Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(17):5153-5163. doi:10.7150/jca.54475 This issue Cite
Research Paper
Development and Validation of a Nomogram based on cell growth-related Biomarkers for Oral Squamous Cell Carcinoma
Department of Maxillofacial & E.N.T oncology, Tianjin Medical University Cancer Institute & Hospital, Key Laboratory of Cancer Prevention and Therapy, Tianjin Cancer Institute, National Clinical Research Center of Cancer, Tianjin, China.
#Co-first authors.
Received 2020-10-14; Accepted 2021-5-25; Published 2021-6-22
Abstract
Purpose: We aimed to develop a prognostic nomogram based on immunohistochemistry (IHC) biomarkers of patients with oral squamous cell carcinoma (OSCC).
Methods: A total of 294 patients were enrolled in the study. The least absolute shrinkage and selection operator (LASSO) Cox regression model was performed to develop a combined IHC score (IHCs) classifier.
Results: Five biomarkers, specifically c-Met, Vimentin, HIF-2α, VEGF-c, and Bcl-2 were extracted. Then, an IHCs classifier was developed, and patients were stratified into high- and low-IHCs groups. In the training cohort, the 5-year overall survival (OS) was 62.1% in low-IHCs group and 28.2% in high-IHCs group (P<0.001). The 5-year OS was 68.6% for the low-IHCs group and 28.4% for the high-IHCs group in the validation cohort (P<0.001). The area under the ROC curve (AUROC) of the combination of the IHCs classifier and TNM stage was 0.746 (95% CI: 0.658-0.833) in the training cohort and 0.735 (95% CI: 0.651-0.818) in the validation cohort, respectively.
Conclusions: The nomogram could effectively predict the prognosis for patients with OSCC and may be employed as a potential tool to guide the individual decision-making process.
Keywords: oral squamous cell carcinoma, immunohistochemistry, biomarkers, nomogram, prognosis
Introduction
Oral cancer is a common malignancy in head and neck cancer. It is the eighth most common cancer in males, and the incidence rate increases annually [1]. Oral squamous cell carcinoma (OSCC) is the predominant histological type, accounting for approximately more than 90% of oral cancers. The histological differentiation of OSCC could be different from that of well-differentiated keratinized to undifferentiated nonkeratinizing carcinoma, which would have a significant difference in prognosis [2, 3]. Although improvements in the treatment have evolved from a surgical-based treatment model to multimodal therapies, the prognosis of patients with OSCC remains poor, with the 5-year overall survival (OS) rate ranging from 15% to 60% [4, 5].
The current American Joint Committee on Cancer (AJCC) TNM staging system and important prognostic indicators are widely used as guidelines in the decision-making process [6]. The depth of invasion (DOI) was added to the surface dimensions and local extent of the tumour as the required parameters for primary tumour staging in the eighth edition of the AJCC staging system, making the TNM staging more accurate for the assessment of the tumour. However, in our clinical practice, it was usually found that the survival outcomes of patients with the same TNM staging could differ greatly even if they received similar treatments, indicating that the current TNM staging system could not provide individualized prognostic information for patients, and this might be related to the tumour heterogeneity of OSCC. Therefore, a supplement to TNM staging system may assist in evaluating the tumour growth, invasiveness, and sensitivity to treatment for better predicting the prognosis of patients with OSCC.
With the recognition of molecular biology, many genes/proteins that play an important role in the process of carcinogenesis have been discovered. A molecular biomarker could assist in the diagnosis of disease, help in the treatment decision making process, provide information about the tumour's sensitivity to the treatment and indicate the possible prognostic effects based on response to therapy. Recently, many studies of the OSCC biomarkers have been published; however, OSCC biomarkers are still in the early stage of development, and most of them have been investigated separately, and are far from being combined with clinical applications.
Therefore, a classifier, which combines the expression of molecular biomarkers with the TNM staging system, is urgently needed to classify patients with different risks of relapse and metastasis. This study included several biomarkers that have been reported to be involved in biological processes during carcinogenesis, such as epithelial-mesenchymal transition (EMT), extracellular matrix disassembly, angiogenesis, and cellular regulatory framework involving activation of crucial signalling pathways and enduring hypoxia. Then, we aimed to develop and validate a prognostic nomogram based on clinicopathological features and IHC biomarkers in patients with OSCC after surgery.
Materials and Methods
Patients and Samples
We retrospectively reviewed the medical records of 301 patients who had undergone surgery with or without induction chemotherapy at Tianjin Medical University Cancer Institute and Hospital between January 2010 and December 2015. A total of 294 patients with complete clinical and follow-up information were enrolled in the study. All patients involved were restaged according to the 8th edition of the AJCC staging system. Using computer-generated numbers, patients were randomly stratified into two groups: a training cohort (147 patients) and validation cohort (147 patients). This study was conducted in accordance with the Declaration of Helsinki. Written-informed consent was obtained from all patients, and the study was approved by the Clinical Research Ethics Committee of Tianjin Medical University Cancer Institute and Hospital.
The inclusion criteria were as follows: (1) patients were primarily diagnosed with OSCC and underwent surgical treatment, (2) patients did not have a history of cancer disease or treatment, (3) patients had complete clinical, pathological and follow-up data, and (4) patients provided written informed consent.
Procedure of IHC Expression
The tissues were formalin-fixed and paraffin-embedded. To assess the expression of the biomarkers, the IHC assay was performed using the following antibodies: c-Met (25849-1-AP, Proteintech), vimentin (5741, Cell Signaling), E-cadherin (3195, Cell Signaling), N-cadherin (13116, Cell Signaling), MMP9 (ab38898, Abcam), HIF-1α (36169, Cell Signaling), HIF-2α (ab199, Abcam), β-catenin (84803, Cell Signaling), pSTAT3 (9145, Cell Signaling), VHL (ab238681, Abcam), Bcl-2 (ab32124, Abcam), VEGF-a (ab39250, Abcam), VEGF-c (22601-1-AP, Proteintech), EZH2 (5246, Cell Signaling), SUZ12 (ab12073, Abcam), H3K27AC (ab4729, Abcam). Sections (4 μm thick) were cut from formalin-fixed, paraffin-embedded blocks. The paraffin sections were placed in an oven at 65 °C and baked for 1 hour; the paraffin was removed and then the sections were immersed in distilled water following routine methods. To block endogenous peroxidase, the sections were blocked with 3% peroxide-methanol at room temperature. Antigens were retrieved by boiling in citrate buffer (pH 6.0) in a pressure cooker. Serum blocking was performed using 10% normal rabbit serum for 30 min, and then the slides were incubated with the primary antibody overnight at 4 °C. Then, the cells were incubated with a response enhancer and enhanced enzyme labelled goat anti-mouse/rabbit IgG polymer for 30 min at room temperature. Then, 0.05% 3,3′-diaminobenzidine tetrahydrochloride (DAB) was used prior to counterstaining with modified Harris hematoxylin, and hydrochloride alcohol was used for differentiation.
Tumour cell staining was assigned a score according to a previous study [7]. For the evaluation of IHC results, all the biomarkers were scored by both the maximal intensity of staining (0, negative; 1, weak; 2, moderate; and 3, strong) and the percentage of positive tumors cells (0%-100%; with any intensity of positive staining) to generate a modified IHC score (IHCs, range: 0-300). All of the biomarkers selected were cytoplasmic and membranous in tumour cells. The IHC results were evaluated by 2 independent practising pathologists who were blinded to the clinical outcome of all cases. A third pathologist was consulted when disagreements occurred between the two pathologists. If the result of the third pathologist agreed with one, then that value was employed. If the opinion of the third pathologist was different, the decision would be cooperatively determined by the three pathologists.
Calculation of the Cut-off Point and Validation of the Combined IHCs Classifier
The cutoff score for each biomarker was selected based on the association with the patients' survival time by R software (version 3.6.1). The “survival” package of R software version was used. Based on the IHCs of each biomarker, patients were classified into two groups and the association between prognosis and biomarker expression was evaluated. The best expression cut-off referred to the IHCs that yielded the maximal difference with respect to survival between the two groups at the lowest log-rank P value. The best cut-off value was selected based on survival analysis.
The least absolute shrinkage and selection operator (LASSO) Cox regression model was used to evaluate high-dimensional data, by shrinking and selecting the biomarkers, that were the most predictive features of patients with OSCC in the training cohort. The LASSO Cox regression model was constructed by R software using the “glmnet” package.
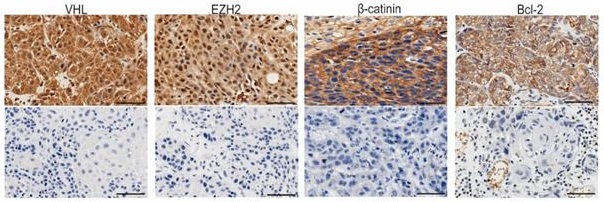
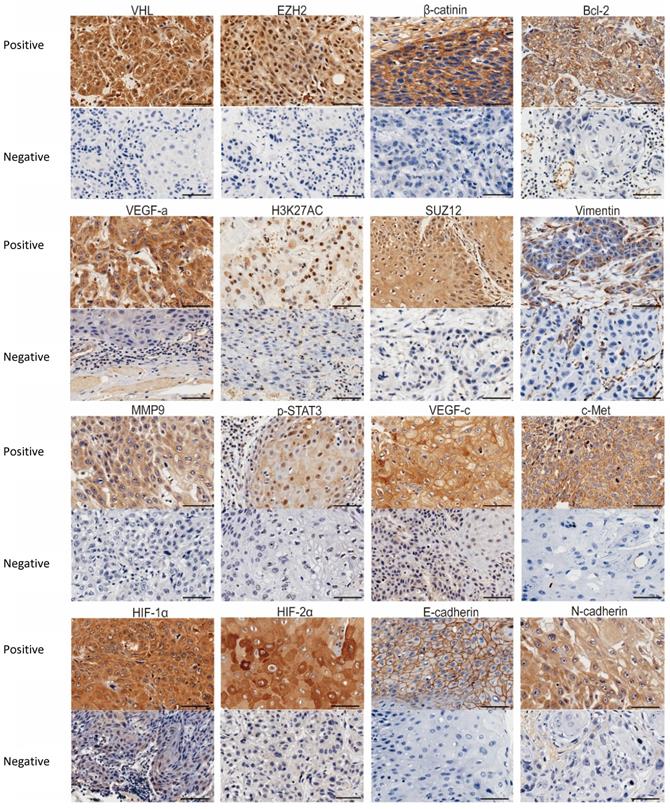
Representative IHC images of the 16 biomarkers expression. Bar, 100 um.
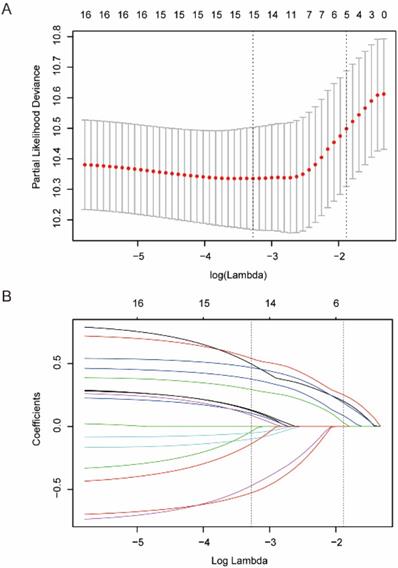
Feature selection using LASSO Cox regression model. (A) LASSO coefficient profiles the 16 biomarkers associated with OSCC. (B) Tuning parameter selection in the LASSO model. We selected λ via 1-SE (standard error) criteria. A value λ = 0.009 with log (λ) = -4.673 was chosen by 10-fold cross-validation via the 1-SE criteria.
Clinical and pathological characteristics of patients according to the IHCs in the training and validation cohorts
| Variables | Training Cohort (n=147) | Validation Cohort (n=147) | ||||||
|---|---|---|---|---|---|---|---|---|
| N | Low-IHCs | High-IHCs | P | N | Low-IHCs | High-IHCs | P | |
| Sex | ||||||||
| Male | 86 | 21 (51.2) | 65 (61.3) | 0.265 | 97 | 37 (68.5) | 60 (64.5) | 0.621 |
| Female | 61 | 20 (48.8) | 41 (51.2) | 50 | 17 (31.5) | 33 (35.5) | ||
| Age | ||||||||
| <60 | 91 | 25 (61.0) | 66 (62.3) | 0.885 | 83 | 34 (63.0) | 49 (52.7) | 0.226 |
| ≥60 | 56 | 16 (39.0) | 40 (37.7) | 64 | 20 (37.0) | 44 (47.3) | ||
| Depth of invasion | ||||||||
| T1+T2 | 92 | 30 (73.2) | 62 (58.5) | 0.099 | 128 | 51 (94.4) | 77 (82.8) | 0.042 |
| T3+T4 | 55 | 11 (26.8) | 44 (41.5) | 19 | 3 (5.6) | 16 (17.2) | ||
| Lymph Node Metastasis | ||||||||
| N0 | 90 | 31 (75.6) | 59 (55.7) | 0.026 | 117 | 47 (87.0) | 70 (75.3) | 0.088 |
| N1+ N2+ N3 | 57 | 10 (24.4) | 47 (44.3) | 30 | 7 (13.0) | 23 (24.7) | ||
| TNM Stage | ||||||||
| I+II | 65 | 25 (61.0) | 40 (37.7) | 0.011 | 103 | 44 (81.5) | 59 (63.4) | 0.021 |
| III+IV | 82 | 16 (39.0) | 66 (62.3) | 44 | 10 (18.5) | 34 (36.6) | ||
| Grade | ||||||||
| G1-G2 | 108 | 31 (75.6) | 77 (72.6) | 0.715 | 106 | 46 (85.2) | 60 (64.5) | 0.007 |
| G3 | 39 | 10 (24.4) | 29 (27.4) | 41 | 8 (14.8) | 33 (35.5) | ||
| Smoke | ||||||||
| Yes | 73 | 21 (51.2) | 52 (49.1) | 0.814 | 66 | 31 (57.4) | 35 (37.6) | 0.023 |
| No | 74 | 20 (48.8) | 54 (50.9) | 81 | 23 (42.6) | 58 (62.4) | ||
| Drink | ||||||||
| Yes | 71 | 20 (48.8) | 51 (48.1) | 0.942 | 55 | 23 (42.6) | 32 (34.4) | 0.347 |
| No | 76 | 21 (51.2) | 55 (51.9) | 92 | 31 (57.4) | 61 (65.6) | ||
Statistical Analysis
The two groups were compared using the t-test for continuous variables and chi-square test for categorical variables. We used the Kaplan-Meier survival analysis and log-rank test to estimate the OS. Univariate and multivariate analyses were performed with the Cox regression model. The nomogram was generated using the “rms” package of R software. Receiver operating characteristic (ROC) analysis was conducted to explore the prognostic performance of the IHCs classifier. Decision curve analysis (DCA) was carried out to evaluate the clinical utility of the nomogram. Statistical analysis was performed with R software (3.6.1) and SPSS software (version 25.0). Statistical significance was two-sided, and P <0.05 was considered statistically significant.
Results
Clinical Characteristics and Selection of Biomarkers
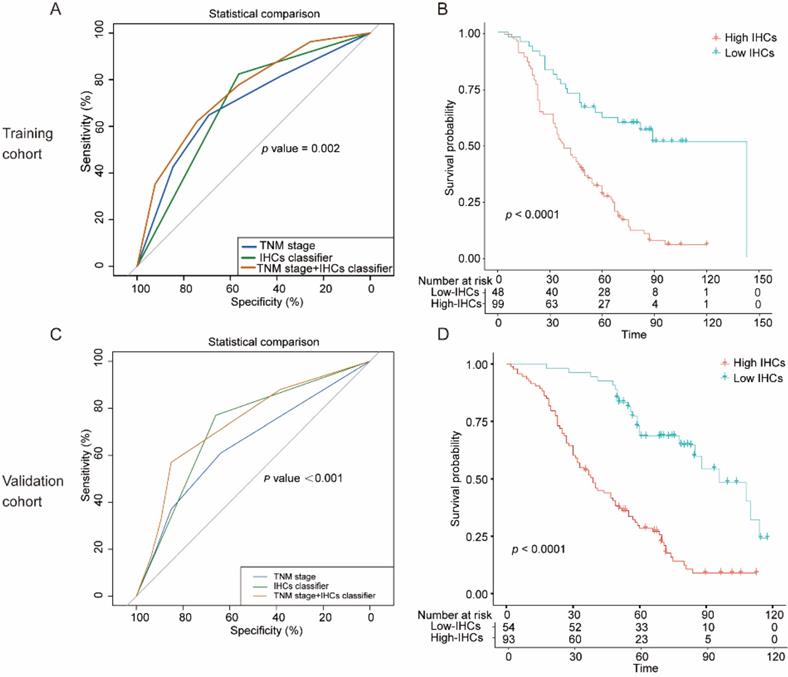
The clinical and pathological characteristics of the patients in the training and validation cohort are listed in Table 1. We used the “survminer” package of R software to determine the cut-off score for the sixteen biomarkers separately (Figure S1). Details are provided in the supplementary materials. We selected five biomarkers of sixteen biomarkers using the LASSO Cox regression model in the training cohort, and these biomarkers were cMet, Vimentin, HIF-2α, VEGF-c, and Bcl-2 (Figure 2). Then, a formula was derived to calculate the score of each patient, based on their individual expression levels of these five biomarkers: IHCs =0.27193962*VEGF-c+0.11069870*HIF-2α +0.22491359*Vimentin+0.06970822*Bcl-2+0.21782503*c-Met. In this formula, low IHC expression equals 0 and high expression equals 1. Then we further divided the 147 patients into a high-IHCs group and a low-IHCs group with an IHCs of 0.383 as the cutoff. In the training cohort, the 5-year OS was 62.1% in low-IHCs group and 28.2% in high-IHCs group (P<0.001, Figure 4B). The 5-year OS was 68.6% for the low-IHCs group and 28.4% for the high-IHCs group in the validation cohort (P<0.001, Figure 4D). Patients with low IHC expression of have a longer 5-year OS in both the training cohort and validation cohort.
Prediction Accuracy of the IHCS Group
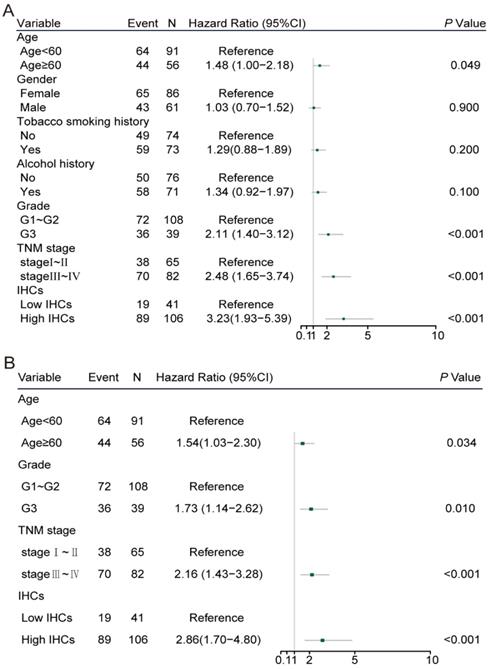
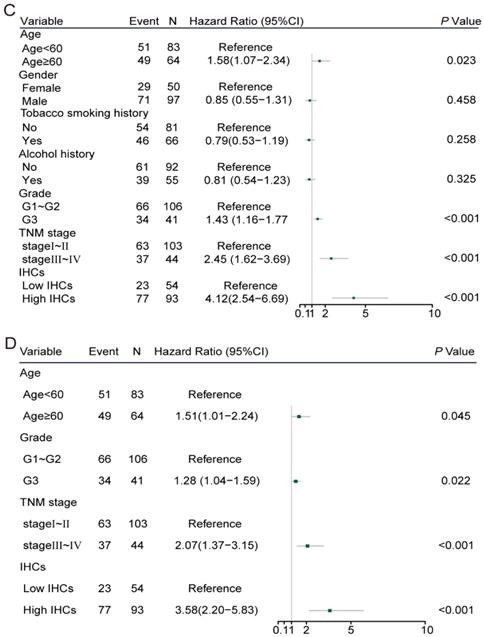
In the univariate Cox regression analysis, age≥60 (hazard ratio [HR]: 1.48, 95% CI: 1.00-2.18, P=0.049), poor tumuor differentiation (HR: 2.11, 95% CI: 1.40-3.12, P<0.001), stage Ⅲ-Ⅳ (HR: 2.48, 95% CI: 1.65-3.74, P<0.001) and high IHC classifier expression (HR: 3.23, 95% CI: 1.93-5.39, P<0.001) were significantly associated with poor prognosis in the training cohort (Figure 3A). The multivariate Cox analysis identified that the age ≥60 (HR: 1.54, 95% CI: 1.03-2.30, P=0.034), poor differentiation (HR: 1.73, 95% CI: 1.14-2.62, P=0.010), stage III-IV (HR: 2.16, 95% CI: 1.43-3.26, P<0.001) and high IHCs classifier (HR: 2.86, 95% CI: 1.70-4.80, P<0.001) were independent prognostic factors for OS in the training cohort (Figure 3B). Similar results were concluded in the validation cohort (Figure 3C, D). The area under the ROC curve (AUROC) of the TNM stage was 0.690 (95% CI: 0.598-0.781), while the AUROC of the combination of the IHCs classifier and TNM stage was 0.746 (95% CI: 0.658-0.833), which showed a better performance for predicting the OS than the TNM stage alone with P=0.002 (Figure 4A). Similar to the results displayed in the validation cohort, the AUROC of the TNM stage combined with the IHCs classifier was 0.735 (95% CI: 0.651-0.818), which was higher than that of the TNM stage alone (0.645, 95% CI: 0.559-0.731) (P<0.002) (Figure 4C). Therefore, the addition of the IHCs classifier to the TNM group could make it a better tool to predict the survival outcomes for patients with OSCC.
Construction of the IHCs-based Nomogram
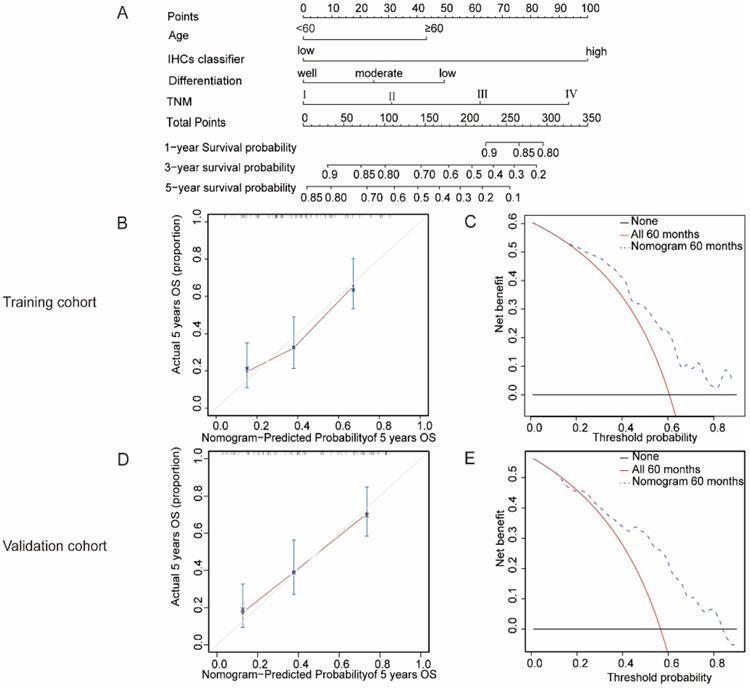
Therefore, a nomogram was built to integrate the IHCs classifier and clinicopathological factors related to the survival outcomes of the patients with OSCC. The C-index was 0.67 and 0.76 in the training cohort and validation cohort, respectively. The calibration curves showed that our nomogram had a good consistency with the ideal model for predicting the 3- and 5-year OS in the training cohort and validation cohort (Figures 5B, 5D and S2). DCA was presented to assess the value of the clinical unity. The results are presented in Figures 5C, 5E and S3; the x-axis of the graph represented the threshold probability, the y-axis measured the net benefit. This method incorporates the clinical consequences of the nomograms by applying a different weight to the true- and false-positive results. These results showed that our nomogram could help gain more net benefits in clinical practice.
Clinicopathological risk factors for patients with OS. (C) Univariate cox regression analysis of patients with OS in the validation cohort. (D) Multivariate cox regression analysis of patients with OS in the validation cohort.
Discussion
The biological propensity for local invasion and the high rate of cervical lymph node metastasis are usually regarded as the main reasons related to the poor prognosis for patients with OSCC [8, 9]. Although the TNM stage system could be used as a tool for predicting the prognosis of patients, it still has some limitations. The different degrees of tumour aggressiveness could be closely related to the molecular subtype, which might result from tumour heterogeneity and indirectly result in different risks of recurrence and metastasis [10]. Hence, an accurate evaluation of patients with different survival outcomes could be regarded as an essential progression, thus systemic therapy could also be formulated.
Along with acknowledging tumour heterogeneity, an increasing number of studies have committed to investigating supplements to the TNM staging system for specifically predicting survival outcomes. Nomograms for the prediction of the prognosis in cholangiocarcinoma, colorectal cancer, nasopharyngeal carcinoma and so on have been constructed and published [11-14]. Among them, a study generated a distant metastasis gene signature for locoregionally advanced nasopharyngeal carcinoma that consisted of 13 genes to divide patients into high-risk and low-risk groups using microarray analysis, which had an accuracy of 74.8% for predicting distant metastasis-free survival [13]. Another study investigated the collagen signature in tumour microenvironment as an indicator of lymph node metastasis in early gastric cancer and validated a nomogram based on the collagen signature and other pathological findings for predicting lymph node metastasis in gastric cancer [15]. Meng J et al. [16] constructed a nomogram combining the IHC biomarkers with TNM stage for the predicting recurrence-free survival of patients with oesophageal squamous cell carcinoma. The combination of the markers and TNM stage was proved to be more predictive than the TNM stage alone (AUROC: 0.751, 95%CI: 0.681-0.882 vs AUROC: 0.678, 95%CI: 0.607-0. 750).
The specificity and significance of the clinical use of the IHCs classier. (A) Comparison of the prognostic value of IHCs with TNM staging in the training cohort. (B) Kaplan-Meier survival analysis of overall survival according to the IHCs classier in the training cohort. (C) Comparison of the prognostic value of IHCs with TNM staging in the validation cohort. (D) Kaplan-Meier survival analysis of overall survival according to the IHCs classier in the validation cohort.
Similarly, nomograms have been constructed to predict prognosis for patients with OSCC. Zhou C [17] developed and validated a seven-immune-feature-based prognostic score for patients with OSCC after curative resection. Some nomograms reported in the previous studies assessed the association of preoperative lymphocyte-to-monocyte ratio or some other clinical features with survival in patients with OSCC [18-20]. Apart from those studies, there have been some other prognostic nomograms based on mRNA expression profiles from the public databases that improved predicting the prognosis of patients with OSCC [21-23]. All of these studies showed promising prospects in clinical applications. However, as far as we know, there have been no reliable molecular profiling features for predicting the prognosis for patients with OSCC. Moreover, some limitations, such as the uncertainty of complicated molecular biology methods, the requirements for fresh tissues and the high cost of examination, exist in gene expression-based methods when applied to the clinical diagnosis. Immunohistochemical staining, as an easy-to-use method, could be applied to conveniently and economically diagnose and determine the expression of tumour-associated proteins. Compared to the previous study, this is the first study to include a relatively large sample size to analyse the IHC expression of biomarkers for patients with OSCC, all of whom received surgical treatment.
Currently, no molecular biomarkers have been clinically used to evaluate the prognosis in patients with OSCC. Researchers have made great efforts on head and neck squamous cell carcinoma before, and found that some biomarkers played an important role in the development of the disease [24-26]. Our group have also found that some biomarkers were associated with the prognosis of patients with OSCC [27, 28]. However, most of the studies have only investigated one or more biomarkers in a single signaling pathway rather than integrating them together. In this study, we analysed the expression of IHC biomarkers in patients from our medical centre, and built an IHC classifier which could divide patients into high-risk group and low-risk group to help to guide clinical decision making. Even though quite a few patients could get benefit from multimodal treatment involving chemotherapy, radiotherapy and targeted-therapeutics [29], the adverse therapeutic effects still cannot be ignored. The suffering could be alleviated and the chance of survival would be increased if patients received a precise and individualized treatment. Thus, the issue of functional preservation can be considered more when formulating surgical procedures for patients in the low-risk group, so that the quality of life of patients with OSCC could be guaranteed. A more positive treatment, such as accepting a radical surgical approach or receiving more adjuvant therapy, should be implemented when patients are classified into high risk group, consequently the risk of recurrence and mortality could be lowered. Moreover, the financial burden could be reduced for patients with OSCC.
The nomogram to predict 1-year, 3-year and 5-year survival probability for OSCC. (A) A nomogram established by the combination of the clinical factors and IHCs. (B)(D) The calibration curve for predicting patients 5-year OS in the training cohort and validation cohort. (C)(E) Evaluation of nomogram using decision curve analysis for the clinical utility of the nomogram in the training cohort and validation cohort.
In this study, we investigated sixteen biomarkers that have been reported to be influential on the prognosis of patients with OSCC, and constructed a model named the IHCs classifier, which integrated the expression of the selected five biomarkers via the LASSO Cox regression model. The LASSO Cox regression model could make the IHCs classifier more accurate to predict the prognosis by shrinking the number of biomarkers and eliminating the influence of gene co-expression on the prognosis [30, 31]. Although the role and mechanism of the five markers in OSCC was reported in the previous studies, a combination role of the biomarkers could be unclear in the present study. The IHCs classifier with the combination of these five biomarkers could be more precise and scientific for predicting prognosis for patients than that with only one biomarker. Moreover, the small number of biomarkers could make this IHCs classifier convenient to use in our clinical work, and may reduce the economic burden of patients. To better improve the predictive accuracy, a nomogram was built by integrating the clinicopathological characteristics with the IHCs classifier. The AUROC of the combination of the IHCs classifier and TNM stage was 0.746 (95% CI: 0.658-0.833) in the primary cohort and 0.735 (95% CI: 0.651-0.818) in the validation cohort, both higher than the AUROC of TNM stage alone respectively in the primary cohort and validation cohort, indicating that the IHCs classifier could be a good supplement to TNM stage for predicting the prognosis of patients and making individualized treatment decisions.
Cancer is a heterogeneous disease. Exploring dysregulated genes associated with carcinogenesis and development may help improve the prognosis and of patients and treatment strategies. In our study, we included five biomarkers that may work in the nomogram for OSCC. The MET gene encodes the receptor for hepatocyte growth factor (HGF), and the HGF/c-Met signalling pathway, which is overexpressed in various human solid tumours [24, 32-34]. A study reported that c-Met positive cells had cancer stem cell properties that could generate tumour cells and make them develop resistance to cisplatin in patients with head and neck squamous cell carcinoma (HNSCC), thereby significantly influencing the prognosis [35]. Hypoxia is one of the characteristics of the microenvironment of malignant solid tumours [36]. Hypoxia-induced factors (HIFs), which have been demonstrated to exert the effects of an oncogene, have a role in angiogenesis and regulate tumorigenesis of cancer cells [37]. Although HIF-2α has been reported to have either a promoting or a suppressing role in different types of tumours [38-40], it has been widely accepted that HIF-2α contributes to the tumour angiogenesis [38, 41]. HIF-2α could influence tumour cell proliferation, and apoptosis or tumour angiogenesis by regulating hypoxia-related genes, such as VEGF, cyclin Dl and erythropoietin (EPO) [39, 42]. The knockdown of HIF-2α could inhibit VEGF expression [41, 43]. In addition, a few studies have shown that HIF-2 may be involved in the tumour resistance to radiotherapy and chemotherapy [44].
Tumour growth and metastasis depend on angiogenesis. VEGF is the most directly related protein influencing angiogenesis. VEGF-C could bind to VEGFR-3, which is mainly expressed in lymphatic endothelial cells, and its affinity for VEGFR-3 is more than three times higher than that for vascular endothelial growth factor receptor-2 (VEGFR-2). Therefore, VEGF-C could be a major inducer of lymphangiogenesis in various malignancies including OSCC [45, 46]. In addition, VEGF-C has been reported to be correlated with lymphatic vessel density and microvessel density [47], and it is an indicator for lymph node metastasis, and tumour recurrence [48]. The Bcl-2 gene is one of the most important genes regulating apoptosis by suppressing apoptosis and prolonging cell survival [49]. Our previous study showed that knocking down Bcl-2 might initiate cascade response and change the EMT process and suppress cell migration and invasion in OSCC [27]. Similarly, Arshad Rahman et al. [50] indicated that the expression of Bcl-2 was higher in patients with stage III/IV or poorly differentiated tumour. It has been demonstrated that EMT plays a critical role in tumour invasion and metastasis [51]. Vimentin, a mesenchymal-specific protein, is generally expressed during the process of EMT, but is not induced in normal epithelial cells. It has also been found that vimentin was one of the most upregulated genes in metastatic OSCC cells, and that high expression of vimentin was correlated with lymph node metastasis and poor prognosis in patients with OSCC [52].
To the best of our knowledge, this is the first study to include 5 biomarkers which were selected from 16 biomarkers that have been reported to be involved in biological processes during carcinogenesis, and construct a nomogram involving a combined multi-biomarker classifier for patients with OSCC. However, our study has some limitations. Our research is a retrospective study in a single cancer centre in North of China. The information being involved in the studies could not be representative for patients from all over the world. This study could be more persuasive if an external validation cohort was included. Another limitation is that the 16 biomarkers involved are based on the previous research of our team and the current recognition of the mechanism of OSCC. As OSCC research develops, more specific and relevant biomarkers need to be investigated. Thus, prospective research including more biomarkers and patients from multiple centres will be needed to validate our findings.
In conclusion, the nomogram showed good predictive accuracy and clinical unity for predicting the prognosis of patients with OSCC, and it could be a potential tool for guiding the individual decision-making process.
Abbreviations
OSCC: oral squamous cell carcinoma; OS: overall survival; AJCC: American Joint Committee on Cancer; DOI: depth of invasion; IHC: Immunohistochemistry; EMT: epithelial-mesenchymal transition; DAB: 3,3′-diaminobenzidine tetrahydrochloride; IHCs: IHC score; LASSO: least absolute shrinkage and selection operator; ROC: Receiver operating characteristic; DCA: Decision curve analysis; HR: hazard ratio; AUROC: area under the ROC curve; HGF: hepatocyte growth factor; HNSCC: head and neck squamous cell carcinoma; HIFs: hypoxia-induced factors; EPO: erythropoietin; VEGFR-2: vascular endothelial growth factor receptor-2.
Supplementary Material
Supplementary figures.
Acknowledgements
This project was supported by the National Natural Science Foundation of China (No. 81672684, No. 81702420 and No. 81902757), the Committee of Tianjin Science and Technology (No. 19JCQNJC10900 and No. 18JCYBJC93500), Science Foundation of Tianjin Cancer Hospital & Institute (No. 1806).
Ethics Committee Approval and Patient Consent
The study was approved by the Clinical Research Ethics Committee of Tianjin Medical University Cancer Institute and Hospital. Written-informed consent was obtained from all patients.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108
2. Huang JF, Wang JW, He BC, Chen F, Liu FP, Yan LJ. et al. [Study of survival factors of oral squamous cell carcinoma]. Zhonghua Yu Fang Yi Xue Za Zhi. 2016;50:880-6
3. Siriwardena B, Rambukewela IK, Pitakotuwage TN, Udagama M, Kumarasiri PVR, Tilakaratne WM. A Predictive Model to Determine the Pattern of Nodal Metastasis in Oral Squamous Cell Carcinoma. Biomed Res Int. 2018;2018:8925818
4. Pruegsanusak K, Peeravut S, Leelamanit V, Sinkijcharoenchai W, Jongsatitpaiboon J, Phungrassami T. et al. Survival and prognostic factors of different sites of head and neck cancer: an analysis from Thailand. Asian Pac J Cancer Prev. 2012;13:885-90
5. Zhong LP, Zhang CP, Ren GX, Guo W, William WN Jr, Sun J. et al. Randomized phase III trial of induction chemotherapy with docetaxel, cisplatin, and fluorouracil followed by surgery versus up-front surgery in locally advanced resectable oral squamous cell carcinoma. J Clin Oncol. 2013;31:744-51
6. Doescher J, Veit JA, Hoffmann TK. The 8th edition of the AJCC Cancer Staging Manual. Updates in otorhinolaryngology, head and neck surgery. Hno. 2017;65:956
7. Howitt BE, Sun HH, Roemer MG, Kelley A, Chapuy B, Aviki E. et al. Genetic Basis for PD-L1 Expression in Squamous Cell Carcinomas of the Cervix and Vulva. JAMA Oncol. 2016;2:518-22
8. Li Y, Liu K, Ke Y, Zeng Y, Chen M, Li W. et al. Risk Factors Analysis of Pathologically Confirmed Cervical Lymph Nodes Metastasis in Oral Squamous Cell Carcinoma Patients with Clinically Negative Cervical Lymph Node: Results from a Cancer Center of Central China. J Cancer. 2019;10:3062-9
9. Bittar RF, Ferraro HP, Ribas MH, Lehn CN. Predictive factors of occult neck metastasis in patients with oral squamous cell carcinoma. Braz J Otorhinolaryngol. 2016;82:543-7
10. Leemans CR, Snijders PJF, Brakenhoff RH. The molecular landscape of head and neck cancer. Nat Rev Cancer. 2018;18:269-82
11. Wang Y, Li J, Xia Y, Gong R, Wang K, Yan Z. et al. Prognostic nomogram for intrahepatic cholangiocarcinoma after partial hepatectomy. J Clin Oncol. 2013;31:1188-95
12. Huang YQ, Liang CH, He L, Tian J, Liang CS, Chen X. et al. Development and Validation of a Radiomics Nomogram for Preoperative Prediction of Lymph Node Metastasis in Colorectal Cancer. J Clin Oncol. 2016;34:2157-64
13. Tang XR, Li YQ, Liang SB, Jiang W, Liu F, Ge WX. et al. Development and validation of a gene expression-based signature to predict distant metastasis in locoregionally advanced nasopharyngeal carcinoma: a retrospective, multicentre, cohort study. Lancet Oncol. 2018;19:382-93
14. Chen T, Ning Z, Xu L, Feng X, Han S, Roth HR. et al. Radiomics nomogram for predicting the malignant potential of gastrointestinal stromal tumours preoperatively. Eur Radiol. 2019;29:1074-82
15. Chen D, Chen G, Jiang W, Fu M, Liu W, Sui J. et al. Association of the Collagen Signature in the Tumor Microenvironment With Lymph Node Metastasis in Early Gastric Cancer. JAMA Surg. 2019;154:e185249
16. Meng J, Zhang J, Xiu Y, Jin Y, Xiang J, Nie Y. et al. Prognostic value of an immunohistochemical signature in patients with esophageal squamous cell carcinoma undergoing radical esophagectomy. Mol Oncol. 2018;12:196-207
17. Zhou C, Diao P, Wu Y, Wei Z, Jiang L, Zhang W. et al. Development and validation of a seven-immune-feature-based prognostic score for oral squamous cell carcinoma after curative resection. Int J Cancer. 2020;146:1152-63
18. Chen L, Qian J, Lin L, Lin J, Chen Q, Zhuang Z. et al. Prognostic value of preoperative lymphocyte-to-monocyte ratio in oral cancer patients and establishment of a dynamic nomogram. Oral Dis. 2020 doi: 10.1111/odi.13629
19. Chang B, He W, Ouyang H, Peng J, Shen L, Wang A. et al. A Prognostic Nomogram Incorporating Depth of Tumor Invasion to Predict Long-term Overall Survival for Tongue Squamous Cell Carcinoma with R0 Resection. J Cancer. 2018;9:2107-15
20. Kao HK, Lofstrand J, Loh CY, Lao WW, Yi JS, Chang YL. et al. Nomogram based on albumin and neutrophil-to-lymphocyte ratio for predicting the prognosis of patients with oral cavity squamous cell carcinoma. Sci Rep. 2018;8:13081
21. Wang F, Zhang H, Wen J, Zhou J, Liu Y, Cheng B. et al. Nomograms forecasting long-term overall and cancer-specific survival of patients with oral squamous cell carcinoma. Cancer Med. 2018;7:943-52
22. Hou C, Cai H, Zhu Y, Huang S, Song F, Hou J. Development and Validation of Autophagy-Related Gene Signature and Nomogram for Predicting Survival in Oral Squamous Cell Carcinoma. Front Oncol. 2020;10:558596
23. Sun W, Cheng M, Zhuang S, Chen H, Yang S, Qiu Z. Nomograms to predict survival of stage IV tongue squamous cell carcinoma after surgery. Medicine (Baltimore). 2019;98:e16206
24. Sun Z, Liu Q, Ye D, Ye K, Yang Z, Li D. Role of c-Met in the progression of human oral squamous cell carcinoma and its potential as a therapeutic target. Oncol Rep. 2018;39:209-16
25. Zhu GJ, Song PP, Zhou H, Shen XH, Wang JG, Ma XF. et al. Role of epithelial-mesenchymal transition markers E-cadherin, N-cadherin, beta-catenin and ZEB2 in laryngeal squamous cell carcinoma. Oncol Lett. 2018;15:3472-81
26. Hussein AA, Forouzanfar T, Bloemena E, de Visscher J, Brakenhoff RH, Leemans CR. et al. A review of the most promising biomarkers for early diagnosis and prognosis prediction of tongue squamous cell carcinoma. Br J Cancer. 2018;119:724-36
27. Duan Y, He Q, Yue K, Si H, Wang J, Zhou X. et al. Hypoxia induced Bcl-2/Twist1 complex promotes tumor cell invasion in oral squamous cell carcinoma. Oncotarget. 2017;8:7729-39
28. Zhang S, Zhou X, Wang B, Zhang K, Liu S, Yue K. et al. Loss of VHL expression contributes to epithelial-mesenchymal transition in oral squamous cell carcinoma. Oral Oncol. 2014;50:809-17
29. Lordick F, Mariette C, Haustermans K, Obermannova R, Arnold D, Committee EG. Oesophageal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2016;27:v50-v7
30. Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med. 1997;16:385-95
31. R T. Regression shrinkage and selection via the lasso: a retrospective. Stat Med. 1997;16:385-95
32. Jiang WG, Martin TA, Parr C, Davies G, Matsumoto K, Nakamura T. Hepatocyte growth factor, its receptor, and their potential value in cancer therapies. Crit Rev Oncol Hematol. 2005;53:35-69
33. Di Renzo MF, Narsimhan RP, Olivero M, Bretti S, Giordano S, Medico E. et al. Expression of the Met/HGF receptor in normal and neoplastic human tissues. Oncogene. 1991;6:1997-2003
34. Vsiansky V, Gumulec J, Raudenska M, Masarik M. Prognostic role of c-Met in head and neck squamous cell cancer tissues: a meta-analysis. Sci Rep. 2018;8:10370
35. Sun S, Wang Z. Head neck squamous cell carcinoma c-Met(+) cells display cancer stem cell properties and are responsible for cisplatin-resistance and metastasis. Int J Cancer. 2011;129:2337-48
36. Li Z, Zhang W. The 2016 Albert Lasker Basic Medical Research Award: Oxygen sensing-a mysterious process essential for survival. Sci China Life Sci. 2016;59:1195-7
37. Beasley NJ, Leek R, Alam M, Turley H, Cox GJ, Gatter K. et al. Hypoxia-inducible factors HIF-1alpha and HIF-2alpha in head and neck cancer: relationship to tumor biology and treatment outcome in surgically resected patients. Cancer Res. 2002;62:2493-7
38. Acker T, Diez-Juan A, Aragones J, Tjwa M, Brusselmans K, Moons L. et al. Genetic evidence for a tumor suppressor role of HIF-2alpha. Cancer Cell. 2005;8:131-41
39. Holmquist-Mengelbier L, Fredlund E, Löfstedt T, Noguera R, Navarro S, Nilsson H. et al. Recruitment of HIF-1α and HIF-2α to common target genes is differentially regulated in neuroblastoma: HIF-2α promotes an aggressive phenotype. Cancer Cell. 2006;10:413-23
40. Covello KL, Simon MC, Keith B. Targeted replacement of hypoxia-inducible factor-1alpha by a hypoxia-inducible factor-2alpha knock-in allele promotes tumor growth. Cancer Res. 2005;65:2277-86
41. Zhu Gq, Tang Yl, Li L, Zheng M, Jiang J, Li Xy. et al. Hypoxia Inducible Factor 1 and Hypoxia Inducible Factor 2 Play Distinct and Functionally Overlapping Roles in Oral Squamous Cell Carcinoma. Clinical Cancer Research. 2010;16:4732-41
42. RatcliffePJ. HIF-l and HIF-2: working alone or together in hypoxia. J Clin Invest. 2007;117(4):862-5
43. Carroll VA, Ashcroft M. Role of hypoxia-inducible factor (HIF)-1alpha versus HIF-2alpha in the regulation of HIF target genes in response to hypoxia, insulin-like growth factor-I, or loss of von Hippel-Lindau function: implications for targeting the HIF pathway. Cancer Res. 2006;66:6264-70
44. Zhao J, Du F, Luo Y, Shen G, Zheng F, Xu B. The emerging role of hypoxia-inducible factor-2 involved in chemo/radioresistance in solid tumors. Cancer Treatment Reviews. 2015;41:623-33
45. Omoto I, Matsumoto M, Okumura H, Uchikado Y, Setoyama T, Kita Y. et al. Expression of vascular endothelial growth factor-C and vascular endothelial growth factor receptor-3 in esophageal squamous cell carcinoma. Oncol Lett. 2014;7:1027-32
46. Woolgar JA, Triantafyllou A. Pitfalls and procedures in the histopathological diagnosis of oral and oropharyngeal squamous cell carcinoma and a review of the role of pathology in prognosis. Oral Oncol. 2009;45:361-85
47. Miyahara M, Tanuma J, Sugihara K, Semba I. Tumor lymphangiogenesis correlates with lymph node metastasis and clinicopathologic parameters in oral squamous cell carcinoma. Cancer. 2007;110:1287-94
48. Kazakydasan S, Rahman ZA, Ismail SM, Abraham MT, Kallarakkal TG. Prognostic significance of VEGF-C in predicting micrometastasis and isolated tumour cells in N0 oral squamous cell carcinoma. Journal of oral pathology & medicine: official publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology. 2017;46:194-200
49. An J, Lv J, Li A, Qiao J, Fang L, Li Z. et al. Constitutive expression of Bcl-2 induces epithelial-Mesenchymal transition in mammary epithelial cells. BMC Cancer. 2015;15:476
50. Rahmani A, Alzohairy M, Babiker AY, Rizvi MA, Elkarimahmad HG. Clinicopathological significance of PTEN and bcl2 expressions in oral squamous cell carcinoma. Int J Clin Exp Pathol. 2012;5:965-71
51. Liu CY, Lin HH, Tang MJ, Wang YK. Vimentin contributes to epithelial-mesenchymal transition cancer cell mechanics by mediating cytoskeletal organization and focal adhesion maturation. Oncotarget. 2015;6:15966-83
52. Liu S, Liu L, Ye W, Ye D, Wang T, Guo W. et al. High Vimentin Expression Associated with Lymph Node Metastasis and Predicated a Poor Prognosis in Oral Squamous Cell Carcinoma. Sci Rep. 2016;6:38834
Author contact
![]() Corresponding authors: Xudong Wang, E-mail: WXD.1133com; Ze Zhang, E-mail: zhangzeedu.cn; Yansheng Wu, E-mail: ywuedu.cn; Department of Maxillofacial and Otorhinolaryngology Oncology, Tianjin Medical University Cancer Institute and Hospital, Key Laboratory of Cancer Prevention and Therapy, Tianjin Cancer Institute, National Clinical Research Center of Cancer, Tianjin 300060, China.
Corresponding authors: Xudong Wang, E-mail: WXD.1133com; Ze Zhang, E-mail: zhangzeedu.cn; Yansheng Wu, E-mail: ywuedu.cn; Department of Maxillofacial and Otorhinolaryngology Oncology, Tianjin Medical University Cancer Institute and Hospital, Key Laboratory of Cancer Prevention and Therapy, Tianjin Cancer Institute, National Clinical Research Center of Cancer, Tianjin 300060, China.

 Global reach, higher impact
Global reach, higher impact