Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(17):5173-5180. doi:10.7150/jca.53708 This issue Cite
Research Paper
E2F4 Promotes the Proliferation of Hepatocellular Carcinoma Cells through Upregulation of CDCA3
1. Key Laboratory of Molecular Biology for Infectious Diseases, Ministry of Education, Department of Infectious Diseases, Institute for Viral Hepatitis, Chongqing Medical University, Chongqing, China.
2. Department of Dermatology and Cosmetology, Chongqing Hospital of Traditional Chinese Medicine, Chongqing, China.
3. College of Laboratory Medicine, Chongqing Medical University, Chongqing, China.
4. Pediatric Research Institute; Ministry of Education Key Laboratory of Child Development and Disorders; National Clinical Research Center for Child Health and Disorders (Chongqing); China International Science and Technology Cooperation base of Child development and Critical Disorders; Children's Hospital of Chongqing Medical University, Chongqing, China.
5. Chongqing Key Laboratory of Pediatrics, Children's Hospital of Chongqing Medical University, Chongqing, China.
6. Department of Clinical Laboratory, Yubei District People's Hospital, Chongqing, China.
*These authors contributed equally to this work.
Received 2020-9-25; Accepted 2021-6-8; Published 2021-6-22
Abstract
Liver cancer, the second most commonly diagnosed cancer, is associated with high mortality rates. E2F4 is a member of the E2F transcription factor family. There are limited studies on the role of E2F4 in hepatocellular carcinoma (HCC). In this study, the expression of E2F4 in HCC tissue samples and cell lines was analyzed using quantitative real-time polymerase chain reaction. E2F4 expression positively correlated with tumor size in patients with HCC. Additionally, E2F4 expression was greater in HCC cells than in normal LO2 cells. Furthermore, overexpression of E2F4 significantly enhanced the proliferation, migration, and invasion of HCC cells. The results of a luciferase assay revealed that E2F4 upregulated the expression of CDCA3 by binding to its promoter region (1863'-ACGCGCGAGAATG-1875') and consequently promoted proliferation and cell cycle progression of HCC cells. Taken together, these results demonstrated that E2F4 might play a vital role in HCC progression and could serve as a potential biomarker for the diagnosis and as a therapeutic target of HCC.
Keywords: hepatocellular carcinoma, E2F4, CDCA3, promoter
Introduction
The incidence of liver cancer, which is the second most commonly diagnosed cancer overall, is higher among men than women. In developed countries, liver cancer is the fifth leading cause of cancer-related deaths among men. The incidence of liver cancer is also high in developing countries. Oftentimes, no symptoms are presented during early stage disease. Hence, liver cancer tends to be diagnosed at an advanced stage, especially in developing countries. The incidence of liver cancer is lowest in South Asia, Northern Europe, Central Europe, and Eastern Europe and highest in East Asia, Southeast Asia, North Africa, and West Africa. Globally, the most common primary liver cancer is hepatocellular carcinoma (HCC), which accounts for 75%-85% of all primary liver cancer cases [1-3].
The E2F transcription factor family is involved in cell proliferation, apoptosis, differentiation, aging, DNA damage response, and DNA repair [4-9]. This family was first identified in 1987 as transcription factors required to activate the E2 adenovirus promoter. Currently, nine members of the family have been identified: E2F1, E2F2, E2F3a, E2F3b, E2F4, E2F5, E2F6, E2F7, and E2F8, which are located on different chromosomes. The genes encoding E2F family transcription factors are highly homologous and encode highly conserved DNA binding, dimerization, and retinoblastoma (RB) protein-binding domains [10]. This family can be divided activator (E2F1, E2F2, and E2F3a) and repressor (E2F3b, E2F4, E2F5, E2F6, E2F7, and E2F8) E2F transcription factors based on their structure and function.
Studies have shown that E2F mRNA expression levels are related to different cancer stages and pathological grades of HCC patients, and E2F4 protein is highly expressed in advanced liver cancer [11]. E2F4 is the most abundantly expressed E2F transcription factor. Additionally, E2F4, which accounts for the majority of E2F activity, is involved in various cellular functions. E2F4 has previously been reported to function as a transcriptional repressor. However, recent studies have reported transcriptional activation functions as well [12,13].
The role of E2F4 in HCC pathogenesis remains unclear. Hence, this study aimed to elucidate the role of E2F4 in HCC. The expression of E2F4 in clinical HCC tissues and adjacent non-tumorous tissues, as well as in HCC and normal liver cell lines, was examined using quantitative real-time polymerase chain reaction (qRT-PCR). The findings of this study indicated that E2F4 might promote HCC carcinogenesis and might function as a candidate biomarker for diagnosis and as a therapeutic target.
Materials and methods
Clinical samples
HCC and adjacent tissues were collected from 40 patients undergoing thoracic surgery in the Department of Hepatobiliary Surgery, the First Affiliated Hospital of Chongqing Medical University, from January 2018 to January 2019. The samples were collected from 33 male and seven female patients. The patients were randomly selected, all showed primary HCC, and had not been treated with radio- or chemotherapy before surgery. Based on the tumor, node, metastasis (TNM) staging, 11, 7, and 22 cases were classified as stage I, stage II, and stage III, respectively. The tissues were immediately placed in RNA preservation solution in liquid nitrogen after surgical removal. The study protocol was approved by the Research Ethics Committee of the First Affiliated Hospital of Chongqing Medical University, and written informed consent was obtained from each patient.
Cell lines and cell culturing
Five human HCC cell lines (Huh7, Hep3B, HepG2, Bel-7402, and SK-HEP-1) and one modified normal liver cell line (LO2) were purchased from the American Type Culture Collection (Manassas, VA, USA). HepG2 cells were cultured in minimal essential medium (MEM) supplemented with 10% fetal bovine serum (FBS). The other cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% FBS. All cells were cultured in a cell incubator at 37 °C and 5% CO2. The pcDNA3.1 plasmid and the adenoviral vector encoding control short interfering RNA (siRNA) and red fluorescence protein (RFP) were obtained from the Infectious Disease Laboratory of Chongqing Medical University. The pcDNA3.1-E2F4 overexpression plasmid and the adenoviral vector encoding si-E2F4 (Ad-siE2F4) were constructed by our group and verified by sequencing. Plasmid transfection was performed using Lipofectamine 3000, according to the manufacturer's instructions. The virus titers used for cell infection were >10× [11].
Materials and reagents
The following materials and reagents were used in this study: DMEM, MEM, and trypsin (Gibco, Waltham, MA, USA); FBS (Natocor Industria Biologica, Cordoba, Argentina); Lipofectamine 3000 kit (Invitrogen, Carlsbad, CA, USA); reverse transcription kit and polyvinyl difluoride (PVDF) membranes (Roche, Basel, Switzerland); SYBR green qPCR master mix (MCE, Monmouth Junction, NJ, USA); bicinchoninic acid (BCA) protein quantitative analysis kit (ThermoFisher Scientific, Waltham, MA, USA); dual-luciferase kit (Promega, Madison, WI, USA); transwell chambers, 96-well plates, cell culture dishes (Corning, Corning, NY, USA); enhanced chemiluminescence (ECL) chromogenic solution (Bio-Rad, Hercules, CA, USA); 30% acrylamide-bis-acrylamide solution (LABTIDE, China); human anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody, goat anti-rabbit secondary antibody, goat anti-mouse secondary antibody, anti-CDCA3 antibody (Proteintech, Wuhan, China); and anti-E2F4 monoclonal antibody (Santa Cruz Biotechnology, Dallas, TX, USA).
RNA extraction, reverse transcription, and qRT-PCR
Total RNA was extracted from tissues and cells using TRIzol following the manufacturer's instructions. cDNA was synthesized using 1 µg of the isolated RNA as a template. The synthesized cDNA was diluted using double distilled water (1:9) and stored at -20 °C until use. qRT-PCR analyses were performed in triplicate. The relative expression levels of E2F4 were calculated using the 2-ΔΔCt method. The primers used for qRT-PCR were listed in Table S1.
Cell proliferation, migration, and invasion assays
Cells were transfected with pcDNA3.1-E2F4 or Ad-siE2F4 for 24 h. The cells were then resuspended and counted using a cow-abalone counting plate. The proliferation of cells (2 × 103/well) seeded in a 96-well plate was examined. When the cells adhered, a long-term dynamic cell imaging system was used to analyze the number of cells in each well and set as an initial value. Cells were counted once every 24 h, and the growth rate was calculated. A colony formation assay was performed using a six-well plate. Cells (500 cells/well) were cultured for one week and stained with crystal violet, and the colonies were counted. To perform the wound-healing assay, 4 × 104 cells/well were plated in a 96-well plate. When the cells adhered to the wall and the density reached at least 90%, the monolayer was scratched to introduce a wound gap. A long-term dynamic cell imaging system was used to capture images of the cells once every 12 h to analyze wound healing. Cell invasion assays were performed using transwell chambers. The upper chamber was coated with Matrigel (1:10). Cells (4 × 104) were resuspended in serum-free DMEM and seeded into the upper chamber. DMEM supplemented with 10% FBS was added to the lower chambers and the cells were incubated for 24 h. Transwell inserts were then removed and the cells were washed three times with phosphate-buffered saline. The cells were fixed in 4% paraformaldehyde at room temperature for 30 min, stained with crystal violet for 10 min, and counted using an optical microscope.
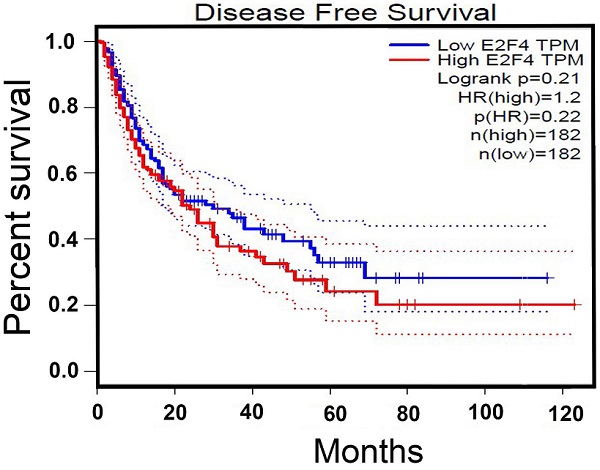
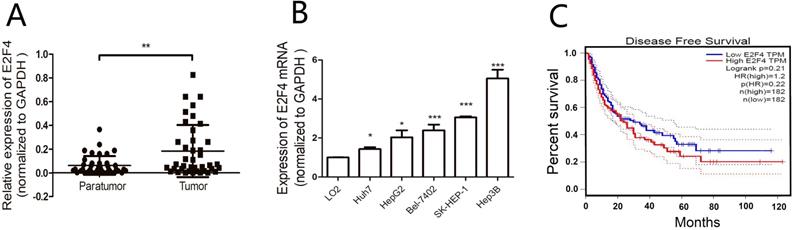
Expression of E2F4 in the hepatocellular carcinoma (HCC) samples and the correlation with clinicopathological features. A. Expression of E2F4 in HCC and adjacent non-tumorous tissues was examined using quantitative real-time polymerase chain reaction (qRT-PCR). B. Expression of E2F4 in Huh7, SK-HEP-1, Hep3B, HepG2, and Bel-7402 cells. The immortalized normal liver cell line (LO2) was used as a control. C. The gene expression profiling interaction analysis (GEPIA; http://gepia.cancer-pku.cn/index.html) database was used to evaluate the correlation between E2F4 expression and the survival rate of patients with HCC.
Dual-luciferase reporter assay
Huh7 cells were co-transfected with pCDNA3.1-E2F4, Renilla Fluorescein TK, pGL3-CDCA3, pGL3-CDCA3-Δ1, pGL3-CDCA3-Δ2, or pGL3-CDCA3-Δ3 constructs. At 48 h or 72 h post-transfection, cells were harvested and lysed. The fluorescence intensity of the cell lysate was measured using a GloMax fluorescence detector (Promega).
Statistical analysis
All statistical analyses were performed using SPSS 22.0. Data were expressed as the mean ± standard deviation from three independent experiments. The means between two groups were analyzed using Student's t-test, while those between multiple groups were analyzed using one-way analysis of variance (ANOVA). The correlation between E2F4 expression and the clinicopathological features of patients was analyzed using the chi-square (χ2) test. Differences were considered significant at P <0.05.
Results
Correlation between E2F4 expression and clinicopathological factors
The mRNA levels of E2F4 in matched HCC and adjacent non-tumorous tissues from 40 HCC patients were analyzed using qRT-PCR. The clinical tissue samples were divided into E2F4-high (21 cases) and E2F4-low groups (19 cases) based on median E2F4 expression (Table 1). Analysis of the correlation between E2F4 expression and clinicopathological features revealed that most patients with tumor sizes > 5 cm (16/24) exhibited upregulated E2F4 expression. This suggests that the expression of E2F4 is positively correlated with tumor size. The absolute expression of E2F4 in the adjacent non-tumorous tissues (0.062 ± 0.0125) was significantly lower than the tumorous tissues (0.183 ± 0.035) (Fig. 1A). Additionally, E2F4 mRNA levels were examined in HCC (Huh7, SK-HEP-1, Hep3B, HepG2, Bel-7402) and normal liver (LO2) cell lines. Among the HCC cell lines, Huh7 and Hep3B exhibited the lowest and highest E2F4 expression, respectively. E2F4 expression in all HCC cell lines was higher than LO2 cells (Fig. 1B). These results suggest that E2F4 functions as an oncogene in liver cancer tissues. Survival analysis using the gene expression profiling interaction analysis (GEPIA; http://gepia.cancer-pku.cn/index.html) database revealed that the survival rate of HCC patients in the E2F4-high cohort was lower than that of patients in the E2F4-low cohort (Fig. 1C), which was consistent with the findings of Huang et al. [11,14]. This suggests that E2F4 is involved in HCC development.
Relationship between E2F4 expression and clinicopathological characteristics in 40 patients with HCC
| Factor | E2F4 expression | χ2 value | P value | |
|---|---|---|---|---|
| High (N=21) | Low (N=19) | |||
| Age | 0.316 | 0.574 | ||
| <40 years | 3 | 4 | ||
| ≤40 years | 18 | 15 | ||
| Gender | 1.948 | 0.163 | ||
| Male | 19 | 14 | ||
| Female | 2 | 5 | ||
| HBsAg | 0.025 | 0.874 | ||
| + | 17 | 15 | ||
| - | 4 | 4 | ||
| HBV DNA | 0.351 | 0.554 | ||
| + | 13 | 10 | ||
| - | 8 | 9 | ||
| AFP | 2.03 | 0.154 | ||
| <400 μg/L | 15 | 17 | ||
| ≥400 μg/L | 6 | 2 | ||
| ALT | 0.422 | 0.516 | ||
| <40 U/L | 10 | 11 | ||
| ≥40 U/L | 11 | 8 | ||
| AST | 0.902 | 0.342 | ||
| <40 U/L | 12 | 8 | ||
| ≥40 U/L | 9 | 11 | ||
| Tumor size | 4.829 | 0.028 | ||
| <5 cm | 5 | 11 | ||
| ≥5 cm | 16 | 8 | ||
| TNM stage | 0.819 | 0.366 | ||
| I | 3 | 8 | ||
| II | 4 | 3 | ||
| III | 14 | 8 | ||
TNM, tumor, node, metastasis; HBsAg, hepatitis B surface antigen; HBV, hepatitis B virus; ALT, alanine aminotransferase; AST, aspartate aminotransferase; AFP, alpha-fetoprotein.
Effect of E2F4 on the HCC cell proliferation, migration, and invasion
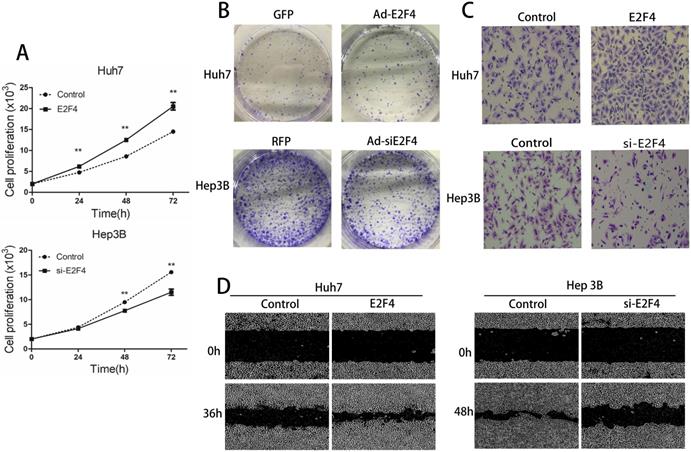
The proliferation, migration, and invasion of Ad-siE2F4-transfected and Ad-E2F4-transfected HCC cells were examined next. As shown in Fig. 2A, the proliferation of Ad-E2F4-transfected Huh7 cells was greater than the control cells. The proliferation of Hep3B cells was significantly decreased upon transfection with Ad-siE2F4. The results of the colony formation assay revealed that the Ad-E2F4-transfected Huh7 cells exhibited an increased number of colonies (Fig. 2B), whereas the Ad-siE2F4-transfected Hep3B cells exhibited a significantly decreased number of colonies. In the wound-healing assay, the migration rate of Ad-E2F4-transfected Huh7 cells significantly increased (Fig. 2C), while for Ad-siE2F4-transfected Hep3B cells, it significantly decreased. The results of the transwell assay (Fig. 2D) revealed that the invasion of Ad-E2F4-transfected Huh7 cells was markedly higher than the control cells. In contrast, the invasion of Ad-siE2F4-transfected Hep3B cells was markedly decreased. These findings further confirmed that E2F4 could enhance the proliferation, migration, and invasion of HCC cells [15].
E2F4 promotes the progression of HCC cell cycle
E2F family transcription factors mediated several cellular functions, including apoptosis and proliferation. E2F-mediated enhancement of cell proliferation could be caused by either inhibition of apoptosis or enhancement of proliferation [16].
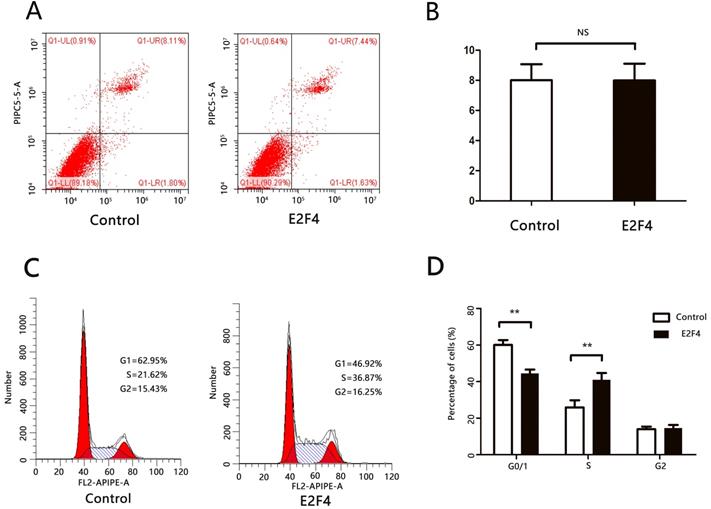
Huh7 cells transfected with either pcDNA3.1 or pcDNA3.1-E2F4 and subjected to flow cytometry analysis to examine apoptosis and cell cycle markers at 48 h or 72 h post-transfection. There was no significant difference in the number of cells at the late apoptotic (upper right quadrant) stage between the pcDNA3.1- and pcDNA3.1-E2F4-transfected groups (Figs. 3A and 3B). This suggests that E2F4 does not promote apoptosis in HCC cells. The pcDNA3.1-E2F4-transfected group exhibited an increased number of cells in the S-phase (Figs. 3C and 3D), indicating that E2F4 promotes the transition from G0/1 to S-phase and contributes to cell cycle progression.
CDCA3 is a target gene for E2F4
E2F4 regulates the expression of various genes by binding to promoters [17]. As E2F4 promotes the progression of the HCC cell cycle, we hypothesized that it modulated the expression of cell cycle regulator genes. Previous studies have reported various target genes of E2F4 using chromatin immunoprecipitation (ChIP) analysis. Among these target genes, CDCA3, CENPI, CDC7, and KIF2C were involved in the regulation of the cell cycle [12,13]. The effect of E2F4 overexpression or knockdown on the expression of these four genes in HCC cells was examined using qRT-PCR. The expression of CDC7 was not detected in both Huh7 and Hep3B cells and expression of CENPI and KIF2C were unaffected by either overexpression of knockdown of E2F4. However, the expression of CDCA3 was positively correlated with that of E2F4 (Fig. 4A). CDCA3, a cell division cycle protein, promoted the proliferation and cell cycle progression of colorectal and gastric cancer cells [14]. The proliferation of CDCA3-transfected cells significantly increased and was significantly inhibited upon co-transfection with siE2F4 (Figs. 4B and 4C). This indicates that E2F4 regulates HCC cell cycle through CDCA3.
Effect of E2F4 overexpression or knockdown on hepatocellular carcinoma (HCC) proliferation, migration, and invasion. E2F4 was overexpressed in Huh7 and knocked down in Hep3B cells. A. Results of the cell proliferation assay. The transfected cells were seeded and imaged using a long-term dynamic cell imaging system once every 24 h and the growth rate was calculated. B. Results of the colony formation assay. The transfected cells were cultured for one week. The cells were stained to examine the growth of colonies. C. Results of the transwell assay. The cells that passed through the membrane were stained and counted using a microscope (Magnification: 200×). D. Results of the wound-healing assay. The cell monolayer was scratched, and the migration of cells into the scratch area was monitored once every 12 h using the long-term dynamic cell imaging system.
E2F4 can promote cell cycle progression of hepatocellular carcinoma (HCC) cells. Huh7 cells were transfected with pCDNA3.1 or pCDNA3.1-E2F4. At 48 h post-transfection, the cells were analyzed via flow cytometry. A. Effect of E2F4 overexpression on apoptosis. B. Quantification of the number of late apoptotic cells in the pCDNA3.1-E2F4- and pCDNA3.1-transfected groups (upper right quadrant). C. Effect of E2F4 overexpression on the cell cycle. D. Quantification of the proportion of cells in the G0/1, S, and G2 phases of the cell cycle in the pCDNA3.1-E2F4- and pCDNA3.1-transfected groups.
Identification of E2F4 target genes. A. E2F4 was overexpressed in Huh7 cells and knocked down in the Hep3B cells. The expression of CDCA3, CENPI, CDC7, and KIF2C was examined using quantitative real-time polymerase chain reaction (qRT-PCR). B-C. The proliferation of Huh7 or Hep3B cells exhibiting overexpression or knockdown of E2F4. D-F. Schematic representation of the full-length and truncated CDCA3 promoters. 1 to 11 represent the predicted E2F4 binding sites. Results of the dual-luciferase reporter assay. Huh7 cells were transfected with CDCA3 full-length promoter [pGL3-CDCA3 (WT)] or three truncated CDCA3 promoter (pGL3-CDCA3-Δ1, pGL3-CDCA3-Δ2, and pGL3-CDCA3-Δ3) constructs. Luciferase activity was examined at 48 h post-transfection.
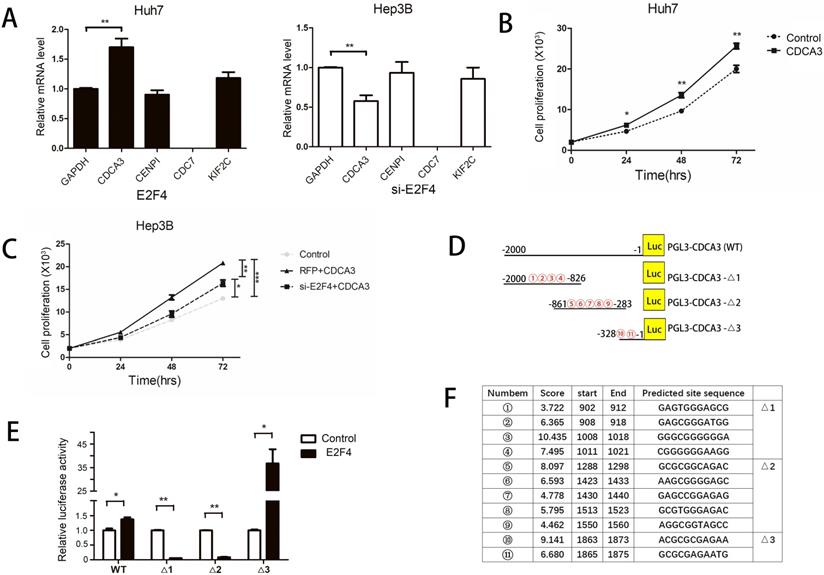
The E2F4 binding sites located in the CDCA3 5'-untranslated region (UTR) comprise a 2000-bp sequence upstream of the transcription start site, which were analyzed using the UCSC database. According to the JASPAR database, 11 candidate sequences in the CDCA3 promoter region were predicted to serve as potential E2F4 binding sites (Figs. 4D and 4F). Therefore, we constructed a vector encoding the full-length CDCA3 promoter (pGL3-CDCA3; pGL3-Basic as the vector) as well as vectors encoding three CDCA3 truncated promoter constructs containing three central high-scoring sequences (with relatively strong binding potential) (pGL3-CDCA3-Δ1, pGL3-CDCA3-Δ2, and pGL3-CDCA3-Δ3) (Fig. 4D). A dual-luciferase reporter assay was performed by co-transfecting Huh7 cells with E2F4 and pGL3-CDCA3, pGL3-CDCA3-Δ1, pGL3-CDCA3-Δ2, or pGL3-CDCA3-Δ3. Huh7 cells co-transfected with E2F4 and PGL3-CDCA3-Δ3 exhibited the highest luciferase activity (Fig. 4E). Two candidate sequences in the pGL3-CDCA3-Δ3 construct, 1863-ACGCGCGAGAA-1873 and 1865-GCGCGAGAATG-1875, were located at the same site. This indicates that E2F4 binds to this 13-bp sequence (1863-ACGCGCGAGAATG-1875) in the CDCA3 promoter.
Discussion
The E2F transcription factor family is involved in various cellular functions [15,16]. Previous studies have reported that E2F1 mediates carcinogenesis in various cancers. However, recent studies have also demonstrated that E2F1 is positively correlated with the tumor cell apoptosis index in cell carcinoma. Thus, E2F1 is a pro-apoptotic regulator that can also activate the tumor suppressor p53 [17,18]. Some studies have reported that E2F4 can arrest the cell cycle at G0 [19,20]. E2F4 is involved in the carcinogenesis of skin tumors, gastrointestinal tumors, and prostate cancer [21,22]. This study reported, for the first time, that E2F4 couldpromote the carcinogenesis of HCC through the upregulation of CDCA3, a cell cycle-related protein.
In the G0 phase, activator E2Fs often form a complex with RB family members (including RB, p107, and p130). The cyclin/CDK complex phosphorylates the RB protein upon stimulation, resulting in the release of E2F. Free E2F can induce the transcription of cell cycle-related proteins and consequently promote cell cycle progression [4,5,23-26]. E2F4, a member of the E2F transcription factor family, may dissociate from phosphorylated RB and upregulate CDCA3 expression by binding to its promoter region (1863-ACGCGCGAGAATG-1875). The upregulated expression of CDCA3 promotes the proliferation and cell cycle progression of HCC cells. However, further studies are required to elucidate the underlying mechanisms.
In this study, we analyzed the expression of E2F4 in HCC tissue samples and cell lines. E2F4 expression positively correlated with tumor size. The expression of E2F4 in HCC cell lines was higher than in LO2 cells. E2F4 overexpression enhanced the proliferation, migration, and invasion of HCC cells. The results of the luciferase reporter assay revealed that E2F4 upregulates CDCA3 expression by binding to its promoter region (1863-ACGCGCGAGAATG-1875) and consequently promotes the proliferation and cell cycle progression of HCC cells.
Abbreviations
HCC: hepatocellular carcinoma; TNM: tumor, node, metastasis; qRT-PCR: quantitative real-time polymerase chain reaction.
Supplementary Material
Supplementary table S1.
Acknowledgements
We would like to thank Editage (www.editage.cn) for English language editing.
Funding
This work was supported by grants from the Municipal Health Commission of Chongqing (2013-2-216) (to ML), the National Science and Technology Major Project (2017ZX10202203 and 2018ZX10732202), the Chongqing Natural Science Foundation (CSTC2018jscx-msybX0032) (to DW) (CSTC2016jcyjA0204) (to ML), and the Natural Science Foundation of Chongqing Yuzhong (20190145) (to DW).
Ethics Committee Approval and Patient Consent
The study protocol was approved by the Clinical Research Ethics Committee of Chongqing Medical University in January 2018 and was in compliance with the Declaration of Helsinki.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Author Contributions
All authors contributed to the conception and design of the study. Material preparation, data collection, and analysis were performed by Lulu Xia, Junye Liu, Shilei Wang, and Miao Luo. The first draft of the manuscript was written by Lulu Xia, Junye Liu, and Baoju Shan, and all authors commented on the previous versions of the manuscript. All authors have read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394-424
2. Fujiwara N, Friedman SL, Goossens N. et al. Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J. Hepatol. 2018;68:526-549
3. Marrero JA, Kulik LM, Sirlin CB. et al. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology. 2018;68:723-750
4. Ren B, Cam H, Takahashi Y. et al. E2F integrates cell cycle progression with DNA repair, replication, and G (2)/M checkpoints. Genes Dev. 2002;16:245-256
5. Dimova DK, Dyson NJ. The E2F transcriptional network: old acquaintances with new faces. Oncogene. 2005;24:2810-2826
6. Attwooll C, Lazzerini DE, Helin K. The E2F family: specific functions and over apping interests. EMBO J. 2004;23:4709-4716
7. Chen HZ, Ouseph MM, Li J. et al. Canonical and atypical E2Fs regulate the mammalian endocycle. Nat. Cell Biol. 2012;14:1192-1202
8. Rowland BD, Bernards R. Re-evaluating cell-cycle regulation by E2Fs. Cell. 2006;127:871-874
9. Lammens T, Li J, Leone G. et al. Atypical E2Fs: new players in the E2F transcription factor family. Trends Cell Biol. 2009;19:111-118
10. Leone G, Nuckolls F, Ishida S. et al. Identification of a novel E2F3 product suggests a mechanism for determining specificity of repression by Rb proteins. Mol. Cell Biol. 2000;20:3626-3632
11. Huang YL, Ning G, Chen LB. et al. Promising diagnostic and prognostic value of E2Fs in human hepatocellular carcinoma. Cancer Manag Res. 2019;11:1725-1740
12. Hsu J, Arand J, Chaikovsky A. et al. E2F4 regulates transcriptional activation in mouse embryonic stem cells independently of the RB family. Nat. Commun. 2019;10:2939
13. Xiaoqin X, Mark B, Victor XJ. et al. A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res. 2007;17:1550-1561
14. Pascale RM, Simile MM, Calvisi DF. et al. Role of HSP90, CDC37, and CRM1 as modulators of P16(INK4A) activity in rat liver carcinogenesis and human liver cancer. Hepatology. 2005;42:1310-1319
15. Yang X, Zu X, Tang J. et al. Zbtb7 suppresses the expression of CDK2 and E2F4 in liver cancer cells: implications for the role of Zbtb7 in cell cycle regulation. Mol Med Rep. 2012;5:1475-1480
16. Hsu J, Sage J. Novel functions for the transcription factor E2F4 in development and disease. Cell Cycle. 2016;15:3183-3190
17. Ferrer-Vicens I, Riffo-Campos ÁL, Zaragozá R. et al. In vivo genome-wide binding of Id2 to E2F4 target genes as part of a reversible program in mice liver. Cell Mol Life Sci. 2014;71:3583-3597
18. Choi M, Lee H, Rho HM. E2F1 activates the human p53 promoter and overcomes the repressive effect of hepatitis B viral X protein (Hbx) on the p53 promoter. IUBMB Life. 2002;53:309-317
19. Moberg K, Starz MA, Lees JA. E2F-4 switches from p130 to p107 and pRB in response to cell cycle reentry. Mol. Cell Biol. 1996;16:1436-1449
20. Bertoli C, Skotheim JM, de Bruin RA. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 2013;14:518-528
21. Waghray A, Schober M, Feroze F. et al. Identification of differentially expressed genes by serial analysis of gene expression in human prostate cancer. Cancer Res. 2001;61:4283-4286
22. Wang D, Russell JL, Johnson DG. E2F4 and E2F1 have similar proliferative properties but different apoptotic and oncogenic properties in vivo. Mol. Cell Biol. 2000;20:3417-3424
23. Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 2002;3:11-20
24. Gaubatz S, Lindeman GJ, Ishida S. et al. E2F4 and E2F5 play an essential role in pocket protein-mediated G1 control. Mol Cell. 2000;6:729-735
25. Wu L, Timmers C, Maiti B. et al. The E2F1-3 transcription factors are essential for cellular proliferation. Nature. 2001;414:457-462
26. Claassen GF, Hann SR. A role for transcriptional repression of p21CIP1 by c-Myc in overcoming transforming growth factor beta -induced cell-cycle arrest. Proc. Natl. Acad. Sci. U S A. 2000;97:979498-9503
Author contact
![]() Corresponding authors: Deqiang Wang, Room 613, College of Life Sciences Building, 1 YiXueYuan Road, YuZhong District, Chongqing, 400016, China. Baoju Shan, Children's Hospital of Chongqing Medical University, No.136 the 2nd Zhongshan Road, Yuzhong District, Chongqing 400014, China. Miao Luo, Yubei District People's Hospital, No.23 North Road of Central Park, Laboratory Medicine, People's Hospital of Yubei district, Chongqing, 401120, China. E-mail: wangdqedu.cn (D Wang); 481080cqmu.edu.cn (B Shan); 536513878com (M Luo).
Corresponding authors: Deqiang Wang, Room 613, College of Life Sciences Building, 1 YiXueYuan Road, YuZhong District, Chongqing, 400016, China. Baoju Shan, Children's Hospital of Chongqing Medical University, No.136 the 2nd Zhongshan Road, Yuzhong District, Chongqing 400014, China. Miao Luo, Yubei District People's Hospital, No.23 North Road of Central Park, Laboratory Medicine, People's Hospital of Yubei district, Chongqing, 401120, China. E-mail: wangdqedu.cn (D Wang); 481080cqmu.edu.cn (B Shan); 536513878com (M Luo).

 Global reach, higher impact
Global reach, higher impact