target of miR-1243-5p. Additionally, rescue experiments suggested that hsa_circ_0091581 regulates the expression and function of RMI1 in a miR-1243-5p dependent manner. Finally, in vivo assays suggested that hsa_circ_0091581 can inhibit glioma growth, and this effect was mediated by miR-1243-5p and RMI1.
In summary, hsa_circ_0091581 can promote glioma proliferation, migration, and invasion via the hsa_circ_0091581/miR-1243-5p/RMI1 axis and may be a novel therapeutic target in glioma.
Acknowledgements
This study was supported by Key Research and Development Program of Anhui Province (No. 202004j07020049).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Jiang Y, Zhou J, Zhao J, Zhang H, Li L, Li H. et al. The U2AF2 /circRNA ARF1/miR-342-3p/ISL2 feedback loop regulates angiogenesis in glioma stem cells. J Exp Clin Cancer Res. 2020;39:182
2. Lou J, Hao Y, Lin K, Lyu Y, Chen M, Wang H. et al. Circular RNA CDR1as disrupts the p53/MDM2 complex to inhibit Gliomagenesis. Mol Cancer. 2020;19:138
3. Zhang X, Wang S, Lin G, Wang D. Down-regulation of circ-PTN suppresses cell proliferation, invasion and glycolysis in glioma by regulating miR-432-5p/RAB10 axis. Neurosci Lett. 2020;735:135153
4. Zhang Y, Lin X, Geng X, Shi L, Li Q, Liu F. et al. Advances in circular RNAs and their role in glioma (Review). Int J Oncol. 2020;57:67-79
5. Huo LW, Wang YF, Bai XB, Zheng HL, Wang MD. circKIF4A promotes tumorogenesis of glioma by targeting miR-139-3p to activate Wnt5a signaling. Mol Med. 2020;26:29
6. Sun J, Li B, Shu C, Ma Q, Wang J. Functions and clinical significance of circular RNAs in glioma. Mol Cancer. 2020;19:34
7. Li Z, Ruan Y, Zhang H, Shen Y, Li T, Xiao B. Tumor-suppressive circular RNAs: Mechanisms underlying their suppression of tumor occurrence and use as therapeutic targets. Cancer Sci. 2019;110:3630-8
8. Paulmurugan R, Malhotra M, Massoud TF. The protean world of non-coding RNAs in glioblastoma. J Mol Med (Berl). 2019;97:909-25
9. Yang Y, Zhang Y, Chen B, Ding L, Mu Z, Li Y. Elevation of circular RNA circ-POSTN facilitates cell growth and invasion by sponging miR-1205 in glioma. J Cell Biochem. 2019;120:16567-74
10. Nie JH, Li TX, Zhang XQ, Liu J. Roles of Non-Coding RNAs in Normal Human Brain Development, Brain Tumor, and Neuropsychiatric Disorders. Noncoding RNA. 2019;5:36
11. Chen Z, Huang J, Feng Y, Li Z, Jiang Y. Screening and bioinformatics analysis of a ceRNA network based on the circular RNAs, miRNAs, and mRNAs in pan-cancer. Cancer Med. 2020;9:7279-92
12. Yao X, Liu M, Jin F, Zhu Z. Comprehensive Analysis of Differentially Expressed Circular RNAs in Patients with Senile Osteoporotic Vertebral Compression Fracture. Biomed Res Int. 2020;2020:4951251
13. Bi J, Liu H, Dong W, Xie W, He Q, Cai Z. et al. Circular RNA circ-ZKSCAN1 inhibits bladder cancer progression through miR-1178-3p/p21 axis and acts as a prognostic factor of recurrence. Mol Cancer. 2019;18:133
14. Yu W, Ma B, Zhao W, Liu J, Yu H, Tian Z. et al. The combination of circRNA-UMAD1 and Galectin-3 in peripheral circulation is a co-biomarker for predicting lymph node metastasis of thyroid carcinoma. Am J Transl Res. 2020;12:5399-415
15. Zhang H, Shen Y, Zhang B, Qian M, Zhang Y, Yang H. Hsa_circ_0003829 serves as a potential diagnostic predictor for oral squamous cell carcinoma. J Int Med Res. 2020;48:300060520936880
16. Liu Y, Li R, Wang X, Yang W. CircTTBK2 Contributes to the Progression of Glioma Through Regulating miR-145-5p/CPEB4 Axis. Cancer Manag Res. 2020;12:8183-95
17. Wang B, Li B, Si T. Knockdown of circ0082374 inhibits cell viability, migration, invasion and glycolysis in glioma cells by miR-326/SIRT1. Brain Res. 2020;1748:147108
18. Xia L, Yi F, Zhai X, Zhang M. [Circular RNA homeodomain-interacting protein kinase 3 (circHIPK3) promotes growth and metastasis of glioma cells by sponging miR-124-3p]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2020;36:609-15
19. Li H, Xia T, Guan Y, Yu Y. Sevoflurane Regulates Glioma Progression by Circ_0002755/miR-628-5p/MAGT1 Axis. Cancer Manag Res. 2020;12:5085-98
20. Zulkarnain S, Yunus N, Kandasamy R, Zun AB, Mat Zin AA. Evaluation Study of Intraoperative Cytology Smear and Frozen Section of Glioma. Asian Pac J Cancer Prev. 2020;21:3085-91
21. KrOl SK, E BE, A SL-B, DmoszyNska-Graniczka M, Boryczka S, Stepulak A. Synthetic Betulin Derivatives Inhibit Growth of Glioma Cells In Vitro. Anticancer Res. 2020;40:6151-8
22. Nitta M, Muragaki Y. [(7)Treatment Strategy for Grade 2 Glioma:Surgery, Radiotherapy, and Chemotherapy]. No Shinkei Geka. 2019;47:147-58
23. He Z, Long J, Yang C, Gong B, Cheng M, Wang Q. et al. LncRNA DGCR5 plays a tumor-suppressive role in glioma via the miR-21/Smad7 and miR-23a/PTEN axes. Aging (Albany NY). 2020;12:20285-307
24. Zhang K, Hu H, Xu J, Qiu L, Chen H, Jiang X. et al. Circ_0001421 facilitates glycolysis and lung cancer development by regulating miR-4677-3p/CDCA3. Diagn Pathol. 2020;15:133
25. Liang Y, Shi J, He Q, Sun G, Gao L, Ye J. et al. Hsa_circ_0026416 promotes proliferation and migration in colorectal cancer via miR-346/NFIB axis. Cancer Cell Int. 2020;20:494
26. Chen X, Xie X, Zhou W. CircCFL1/MiR-107 Axis Targeting HMGB1 Promotes the Malignant Progression of Diffuse Large B-Cell Lymphoma Tumors. Cancer Manag Res. 2020;12:9351-62
27. Wang X, Yao Y, Jin M. Circ-0001068 is a novel biomarker for ovarian cancer and inducer of PD1 expression in T cells. Aging (Albany NY). 2020;12:19095-106
28. Ji C, Hong X, Lan B, Lin Y, He Y, Chen J. et al. Circ_0091581 Promotes the Progression of Hepatocellular Carcinoma Through Targeting miR-591/FOSL2 Axis. Dig Dis Sci. 2020 (Epub: doi: 10.1007/s10620-020-06641-4)
29. Wei X, Zheng W, Tian P, He Y, Liu H, Peng M. et al. Oncogenic hsa_circ_0091581 promotes the malignancy of HCC cell through blocking miR-526b from degrading c-MYC mRNA. Cell Cycle. 2020;19:817-24
30. Li Z, Li X, Xu D, Chen X, Li S, Zhang L. et al. An update on the roles of circular RNAs in osteosarcoma. Cell Prolif. 2020: e12936.
31. Zhou P, Zheng G, Li Y, Wu D, Chen Y. Construction of a circRNA-miRNA-mRNA Network Related to Macrophage Infiltration in Hepatocellular Carcinoma. Front Genet. 2020;11:1026
32. Liu T, Zhang G, Wang Y, Rao M, Zhang Y, Guo A. et al. Identification of Circular RNA-MicroRNA-Messenger RNA Regulatory Network in Atrial Fibrillation by Integrated Analysis. Biomed Res Int. 2020;2020:8037273
33. Li Z, Zheng J, Lin W, Weng J, Hong W, Zou J. et al. Circular RNA hsa_circ_0001785 inhibits the proliferation, migration and invasion of breast cancer cells in vitro and in vivo by sponging miR-942 to upregulate SOCS3. Cell Cycle. 2020:1-15
34. Shen D, Zhao H, Zeng P, Song J, Yang Y, Gu X. et al. Circular RNA hsa_circ_0005556 Accelerates Gastric Cancer Progression by Sponging miR-4270 to Increase MMP19 Expression. J Gastric Cancer. 2020;20:300-12
35. Wu M, Sun T, Xing L. Circ_0004913 Inhibits Cell Growth, Metastasis, and Glycolysis by Absorbing miR-184 to Regulate HAMP in Hepatocellular Carcinoma. Cancer Biother Radiopharm. 2020 (Epub: doi: 10.1089/cbr. 2020 3779)
36. Liu G, Zhou S, Li X, Ding X, Tian M. Inhibition of hsa_circ_0002570 suppresses high-glucose-induced angiogenesis and inflammation in retinal microvascular endothelial cells through miR-1243/angiomotin axis. Cell Stress Chaperones. 2020;25:767-77
37. Chen H, You MJ, Jiang Y, Wang W, Li L. RMI1 attenuates tumor development and is essential for early embryonic survival. Mol Carcinog. 2011;50:80-8
38. Suwa A, Kurama T, Shimokawa T. Adipocyte hyperplasia and RMI1 in the treatment of obesity. FEBS J. 2011;278:565-9
39. Hoadley KA, Xu D, Xue Y, Satyshur KA, Wang W, Keck JL. Structure and cellular roles of the RMI core complex from the bloom syndrome dissolvasome. Structure. 2010;18:1149-58
Author contact
![]() Corresponding authors: Jin Qian, M.D., Department of Neurosurgery, People's Hospital of Xuancheng City, Xuancheng, Anhui, China. Tel: 18705636336 E-mail: joshuajinqiancom; Chunfa Qian, M.D., Department of Neurosurgery, The Affiliated Brain Hospital of Nanjing Medical University, Nanjing, Jiangsu, China. Tel: 13813965019, E-mail: njnkqcfedu.cn.
Corresponding authors: Jin Qian, M.D., Department of Neurosurgery, People's Hospital of Xuancheng City, Xuancheng, Anhui, China. Tel: 18705636336 E-mail: joshuajinqiancom; Chunfa Qian, M.D., Department of Neurosurgery, The Affiliated Brain Hospital of Nanjing Medical University, Nanjing, Jiangsu, China. Tel: 13813965019, E-mail: njnkqcfedu.cn.

 Global reach, higher impact
Global reach, higher impact