Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(17):5136-5143. doi:10.7150/jca.57460 This issue Cite
Research Paper
Methylation of PAX1 gene promoter in the prediction of concurrent chemo-radiotherapy efficacy in cervical cancer
1. Department of Oncology, Xiangya Hospital, Central South University, Hunan 410008 China.
2. Department of Obstetrics and Gynecology, Xiangya Hospital, Central South University, Hunan 410008 China.
3. Department of Clinical Pharmacology, Xiangya Hospital, Central South University, Hunan 410008 China.
Received 2020-12-22; Accepted 2021-6-13; Published 2021-6-22
Abstract
Objectives: Cervical cancer is the fourth leading cause of cancer death among women worldwide. In currently, aberrant methylation of PAX1 is found in variety of solid tumors, including cervical cancer. In addition, the role of PAX1 gene methylation in cervical cancer and precancerous lesions screening has been confirmed in previous study. Here, we evaluated the predictive value of PAX1 methylation in concurrent chemo-radiotherapy (CCRT) outcomes in cervical cancer.
Methods: This study enrolled 82 cervical cancer patients from August 2018 to August 2020. We compared the clinical results between different PAX1 methylation status. Hyper-methylation patients were subjects to MRI and quantitative methylation-specific PCR (QMSP) for PAX1 before, in the middle, immediately after, 1 month and 3 months after CCRT. The changes in PAX1 methylation during CCRT were analyzed.
Results: The lower PAX1 methylation status were related to a poor tumor response. Based on the MRI findings three months post-treatment, the hypermethylated patients were classified into the complete response (CR; n=50) and partial remission (PR; n=18) groups. The average PAX1 △Cp value of CR and PR groups before radiotherapy was 5.08±1.98 and 4.32±2.00 respectively, and after concurrent chemo-radiotherapy was significantly increased to 17.35±4.96 and 16.99±6.17, respectively (P<0.05). Furthermore, the PAX1 △Cp value between CR and PR groups were significantly different at mid-treatment and performed well in predicting short-term efficacy (AUC 0.84) in this period, and its sensitivity and specificity for predicting PR were 0.72 and 0.88, respectively.
Conclusion: The PAX1 methylation level may predict the sensitivity and efficacy of CCRT in cervical cancer.
Keywords: Cervical cancer, PAX1, DNA methylation, Current chemo-radiotherapy, Biomarkers
Introduction
Cervical cancer ranks fourth among gynecological malignancies in terms of both incidence and mortality rates, and 80% of the newly diagnosed cases are from developing countries [1]. Radiotherapy is a common treatment modality for cervical cancer, and has significantly improved patient prognosis, prolonged survival rates, and reduced the risk of recurrence and distant metastasis. However, some patients are resistant to radiotherapy, and its side effects such as radiation enteritis and bone marrow suppression significantly reduce the quality of life. Therefore, early assessment of tumor response is critical to avoid exposing the potentially unresponsive patients to radiotoxicity. Currently, radiotherapeutic effects on cervical tumors are monitored by computed tomography (CT), magnetic resonance imaging (MRI) and other imaging techniques. However, they may take weeks or even months to ascertain treatment efficacy [2]. Therefore, the high costs, long duration and increased potential radiation burden significantly limit the utility of MRI during clinical follow-up. On the other hand, the lag of tumor histological regression, oedema and inflammation of surrounding normal tissue caused by radiation changes may hide the morphological changes of tumor to a certain extent, which may lead to deviation in evaluating tumor measurement [3]. In contrast, molecular biomarkers can enable real-time monitoring and early prediction of treatment efficacy and help optimize individualized treatment methods for cervical cancer.
Ionizing radiations often lead to epigenetic changes such as DNA methylation, which alter gene expression levels without any transcriptional changes [4]. The DNA methylation status of normal and malignant cells is considerably different, which indicates its biological significance in tumor development. Aberrant methylation of CpG islands in the promoter region of tumor suppressor genes is an early event in carcinogenesis, and induces malignant transformation via decreased expression and even loss of function of these genes [5]. The progression of cervical cancer is dependent on multiple tumor suppressors including PAX1, SOX1, LMX1A and ZNF582 [6-8].
The PAX1 gene is located on chromosome 20p11 and consists of a paired domain (PD) and an octapeptide domain (OP). It plays an important role in the growth and development of bone, spine, thymus and parathyroid gland [9, 10]. Lai et al. [11] first reported that the PAX1 gene is significantly hypermethylated (PAX1m) in cervical cancer tissues compared to normal cervical tissues, and the methylation level correlated positively with the tumor grade. Subsequent studies demonstrated that the PAX1 methylated level could accurately distinguish high-grade CIN lesions and cervical cancer. For instance, PAX1m can detect CIN3+ lesions with sensitivity and specificity of 86% and 85%, respectively [12, 13]. In addition, PAX1m shows a better diagnostic performance than HPV-DNA for the triage of patients with atypical squamous cells of undetermined significance (ASCUS) [14].
Therefore, PAX1 methylation level is a promising biomarker for cervical cancer screening and early diagnosis. However, few studies have evaluated the changes in PAX1 gene methylation status during radiotherapy, and it is unclear whether it can predict the therapeutic response in cervical cancer. In this study, we investigated the predictive value of PAX1 methylation status in monitoring the early response to radiotherapy in cervical cancer.
Methods
Patients and study design
This study was approved by the Ethics Committee of Xiangya Hospital (202010138), and all treatment procedures were carried out in accordance with the relevant guidelines. The inclusion criteria were as follows: (1) cervical cancer diagnosis by pathological biopsy and (2) no previous history of surgery and chemoradiotherapy for cervical cancer. Patients that did not complete the treatment regimen, showed recurrence, underwent non-primary treatment, or lost follow-up were excluded.
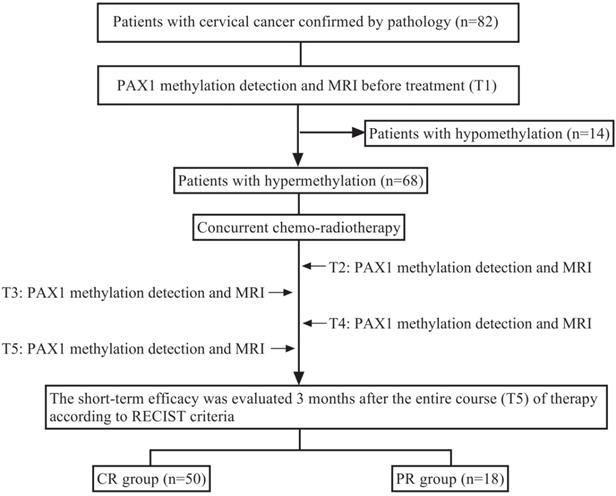
A total of 82 cervical cancer patients were enrolled in this cohort. The patients with hypermethylation before concurrent chemo-radiotherapy (CCRT) were further monitored during radiotherapy by methylation tests and MRI at the following time points: T1 (baseline assessment before treatment), T2 (middle stage of radiotherapy), T3 (end of radiotherapy), T4 (1 month after radiotherapy) and T5 (3 months after radiotherapy). Early tumor response was determined by comparing the baseline MRI results with that of three months post-radiotherapy. The research design is illustrated in Figure 1.
Treatment
All patients were treated with a combination of external beam radiotherapy (EBRT) and intracavitary brachytherapy (ICR). The total dose of EBRT was 45~50Gy across the entire course of treatment, and accompanied with 35~40mg/m2 cisplatin once a week. The treatment plan was adjusted according to the condition of the patients. ICR was initiated after 15 rounds of EBRT.
MR imaging and evaluation
Baseline tumor assessment was performed before CCRT, and the patients were staged based on MRI and gynecological examination. The final treatment response was determined 3 months post-CCRT by MRI, visual diagnosis and physical examination, and compared to baseline. The clinical efficacy was evaluated according to RECIST criteria as complete remission (CR), partial remission (PR), stable disease (SD) or progressive disease (PD) [15]. The width, height and thickness of the tumor were measured based on T2-weighted images, and tumor volume was calculated as 1/6*π*w*h*t [16]. The change in tumor size (%) was calculated as (pre-treatment volume - post-treatment volume) / pre-treatment volume × 100 (%).
Research design. Before treatment (T1), PAX1 was detected in patients with cervical cancer who were scheduled to receive CCRT. Hypermethylated patients were detected by MRI and methylation test at T2 (middle stage of radiotherapy), T3 (end of radiotherapy), T4 (1 month after radiotherapy), T5 (3 months after radiotherapy) time points during treatment, and were grouped with MRI.
Quantitative methylation-specific PCR
PAX1 methylation was evaluated at the aforementioned time points. Cervical exfoliated cells were obtained by cervical brush during gynecological examination, centrifuged and stored in phosphate-buffered saline at -20°C. Genomic DNA was extracted using standard protocols, and converted to bisulfite form using the EZ DNA Methylation-Gold kits (Zymo Research, Irvine, CA, USA) according to the manufacturer's instructions. Methylation-specific PCR was performed on the Light Cycler LC480 system (Roche Applied Science, Penzberg, Germany) to determine the methylation level of PAX1. Type II collagen gene (COL2A) was used as an internal reference. The △Cp is the difference between the △Cp values for PAX1m and COL2A. The methylation level (△Cp) was assessed by the following formula: △Cp=Cptarget gene - CpCol2A [12]. The smaller △Cp value denote a higher degree of methylation detected in the collected samples. Accordingly, if △Cp≤9 (the cut-off value), PAX1 was considered hypermethylated or positive. A decrease of △Cp value appeared indicating an increased methylation level [17]. The change in △Cp value (%) was calculated using the following equation: (post-treatment △Cp - pre-treatment △Cp) / pre-treatment △Cp × 100 (%).
Statistical analysis
SPSS software version 25.0 was used for statistical analysis. Quantitative data were expressed by means ± standard deviation (SD). Student's t test, Chi-square test or Fisher exact test was used to evaluate the relationship between PAX1 methylation status and the clinical parameters and treatment responses. The multiple comparisons of PAX1m level, change of △Cp value, tumor size and tumor size regression rate between two groups at each time were performed using repeated measures analysis of variance (ANOVA) and Student's t test. Receiver operating characteristic (ROC) analysis was performed to investigate the discriminatory capacity of PAX1 and tumor size for early response to radiotherapy. The area under the curve (AUC) was computed for the mean ROC curves. The threshold obtained from the Youden's index was used to calculate the sensitivity and specificity. All tests were performed using bilateral 95% confidence intervals (CI). P < 0.05 was considered statistically significant.
Results
Relationship between PAX1 methylation status and clinical parameters
In this study, we performed QMSP to detect PAX1 methylation level in 82 patients. We explored the correlation of baseline PAX1 methylation status with clinical outcomes including age, FIGO stage, pathological type, HPV status, lymph node metastasis, tumor size and short-term efficacy. From the result, we found that the hypermethylation before CCRT was related to a superior treatment response (P=0.025). However, the patient age, FIGO stage, pathological type, HPV status, lymph node metastasis and tumor size were not statistically significant (P>0.05) (Table 1).
We also explored that 68 patients exhibiting high pre-treatment PAX1 methylation level. We further investigated the PAX1 methylation level for the hypermethylated group before, in the middle, immediately after, 1 month and 3 months after the CCRT. Among them, 50 patients were classified as CR and 18 patients were PR according to the 3 months post-CCRT MRI evaluation.
Changes of PAX1 methylation levels during treatment
We monitored the PAX1 methylation changes of the 68 hypermethylated patients during radiotherapy. According to the MRI results, we further grouped 68 patients into CR (n=50) and PR (n=18) group. The average PAX1 △Cp values of the CR and PR groups before radiotherapy were 5.08±1.98 and 4.32±2.00, respectively, and the △Cp of the CR group was higher than that of the PR group, not reaching significance (P=0.17). At the end of CCRT (T3), the △Cp values of the CR and PR groups increased significantly to 17.35±4.96 and 16.99±6.17, respectively (P< 0.05).
After the initiation of CCRT, the most dramatic change occurred in two groups (change rate, 183.80±163.18% vs 62.13±99.04%, P=0.00) at T2. For the CR group, △Cp values of PAX1 methylation increased sharply from T1 to T2 and gradually thereafter. The PAX1 △Cp values increased at T2 and T3 were significantly different from those before CCRT (P<0.05) (Table 2). In contrast, the PAX1 △Cp values increased at a slower rate after treatment initiation in the PR group and slightly higher at T2 compared to baseline level (Table 2). A significant change in PAX1 methylation level was observed only at the end of the treatment for the PR group. Finally, the values of △Cp was significantly less in the PR group compared to the CR group at the treatment mid-point (Table 2). Thus, results showed that the trend of △Cp value was different in patients with different therapeutic effects, especially in the middle of the stage of radiotherapy.
Relationship between PAX1 promoter methylation and clinical parameters.
| Characteristics | PAX1 hypo-(n=14) | PAX1 hyper-(n=68) | P | |
|---|---|---|---|---|
| Age (years) | 57.21±6.267 | 54.77±8.75 | 0.32# | |
| FIGO stage | 0.62Δ | |||
| <ⅡB | 2(14.29) | 6(8.82) | ||
| ≥ⅡB | 12(85.71) | 62(91.18) | ||
| Pathology | 0.07Δ | |||
| SCC | 12(85.71) | 67(98.53) | ||
| AC | 2(14.29) | 1(1.47) | ||
| Lymph node metastasis | 0.16* | |||
| No | 6(42.86) | 43(63.24) | ||
| Yes | 8(57.14) | 25(36.76) | ||
| HPV | 0.17* | |||
| Negative | 7(50.00) | 21(30.88) | ||
| Positive | 7(50.00) | 47(69.12) | ||
| Size of tumor (cm3) | 21.50±24.15 | 23.41±22.74 | 0.78# | |
| Tumor response | 0.025* | |||
| CR | 6 | 50 | ||
| PR | 8 | 18 |
Note: Data presented as mean ± standard deviation or n (%). SCC: squamous cell carcinoma; AC: adenocarcinoma; FIGO: International Federation of Gynecology and Obstetrics; HPV: human papilloma virus. CR: complete response; PR: partial response. #T test; *Chi-square test; ΔFisher's test.
Difference of PAX1 methylation level between CR and PR groups at each time points of treatment for cervical cancer.
| Time points | △Cp | △Cp change rate (%) | ||||
|---|---|---|---|---|---|---|
| CR (n=50) | PR (n=18) | P | CR (n=50) | PR (n=18) | P | |
| T1 | 5.08±1.98 | 4.32±2.00 | 0.17 | - | - | - |
| T2 | 12.57±5.88 | 6.15±4.09 | 6.50×10-5 | 183.80±163.18 | 62.13±99.04 | 0.004 |
| T3 | 17.35±4.96 | 16.99±6.17 | 0.80 | 315.82±258.21 | 381.19±194.96 | 0.38 |
| T4 | 15.60±4.76 | 14.88±4.94 | 0.59 | 274.82±220.35 | 302.69±200.91 | 0.64 |
| T5 | 16.66±4.95 | 17.09±5.67 | 0.76 | 300.49±235.44 | 377.33±280.21 | 0.26 |
Note: T1 (pre-radiotherapy), T2 (middle stage of radiotherapy), T3 (end of radiotherapy), T4 (1 month after radiotherapy) and T5 (3 months after radiotherapy). CR: complete response; PR: partial response.
Changes of tumor size during treatment
Tumor size is one of the important factors that affect efficacy. In 68 patients with high PAX1 methylation level, we analyzed the change of tumor size (n=68) and during radiotherapy to explore the correlations between therapeutic response and the change rate of tumor size during CCRT. The mean baseline tumor size of hypermethylated patients was 23.41 ± 22.74 cm3 (Table 1). The mean baseline tumor size of CR and PR group were 16.08 ± 13.55 cm3, 43.79±30.19 cm3, respectively, and were remarkably decreased after radiotherapy in the two groups (P<0.05, Table 3). At each time point, tumor size in the PR group were significantly larger than those in the CR group. However, no significant difference in tumor regression rate was observed between the groups at T2 and T3 (All P<0.05, Table 3). Up to T4 and T5 time point, the tumor regression rate in CR group was significantly higher than that in the PR group. This indicated that the change trend is not obvious at the early stage of treatment.
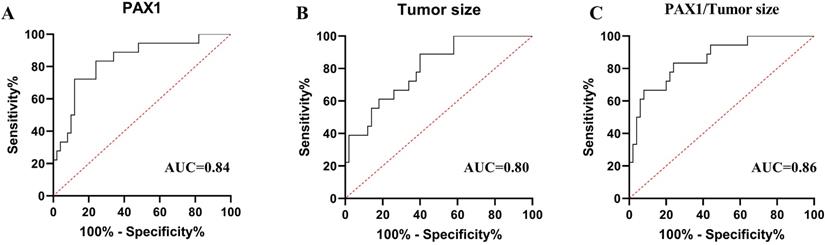
Receiver operating characteristic (ROC) curve analysis of PAX1m (A), tumor size (B) and combined factors (C) during CCRT. The area under the curve (AUC)of each parameter's ROC curve was calculated for the predicting tumor residual group.
Comparison of change rate of tumor size during radiotherapy between CR and PR groups.
| Time points | Tumor size (cm3) | Tumor size regression (%) | ||||
|---|---|---|---|---|---|---|
| CR (n=50) | PR (n=18) | P | CR (n=50) | PR (n=18) | P | |
| T1 | 16.08±13.55 | 43.79±30.19 | 2.00×10-6 | - | - | - |
| T2 | 5.00±4.74 | 12.73±8.40 | 1.10×10-5 | 65.16-25.27 | 63.82-20.94 | 0.83 |
| T3 | 1.05±1.45 | 3.73±2.43 | 5.51×10-7 | 92.25-10.34 | 87.51-13.99 | 0.14 |
| T4 | 0.23±0.53 | 1.83±1.56 | 2.05×10-8 | 98.30-4.34 | 94.28-5.57 | 3.20×10-5 |
| T5 | 0.00±0.00 | 0.77±0.71 | 7.72×10-11 | 100.00-0.00 | 97.67-2.51 | 5.83×10-22 |
Note: T1 (pre-radiotherapy), T2 (middle stage of radiotherapy), T3 (end of radiotherapy), T4 (1 month after radiotherapy) and T5 (3 months after radiotherapy).CR: complete response; PR: partial response.
The performance of two detection methods at the middle stage of treatment.
| Variable | Cut-off | AUC | Sensitivity | Specificity | PPV | NPV | 95%CI | P value |
|---|---|---|---|---|---|---|---|---|
| PAX1 | 6.38 | 0.84 | 0.72 | 0.88 | 0.68 | 0.90 | 0.73-0.95 | 2.50×10-5 |
| Tumor size (cm3) | 4.59 | 0.80 | 0.89 | 0.60 | 0.44 | 0.94 | 0.69-0.91 | 1.65×10-4 |
| Combined factors | - | 0.86 | 0.83 | 0.76 | 0.56 | 0.93 | 0.75-0.93 | 7.00×10-6 |
Predictive value of PAX1 methylation level and tumor size in mid-treatment
To evaluated and compared the diagnostic performances of PAX1 methylation and tumor size in predicting the tumor partial response at T2 stage, we generated the ROC analysis curves (Figure 2). The AUCs of PAX1 methylation and tumor size for predicting PR at T2 were 0.84 (P<0.05, 95% CI:0.73-0.95) and 0.80 (P<0.05, 95% CI: 0.69-0.91). The cutoff PAX1 methylation △Cp value was 6.38 and the sensitivity, specificity, PPV and NPV for predicting PR were 0.72, 0.88, 0.68 and 0.90, respectively. Tumor size ≥ 4.59 cm³ in the middle of the treatment regimen predicted PR with the specificity of 0.60, sensitivity of 0.89, PPV of 0.44, NPV of 0.94 (Table 4).
To test the adjunct role of DNA methylation for MRI, we calculated the combined diagnostic performances of PAX1 methylation and tumor size. As shown in Table 4, the combination of two factors at T2 had an average AUC of 0.86 (P<0.05, 95% CI:0.75-0.93), with a specificity of 0.76, a sensitivity of 0.83, PPV of 0.56, NPV of 0.93.
Discussion
The role of PAX1 gene detecting in cervical cancer screening and triage has been demonstrated. We analyzed the methylation level of PAX1 gene in cervical cancer cells during different periods of CCRT using qMSP-PCR and found that PAX1 methylation level can predict and monitor early therapeutic response.
DNA methylation is a key event in tumor genesis and progression. The tumor suppressor PAX1 is aberrantly methylated in various human malignancies, such as oral cancer [18], esophageal cancer [19], ovarian cancer [20] etc. PAX1 inhibits cancer cell growth by forming complexes with SETIB and WDR5 to activate phosphatase that inhibit the oncogenic MAPK and SRC pathways [21]. Thus, hypermethylation of PAX1 promoter silences its expression and promotes cancer progression. Recent studies show that PAX1 methylation level is a reliable marker for the differential diagnosis of benign and malignant cervical growth, and has been approved in Taiwan to supplement cytological examination. Lai et al. [11] reported that the methylation rates of PAX1 in cervical cancer were as high as 94.5%. Huang et al. showed that the level of PAX1 methylation was associated with TNM staging in colorectal cancer [22]. In present study, we detected 83% methylation rate of PAX1 in the cervical cancer tissues. However, there were no significant differences in the age, tumor size, FIGO stage, histological types, HPV and lymphatic metastasis between the PAX1 hypermethylated and PAX1 hypomethylated groups, suggesting that PAX1 methylation status may not be affected by other clinical factors.
Many researchers have previously found the influence of gene methylation status to evaluate the radiotherapy sensitivity for cervical cancer. David Guerrero-Setas et al. [23] reported that a significant correlation between RASSF2 hypermethylation and bad prognosis of cervical cancer. Wu et al. [24] demonstrated that ZNF582 negative could increase resistance to radiation and affect the prognosis. Meanwhile, Ph Su et al. [21] analyzed DNA methylation from TCGA database that disease-free survival (PFS) and overall survival (OS) of patients with the hypermethylated PAX1 gene was significantly shorter than the hypomethylated patients in uterine cervical cancer. Interestingly, our data show that PAX1 gene hyper-methylation was associated with an excellent therapeutic response, lower methylation level or negative methylation indicates partial response. The explanation might be that therapy difference might be the main reason for this result because our patients underwent CCRT compared with other studies that involved patients underwent surgery or neoadjuvant chemotherapy. Therefore, we infer that the PAX1 hypomethylation may be insensitive to radiation, which may lead to a poor result.
DNA methylation status is reversible and can be altered in response to external stimuli. Antwih et al.[25] found that ionizing radiations significantly altered the level of gene methylation by downregulating DNMT2 in breast cancer cells. Wu et al.[24] reported that ZNF582 methylation levels decreased in patients receiving neoadjuvant radiotherapy. However, few studies have reported the functional of PAX1 methylation during CCRT. In our study as well, PAX1 △Cp value increased after treatment and stabilized after radiotherapy, indicating that PAX1 methylation levels decreased steadily during CCRT. The gradual decrease in PAX1 methylation level was consistent with the change in tumor size in radiotherapy. Effective anticancer therapy leads to the destruction of tumor cell membrane integrity and tumor dissolution, thus reducing the density of tumor size. The decreased methylation levels may suggest fewer cancer cell after treatment. Therefore, the fluctuation of the methylation level is not obvious after CCRT, which may be related to the difficulty in obtaining effective cells. According to our results, PAX1 methylation levels decreased significantly by the mid-point of CCRT in the CR but not PR groups.
Tumor size is closely related to the prognosis of cervical cancer patients [26-28]. However, in our study, the mid-RT tumor regression rates were similar between the CR and PR group, and significant differences were seen only after the end of treatment. The unsatisfactory outcome of short-term treatment may be lagging at the early reflection. Furthermore, changes in tumor size varied only at the end of radiotherapy. These data indicate that changes in PAX1 methylation levels precede tumor size because epigenetic changes in tumor cell may have occurred in the early stages of treatment. Therefore, monitoring the dynamic change in PAX1 methylation can help optimize individualized therapeutic strategies. Early decrease of PAX1 level indicates a better response to radiotherapy and a favorable prognosis in cervical cancer, whereas minute fluctuations may reflect a therapeutically unresponsive tumor.
In present study, PAX1 methylation level showed slightly better performance than tumor size for distinguishing between CR or residual tumor cells (AUC 0.84 vs 0.80). In addition, at the △Cp threshold of 6.38, PAX1 methylation distinguished CR and PR with the high specificity of 0.88 compared to only 0.60 for tumor size. Therefore, the reasonable sensitivity (0.72 vs 0.89) and high specificity (0.88 vs 0.60) of PAX1 methylation make it a promising predictive marker of the short-term efficacy of CCRT in cervical cancer, and combining it with MRI may further enhance the accuracy of therapeutic monitoring. Radiotherapy significantly improves the treatment outcomes in cervical cancer in a dose-dependent manner. It is critical to rapidly evaluate the early treatment response, precisely and sensitively, to adjust the treatment plan and reduce the risk of adverse reactions. Taken in aggregate, PAX1 methylation level is a better indicator of the early molecular changes in the tumor during treatment than MRI parameters to timely detect patients who are not sensitive to radiotherapy and adjust treatment regimen. In addition, considering the second pathological biopsy may bring trauma and subjective pain to patients because the tumor shrinks after treatment, so cytology has the advantage of more straightforward, convenient and non-invasive compared pathological examination when needed to collect samples at multiple time points. Methylation PCR can be easily performed using cervical exfoliated cells and does not require cervical tissue.
This study has several limitations that ought to be considered. First, the small sample size limits the generalizability of the results, although each patient underwent five tests that increased the data pool. Nevertheless, our findings need to be validated on larger cohorts. Second, we only analyzed the effect of PAX1 methylation on short-term therapeutic efficacy, and its effect on long-term survival will need to be confirmed by long-term follow-up. Third, in the absence of histological confirmation, it may be impractical to judge through magnetic resonance imaging, and future animal experiments will help us to understand the early tumor response better.
In conclusion, we found that PAX1 gene methylation status will change under the influence of radiation, and may predict early treatment response of cervical cancer patients post-radiotherapy. Our result indicates that PAX1 methylation as a promising biomarker plays an important role in monitoring and treatment following up of cervical cancer. Therefore, by combining methylation detection with traditional imaging methods, it may provide a new method for monitoring the effect of treatment.
Acknowledgements
This study was supported by the Natural Science Foundation of Hunan province (grant number 2019JJ40490 and 2020JJ4903), Clinical Research Project of Xiangya Hospital (grant number 2016L06) and the 12th Five-Year Plan of Education Science in Hunan Province (XJKO11BGD032).
Author Contributions
Xuanxuan Li: Performed collected data, statistical analysis and drafted the manuscript.
Xue Zhou: Participated in data collection.
Manting Zeng: Participated in data collection and statistical analysis.
Yangying Zhou: Participated in the discussion of article revision and provided advice during manuscript modification.
Yu Zhang: Participated in designing and developed of study.
Yu-Ligh Liou: Designed and developed of study and provided technical assistance.
Hong Zhu: Conceived and designed the study, supervised the research program and manuscript review and provided advice during the study.
All authors: Read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA-Cancer J Clin. 2018;68:394-424
2. Harry VN, Semple SI, Parkin DE, Gilbert FJ. Use of new imaging techniques to predict tumour response to therapy. Lancet Oncology. 2010;11:92-102
3. Hricak H, Swift PS, Campos Z, Quivey JM, Gildengorin V, Goranson H. Irradiation of the cervix uteri: value of unenhanced and contrast-enhanced MR imaging. Radiology. 1993;189:381-388
4. Pogribny I, Raiche J, Slovack M, Kovalchuk O. Dose-dependence, sex- and tissue-specificity, and persistence of radiation-induced genomic DNA methylation changes. Biochemical and Biophysical Research Communications. 2004;320:1253-1261
5. Montenegro MF, Sanchez-del-Campo L, Fernandez-Perez MP, Saez-Ayala M, Cabezas-Herrera J, Rodriguez-Lopez JN. Targeting the epigenetic machinery of cancer cells. Oncogene. 2015;34:135-143
6. Rogeri CD, Santejo Silveira HC, Causin RL, Villa LL, Stein MD, de Carvalho AC. et al. Methylation of the hsa-miR-124, SOX1, TERT, and LMX1A genes as biomarkers for precursor lesions in cervical cancer. Gynecologic Oncology. 2018;150:545-551
7. van Leeuwen RW, Ostrbenk A, Poljak M, van der Zee AGJ, Schuuring E, Wisman GBA. DNA methylation markers as a triage test for identification of cervical lesions in a high risk human papillomavirus positive screening cohort. International Journal of Cancer. 2019;144:746-754
8. Lin H, Chen T-C, Chang T-C, Cheng Y-M, Chen C-H, Chu T-Y. et al. Methylated ZNF582 gene as a marker for triage of women with Pap smear reporting low-grade squamous intraepithelial lesions - A Taiwanese Gynecologic Oncology Group (TGOG) study. Gynecologic Oncology. 2014;135:64-68
9. Yamazaki Y, Urrutia R, Franco LM, Giliani S, Zhang K, Alazami AM. et al. PAX1 is essential for development and function of the human thymus. Science Immunology. 2020;5:eaax1036
10. Paixao-Cortes VR, Salzano FM, Bortolini MC. Origins and evolvability of the PAX family. Seminars in Cell & Developmental Biology. 2015;44:64-74
11. Lai H-C, Lin Y-W, Huang THM, Yan P, Huang R-L, Wang H-C. et al. Identification of novel DNA methylation markers in cervical cancer. International Journal of Cancer. 2008;123:161-167
12. Kan Y-Y, Liou Y-L, Wang H-J, Chen C-Y, Sung L-C, Chang C-F. et al. PAX1 Methylation as a Potential Biomarker for Cervical Cancer Screening. International Journal of Gynecological Cancer. 2014;24:928-934
13. Luan T, Hua Q, Liu X, Xu P, Gu Y, Qian H. et al. PAX1 Methylation as a Potential Biomarker to Predict the Progression of Cervical Intraepithelial Neoplasia: A Meta-analysis of Related Studies. International Journal of Gynecological Cancer. 2017;27:1480-1488
14. Li S-R, Wang Z-M, Wang Y-H, Wang X-B, Zhao J-Q, Xue H-B. et al. Value of PAX1 Methylation Analysis by MS-HRM in the Triage of Atypical Squamous Cells of Undetermined Significance. Asian Pacific journal of cancer prevention: APJCP. 2015;16:5843-5846
15. Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R. et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). European Journal of Cancer. 2009;45:228-247
16. Fischerova D, Cibula D, Stenhova H, Vondrichova H, Calda P, Zikan M. et al. Transrectal ultrasound and magnetic resonance imaging in staging of early cervical cancer. International Journal of Gynecological Cancer. 2008;18:766-772
17. Liou YL, Zhang TL, Yan T, Yeh CT, Kang YN, Cao L. et al. Combined clinical and genetic testing algorithm for cervical cancer diagnosis. Clin Epigenetics. 2016;8:66
18. Cheng S-J, Chang C-F, Lee J-J, Chen H-M, Wang H-J, Liou Y-l. et al. Hypermethylated ZNF582 and PAX1 are effective biomarkers for detection of oral dysplasia and oral cancer. Oral Oncology. 2016;62:34-43
19. Huang J, Wang G, Tang J, Zhuang W, Wang LP, Liou YL. et al. DNA Methylation Status of PAX1 and ZNF582 in Esophageal Squamous Cell Carcinoma. Int J Environ Res Public Health. 2017;14:216
20. Hassan ZK, Hafez MM, Kamel MM, Zekri ARN. Human Papillomavirus Genotypes and Methylation of CADM1, PAX1, MAL and ADCYAP1 Genes in Epithelial Ovarian Cancer Patients. Asian Pacific journal of cancer prevention: APJCP. 2017;18:169-176
21. Su PH, Lai HC, Huang RL, Chen LY, Wang YC, Wu TI. et al. Paired Box-1 (PAX1) Activates Multiple Phosphatases and Inhibits Kinase Cascades in Cervical Cancer. Sci Rep. 2019;9:9195
22. Huang J, Tan Z-R, Yu J, Li H, Lv Q-L, Shao Y-Y. et al. DNA hypermethylated status and gene expression of PAX1/SOX1 in patients with colorectal carcinoma. Oncotargets and Therapy. 2017;10:4739-4751
23. Guerrero-Setas D, Perez-Janices N, Blanco-Fernandez L, Ojer A, Cambra K, Berdasco M. et al. RASSF2 hypermethylation is present and related to shorter survival in squamous cervical cancer. Modern Pathology. 2013;26:1111-1122
24. Wu NY, Zhang X, Chu T, Zhu S, Deng Y, Zhou Y. et al. High methylation of ZNF582 in cervical adenocarcinoma affects radiosensitivity and prognosis. Ann Transl Med. 2019;7:328
25. Antwih DA, Gabbara KM, Lancaster WD, Ruden DM, Zielske SP. Radiation-induced epigenetic DNA methylation modification of radiation-response pathways. Epigenetics. 2013;8:839-848
26. Kidd EA, Siegel BA, Dehdashti F, Rader JS, Mutch DG, Powell MA. et al. Lymph Node Staging by Positron Emission Tomography in Cervical Cancer: Relationship to Prognosis. Journal of Clinical Oncology. 2010;28:2108-2113
27. Grigiene R, Valuckas K, Aleknavicius E, Kurtinaitis J, Letautiene SJBc. The value of prognostic factors for uterine cervical cancer patients treated with irradiation alone. 2007; 7: 234.
28. Raut A, Chopra S, Mittal P, Patil G, Mahantshetty U, Gurram L. et al. FIGO Classification 2018: Validation Study in Patients With Locally Advanced Cervix Cancer Treated With Chemoradiation. International Journal of Radiation Oncology Biology Physics. 2020;108:1248-1256
Author contact
![]() Corresponding authors: Prof. Hong Zhu, Department of Oncology, Xiangya Hospital, Central South University, Hunan 410008 China (E-mail: zhuhong0719gcom; telephone: +86-13507488416); Yu-Ligh Liou, Department of Clinical Pharmacology, Xiangya Hospital, Central South University, Hunan 410008 China (E-mail: 126567657com)
Corresponding authors: Prof. Hong Zhu, Department of Oncology, Xiangya Hospital, Central South University, Hunan 410008 China (E-mail: zhuhong0719gcom; telephone: +86-13507488416); Yu-Ligh Liou, Department of Clinical Pharmacology, Xiangya Hospital, Central South University, Hunan 410008 China (E-mail: 126567657com)

 Global reach, higher impact
Global reach, higher impact