Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(17):5206-5219. doi:10.7150/jca.58182 This issue Cite
Review
The Emerging Role of the Interactions between Circular RNAs and RNA-binding Proteins in Common Human Cancers
Department of General Surgery, the First Affiliated Hospital of Nanjing Medical University, Nanjing, Jiangsu, China.
#These authors contributed equally to this article.
Received 2021-1-14; Accepted 2021-6-9; Published 2021-6-26
Abstract
Circular RNAs (circRNAs) are a unique family of noncoding RNAs that could regulate multiple biological processes, which play a crucial role in carcinogenesis, progression and chemotherapy resistance of cancers. Growing studies have demonstrated that circRNAs act as novel biomarkers and therapeutic targets for cancers by sponging microRNAs (miRNAs). Up to date, another function of circRNAs, combining with RNA-binding proteins (RBPs), was uncovered. However, there is limit studies illustrating the underlying mechanism of circRNAs-RBPs interactions, as well as showing its roles in diverse types of cancers. In this review, we collected the biogenesis, properties of circRNAs, and then synthesize the connection between circRNAs and RBPs, and try to clarify its molecular mechanisms involving in the pathogenesis and progression of several common cancers, aiming to provide a brand-new insight to the prognosis and treatment strategy for cancers.
Keywords: circular RNAs (circRNAs), RNA binding proteins (RBPs), biogenesis, transcription, cancers
Introduction
Circular RNAs (CircRNAs), with covalently closed loop structure, are a vital type of noncoding RNAs and have attracted plenty of researchers' attention in recent years. Actually, the incidental discovery of the first circRNAs, as a class of infectious plant viroids, could date back to as early as the 1970s [1]. Owing to remarkable transition from traditional transcriptome analyses to accurate sequencing and bioinformatics, a large amount of circRNAs have widely identified in eukaryotic cells but viewed as transcription byproducts without function [2]. Dramatically, it was first proposed that circRNAs functioned as miRNA sponge to regulate gene expression at post-transcriptional level [3, 4]. Afterwards, emerging researches revealed other essential roles of circRNAs in diverse biology process: binding partners of RNA binding proteins (RBPs), regulators of transcription, and templates for translated protein [5-7]. Nowadays, it has been found that abundant circRNAs to a large extent participate in development of human diseases, especially cancer, promoting to increase researches on underlying mechanism and providing potential candidates for effective disease diagnosis and treatment.
Although circRNAs generally act as a miRNA sponge to perform biological function, studies have shown circRNAs are involved in diverse pathological processes by binding to RBPs [7]. RBPs are a class of proteins containing RNA-binding domains that integrate with and control target RNAs at the post-transcriptional level, including transcription, splicing, stabilization, localization, translation, translation [8]. Previous studies have shown that hundreds of RBPs identified in the human genome are widely expressed in tissues, some of which exhibit aberrant expression under disease conditions [9].
With updated techniques like RNA pull-down and RNA immunoprecipitation (RIP), the physical interactions between circRNAs and RBPs are recognized and play an irreplaceable role in human pathogeneses, particularly in progression of tumors. Nevertheless, the number of reviews related to circRNAs-RBPs complexes and detailed mechanism is limited [7, 10].
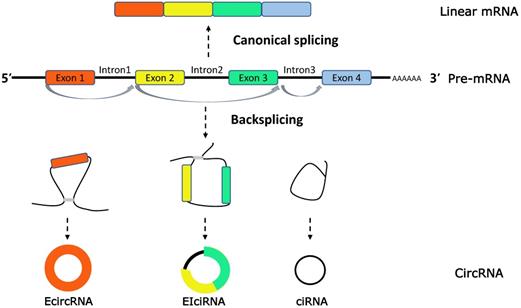
CircRNAs biogenesis. CircRNAs are generated in the process of splicing of pre-mRNA and compete with the counterparts, linear mRNA. CircRNAs are generally classified into three types (1) exonic RNAs (EcircRNAs), generated by exons only. (2) EIciRNAs, generated by introns 'retained' between the exons. (3) circular intronic RNAs (ciRNAs), generated by two or more connected introns.
In our review, we aim to summarize the intimate relationship between circRNAs and RBPs, meanwhile we focus on the influence of circRNAs-RBPs interactions in several common types of tumors, providing new sight into diagnostic and therapeutic tools for incurable human cancers in the future.
The biogenesis of circRNAs
Extensive studies on circRNAs have revealed that circRNAs are characterized with a ring covalently bound by a 5′ cap and 3′ poly (A) tails, produced by precursor-mRNA (pre-mRNA) back-splicing, which is distinct from the mature mRNAs generated by pre-mRNA through canonical or alternative splicings [11, 12]. On the basis of the source of internal sequence, circRNAs can be classified into three categories: exonic circRNAs (EcircRNAs), intronic circRNAs (ciRNAs) and exon-intron circRNAs (EIciRNAs) [13] (Figure 1).
By collecting published researches, circRNA biogenesis have been demonstrated as a complex process regulated by three main biological regulatory mechanisms. As we concluded: 1) intronic repeat sequences. For example, the intronic repeats, in the Sry circular RNA biogenesis model, may base pair to one another, bringing the splice sites into close proximity to facilitate backsplicing [14]. 2) Exon skipping and resplicing of the lariat RNA. Thorough mechanistic evidence was shown that a large lariat containing the skipped exon, followed by exons skipping during alternative splicing, is a common intermediate step before the production of a circular RNA [15]. 3) RNA-binding proteins. Recently, it was reported that trans-acting splicing factors, such as hnRNPs and SR proteins, act to co-regulate pre-mRNA splicing patterns through site-specific binding to target RNAs [16].
Taken together, although there is increasing studies focusing on the formation of circRNAs at gene level, more detailed molecular mechanisms still need to be explore.
The properties of circRNAs
Increasing studies have manifested that circRNAs have some predominant properties. First of all, the high stability of circRMAs has been widely acknowledged, mainly attributing to the covalent looping structure and resistance against RNase R [17]. There is supporting evidence showing a longer average half-life of circRNAs, over 48 h, in comparison to the average 10 h linear RNA counterparts [18].
Followingly, circRNA is highly abundant in human tissues and cells. Thanks to RNA deep sequencing and computational algorithms, researchers have managed to identify over 25,000 presumptive circular RNAs stemed from more than 15% of encoding gene transcripts in human fibroblasts [11]. In general, circRNAs still maintain a low expression as comparing to their host mRNAs.
Another characteristic of circRNAs is specificity in various tissues and developmental stage. For instance, an early study has demonstrated a tissue-specifically expression of circRNAs, with an enrichment in brain tissues [19]. In virtue of circRNA sequencing, most of about 10,000 noval circRNAs are development-specific expressed in preimplantation human embryos [20].
Evolutionary conservation is a crucial property for circRNAs. For example, Xia et al. found approximately 700 homologous circRNAs between mouse and fetal human tissues, especially in brain [21]. In addition, a mass of circRNAs has been identified in fungi, plants, and protists, indicating the feature of evolutionarily conserved circRNAs [22].
The connection between circRNAs and RNA-binding proteins
As one of circRNAs' functions, previous bioinformatic analyses suggested that circRNAs were predicted to own abundant binding motifs for proteins, allowing circRNAs to combine with proteins, including RBPs. In recent studies, investigators are trying to uncover concrete contact patterns for circRNAs-proteins interactions, especially the connection between circRNAs and RBPs.
Firstly, accumulating studies have shown the existence and importance of RBPs participating in the formation of circRNAs in vivo or in vitro models. The most representative example is the RNA binding protein QKI, one of the members of the STAR family of KH domain-containing RNA binding proteins, was identified as a pre-mRNA splicing factor and a chief modulator of circRNA biogenesis [23, 24]. Previously, it has been shown that QKI enhance circRNA formation via pre-mRNAs splicing regulation through binding to recognition elements within introns, in the vicinity of the circRNA-forming splice sites, and promoting circRNA-forming exons into close proximity [25]. Lately, estrogen receptor α (ERα), a protein associated with progression of hepatocellular carcinoma (HCC), was found to directly bind to the 5′ promoter region of its host gene SMG1, thereby suppressing the circular RNA-SMG1.72 expression [26]. In recent years, many RBPs, such as RNA helicase DHX9 [27], RBM3 [28], NF90/NF110 [29], MBL [30], have been shown to a substantial contribution to the regulation of circRNA production.
While RBPs are able to regulate circRNAs biogenesis, circRNAs can relatively modulate the expression of RNA-binding proteins. A canonical example involves the tumor suppressor gene TP53-coded protein, p53 acts as a RBP to directly interact with circRNAs, which exerts joint function in the occurrence and progression of tumors [31]. Additionally, it was shown that the expression of p53 can be regulated by some circRNAs. Among them, Circ-DNMT1 was reported to regulate p53 transcriptional activity by forming a heterogeneous nuclear ribonucleoprotein and translocating into the nucleus via the interaction with p53 itself and the other AUF1 [32]. A growing number of studies have indicated that other circRNAs are involved in the process of RBP transcription via a direct or indirect manner, including Circ-DNMT1 [32], circ-Ccnb1 [33], circ-MDM2 [34], and so on.
Moreover, RBPs are more likely to directly or indirectly participate in circRNAs-mediated transcription. CircRNA function is also elicited by regulating the related gene transcription and gene expression. Moreover, it was evident that EIciRNAs hold U1 snRNP through interaction with U1 snRNA, and then further bind to Pol II transcription complex at the promoters of parental genes to enhance gene transcription [35]. Additional support for an important role of RBPs on circRNAs modulating gene expression comes from bioinformatics and functional studies of HuR. It has extensively been shown to affect selectively the expression of certain mRNAs at similar post-transcriptional levels by binding to circ-PABPN1 rather than PABPN1 mRNA [36]. Further studies have revealed that RBPs play a vital role in the transcription regulation of circRNAs.
Furthermore, increasing lines of evidence suggest that some RBPs are capable of facilitating translation of circRNAs. According to previous studies, N6-methyladenosine (m6A) located on circRNAs seems to a main mechanism responsible for the regulation of circRNA translation. The m6A is an adenosine methylation modification of RNA bases that can promote the efficient initiation of translation from circRNAs [37]. Therefore, with the assistant of two RBPs, m6A demethylase FTO and adenosine methyltransferase METTL3/14, we could affect the translation efficiency through selectively altering m6A modification. In addition, different from linear mRNA translation, m6A-initiated circRNA translation needs to be initiated by protein factors, such as eukaryotic translation initiation factor eIF4G2 and m6A reader YTHDF3 [38]. To sum up, RBPs are essential regulators of the circRNAs translation.
CircRNAs-RBPs interaction and cancers
Cancer is a common public health problem and the treatment of tumor and its internal mechanism are the hotpots of medical research. And, a large number of researchers have identified extensive circRNAs functioning as miRNAs sponge, emphasizing its important role in cancer development. Additionally, circRNAs-RBPs interaction is emerging as a new mechanism in several hallmarks of cancer, such as cell death and survival, invasion, and metastasis. To date, only a few reports have also shown that circRNAs may associate with specific proteins to exert important functions in cancer. Next, we mainly summarize some significant associations between RBPs and circRNAs in the following several types of tumors (Table 1).
Breast cancer
Breast cancer (BC) is the most common female cancer worldwide and one of the leading cause of cancer-related death among women [39]. Early breast cancer can be better improved by precise and individualized treatment, largely due to comprehensive therapies, including surgery, radiotherapy, chemotherapy and endocrinotherapy and targeted therapy. Despite this, treatment for advanced breast cancer remains a huge challenge, with a five-year survival rate of 26% [40]. Therefore, valuable biomarkers deserve further exploration for the early detection, treatment and prognosis for BC.
CircRNAs have been proposed to have a key role in BC development through interacting with RNA binding proteins. Circ Foxo3 was a classic candidate that had a significant low expression in BC tumors and cancer cells as comparing to normal tissue and cells, respectively. Previously, it was reported that poly-ubiquitination function of MDM2 protein was essential in the degradation of p53 and Foxo3 [41]. Further study indicated that circFOXO3 directly interacted with p53 and MDM2, providing a platform for p53 degradation in MDM2-mediated manner. Of note, Foxo3 was identified to compete with p53 for binding circFOXO3, but up-regulated circFOXO3 had a higher affinity for p53, leaving FOXO3 activate downstream Puma-mediated apoptosis [42].
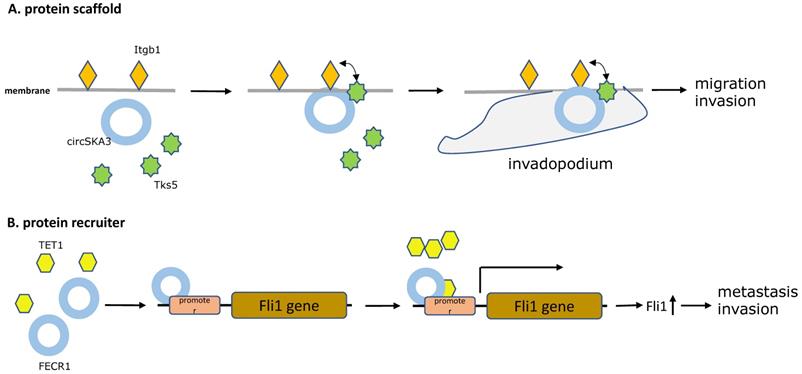
A similar example, by microarray analysis, overexpressed circSKA3 was found to promote breast cancer cell migration and invasion in breast cancer cells and human breast cancer tissues. Mechanistically, providing evidence showed that the circSKA3 induced the formation of invadopodium to promote tumor progression by directly complexing with Tks5 and integrin b1, which was detected by immunoprecipitation of cell lysates [43]. As in the previous article, Itgb1 is known as a kind of cell-surface receptor involved in the process of cell adhesion, detachment, and migration [44]. Tks5 is an adaptor protein and its phosphorylation is critical for invadopodium formation [45]. Notably, William Du et al. demonstrated that circSKA3 functioned as protein scaffolds and provided a bridge for Tks5 and integrin b1 on the cell membrane, which is the key complex in formation of invadopodia (Figure 2A).
In an another study in 2018, a novel circular RNA FECR1 derived from FLI1 exons, was illustrated to have a positive correlation with tumor invasion in breast cancer cell lines, suggesting its role as an oncogenic driver in metastasis of breast cancer. Furthermore, a novel epigenetic pathway was discovered that FECR1 could bind to the parental gene FLI1 promoter in cis and acts as a protein recruiter for TET1 demethylase which demethylated the promoter CpG islands, thereby activating transcription of the oncogene FLI1 to promote tumor metastasis [46] (Figure 2B).
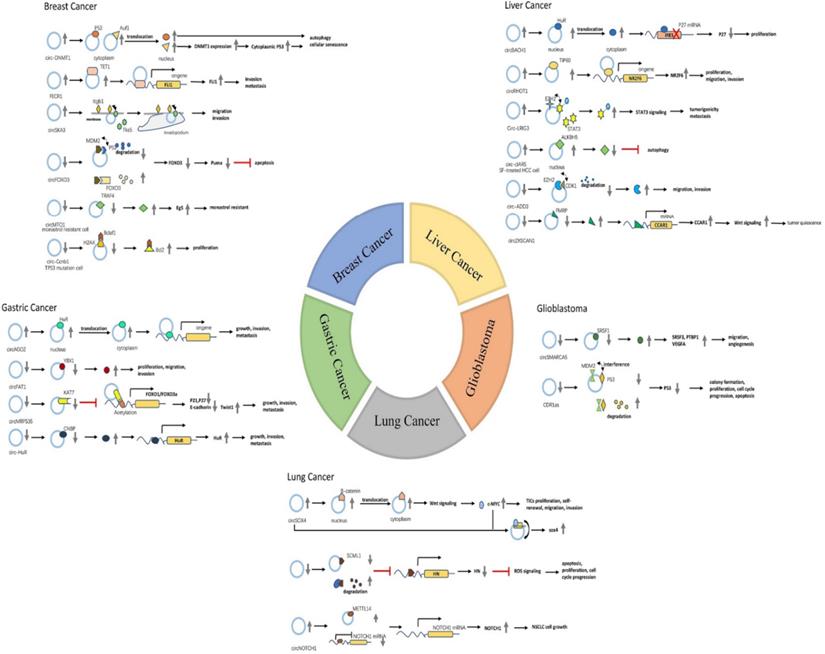
Known circRNAs-RBPs interactions in several common types of tumors
| Cancer | CircRNA | Dysregulation | Type of tissue/cell lines | RBPs | CircRNAs-RBPs interaction | Participation | Ref. |
|---|---|---|---|---|---|---|---|
| Breast cancer | circFOXO3 | down | BC tissues and cells | MDM2,p53 | protein scaffold | apoptosis | 2017 [42] |
| FECR1 | up | BC tissues | TET1 | protein recruiter | invasion, metastasis | 2017 [46] | |
| circ-Ccnb1 | down | BC cell lines | H2AX,p53; H2AX,Bcla1 | protein scaffold | proliferation, apoptosis | 2017 [33] | |
| circDNMT1 | up | BC cell lines | p53,AUF1 | protein translocation | autophagy mediated cell proliferation, survival, and tumor growth | 2018 [32] | |
| circMTO1 | down | monastrol-resistant cell lines | TRAF4 | protein decoy | monastrol resistance | 2018 [49] | |
| circSKA3 | up | BC tissues and cells | Interinβ1,Tks5 | protein scaffold | migration, invasion | 2020 [43] | |
| Liver cancer | circRHOT1 | up | HCC tissues | TIP60 | protein recruiter | proliferation, migration, invasion | 2019 [54] |
| circ-ADD3 | down | HCC tissues | EZH2,CDK1 | protein scaffold | migration, invasion, metastasis | 2019 [38] | |
| circZKSCAN1 | down | HCC tissues | FMRP | protein decoy | tumor quiescence | 2019 [57] | |
| circ-cIARS | up | SF-treated HCC cells | ALKBH5 | protein decoy | autophagy | 2020 [60] | |
| circ-LRIG3 | up | HCC tissues, cells and plasma | EZH2,STAT3 | protein scaffold | tumorigenicity, metastasis | 2020 [64] | |
| circBACH1 | up | HCC tissues | HuR | protein translocation | proliferation | 2020 [63] | |
| Gastric cancer | circAGO2 | up | GC tissues and cells | HuR | protein translocation | growth, invasion, metastasis | 2019 [66] |
| circFAT1(e2) | down | GC tissues and cells | YBX1 | protein decoy | proliferation, migration, invasion | 2019 [69] | |
| circMRPS35 | down | GC tissues | KAT7 | protein recruiter | growth, invasion, metastasis | 2020 [72] | |
| circ-HuR | down | GC tissues and cells | CNBP | protein decoy | growth, invasion, metastasis | 2020 [75] | |
| Lung cancer | circNOL10 | down | LC tissues | SCML1 | protein defender | apoptosis, proliferation, cell cycle progression | 2019 [79] |
| circ-SOX4 | up | CD133+ NSCLC cells | β-catenin, c-MYC | protein translocation, protein defender | TICs proliferation, self-renewal, migration, invasion | 2020 [81] | |
| circNOTCH1 | up | NSCLC cell lines | METTL14 | protein decoy | NSCLC cell growth | 2020 [84] | |
| Glioblastoma | circSMARCA5 | down | GBM tissues | SRSF1 | protein decoy | migration, angiogenesis | 2018 [87], 2019 [88] |
| CDR1as | down | GBM tissues | MDM2,p53 | protein defender | colony formation, proliferation, cell cycle progression, apoptosis | 2020 [90] |
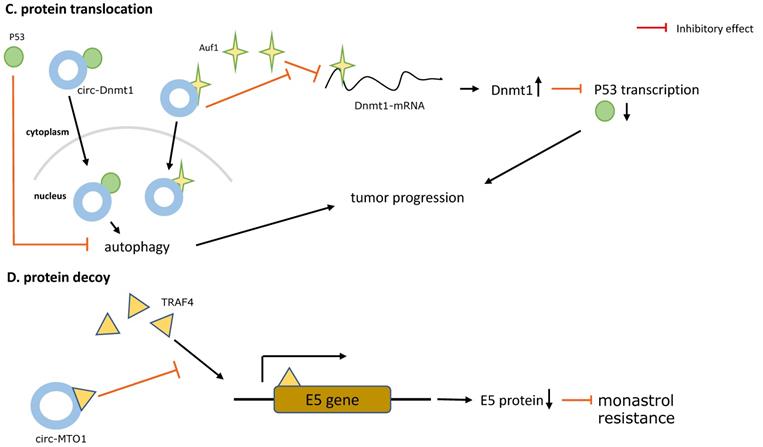
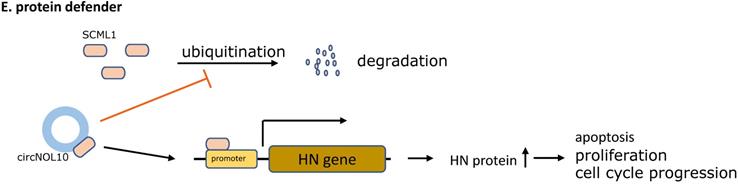
The five main types of CircRNAs-RBPs interactions. A. Protein scaffold.CircSKA3 binds to Tks5 and integrin b1, and serves as a protein scaffold for interaction of them on the cell membrane, inducing the production of invadopodia in breast cancer. B. Protein recruiter. FECR1 attaches to the parental gene FLI1 promoter and recruiters for TET1 demethylase to demethylate the promoter, stimulating oncogene FLI1 transcription to enhance tumor metastasis and invasion. C. Protein translocation.Circ-Dnmt1 promotes translocation of p53 and AUF1 from cytoplasm to nucleus, which enhances cellular autophagy and reduces p53 expression in the cytoplasm, thereby accelerating tumor progression. D. Protein decoy. CircRNA-MTO1 combines with TRAF4 and blocks its positive regulation of E5 translation, and finally reverses monastrol resistance. E. Protein defender. CircNOL10 physically connects with SCML1 and protects SCML1 from degradation induced by ubiquitination, thus the increased SCML1 expression activating transcription of the HN polypeptide family related to apoptosis, proliferation, and cell cycle progression in lung cancer.
The detailed mechanism of CircRNAs-RBPs interactions in five common human cancers.
Additionally, another oncogenic circRNA in breast cancer is circ-Dnmt1. Du et al. used microarrays analysis to identify circ-Dnmt1 upregulated in BC cell lines and tumors and that is link to breast cancer progression. Of note, circ-Dnmt1 utilized an autophagy mechanism to inhibiting cellular senescence and enhancing cell proliferation, survival, and tumor growth, which was induced by nuclear translocation of p53 and AUF1 via circ-Dnmt1 directly binding to its two oncogenic protein partners. Nuclear translocation of p53 enhanced cellular autophagy, in contrast to p53 in the cytoplasm, while the AU-rich RNA-binding protein AUF1 was transported into the nucleus increasing Dnmt1 translation and thus inhibiting p53 transcription [32] (Figure 2C).
According to another study published in 2018, a circRNA called circRNA-MTO1 was expressed at abnormally low levels in monastrol resistant cells and negatively regulated cell viability and monastrol resistance. In order to uncover the mechanism of circRNA-MTO1, Liu et al. used mass spectrometry and RNA-pull down assay to confirm the direct interactions between circRNA-MTO1 and tumor necrosis factor receptor associated factor 4 (TRAF4). TRAF4 is an (A+U)-rich elements (AREs)-binding protein, and can interact with ARE areas and functions as a potential oncogenic protein due to the high expression levels in human carcinomas [47, 48]. Collectively, the findings of this study indicated that overexpressed circRNA-MTO1 could bind to TRAF4, blocking the TRAF4-mediated E5 translation and finally inhibiting viability and reversing monastrol resistance [49] (Figure 2D).
Hepatocellular carcinoma
According to World Health Organization statistics, hepatocellular carcinoma (HCC) is the fourth most prevalent cause of cancer-related deaths globally. Due to aggressive tumor biological characteristics, HCC patients suffer a high rate of mortality, with ~841,000 new cases and 782,000 deaths every year [39]. Currently curative liver surgery benefits patients at early stage, but exerts only limited effects on advanced HCC patients. Thus, it is urgent to elucidate the mechanisms underlying the development of HCC and explore the effective biomarker for early diagnosis and prognosis.
Accumulating evidence suggests that both of circRNAs and some certain RBPs have a crucial role in HCC.TIP60 is a classic cancer-related RNA-binding protein, contributing to tumorigenesis, mesothelioma malignance, cancer growth in various types of cancer [50-52]. The main oncogenic effects of TIP60 appeared to trigger target gene transcription through governing histone acylation modification [53]. To explore the role of TIP60 in liver cancer, Wang et al. used ChIP and FISH assays to indicate that TIP60 was recruited by circRHOT1 to combine with the NR2F6 promoter, and next actively recruited other components of NuA4 complex to finally enhance target gene NR2F6 expressions, resulting in suppressing HCC development and progression. Of them, circRHOT1 was also illustrated as a potential prognosis biomarker for HCC, depending on that patients' with high circRHOT1 was more likely related with poor prognosis [54].
Although FMRP serves as a RBP and exerts its function of regulating translation of target mRNAs mostly studied in the nervous system [55], its expression appeared to be significantly low in HCC [56]. In the previous published study, circZKSCAN1 was confirmed to interact with FMRP and prevent tumor development by inhibiting cell stemness, proliferation, and metastasis in HCC. Mechanistical results indicated that the combination of circZKSCAN1 and FMRP sequestered FMRP from binding to β-catenin-binding protein-cell cycle and apoptosis regulator 1 (CCAR1) mRNA, which induced tumor quiescence by blocking the Wnt/β-catenin signaling pathway [57].
It was recently reported that cIARS appeared to be ectopic expression by using RNA-seq during sorafenib (SF) treatment in HCC cells. Functional experimental analysis suggested that cIARS was demonstrated as a positive regulator of SF-induced ferroptosis through activating autophagy and ferritinophagy. By RNA pulldown and RNA EMSA assays, the authors validated ALKBH5 as a potential interacting protein of cIARS, whose effect on autophagy regulation is reflected in a variety of tumor cells [58, 59]. In this study, cIARS was proven to involve in SF-induced ferroptosis by bounding to its protein couple ALKBH5 and abolishing the autophagy inhibitor role of ALKBH5 in HCC cells [60].
As a recent research indicated, circBACH1 expression level was increased specifically in HCC tissues and cell lines. An analysis of clinical data revealed that the HCC patients with high circBACH1 expression may have poor prognosis. Moreover, based on cell function experiments, circBACH1 was confirmed to facilitate cell proliferation by negatively modulating p27 expression. In addition, RIP assays, pull‐down assays and EMSAs was performed to identify HuR as circBACH1 binding partner, which had been found to target p27 mRNA and repress its translation [61]. P27, belongs to cyclin‐dependent kinase inhibitor family, is a tumor suppressor responsible for cell cycle arrest at G1‐S stage [62]. In brief, Liu et al. illustrated that HuR could combined with circBACH1 in the nucleus and transported into the cytoplasm, where HuR inhibited p27 expression at translational levels [63].
Circ-LRIG3, an overexpressed nuclear circRNA in HCC, was found to enhance HCC tumorigenicity and progression. In mechanism, Sun et al. indicated that Circ-LRIG3 acted as a protein scaffold for EZH2 and STAT3 and activated STAT3 signaling through methylating and phosphorylating STAT3 induced by EZH2. In turn, a positive feedback loop was uncovered that activated STAT3 could directly interact with circ-LRIG3 promoter to facilitate circ-LRIG3 transcription and increase its expression [64]. EZH2 is a notable oncogenic RBP and was previously reported to directly bind to another tumor suppressor, circ-ADD3, in HCC. Other than circ-LRIG3-EZH2-STAT3 ternary complex, EZH2 was able to bind to CDK1 in the presence of circ-ADD3 and increased phosphorylation and ubiquitination of EZH2, resulting in EZH2 degradation. Subsequently, low EZH2 protein expression weakens its tumor metastasis-promoting effect in HCC [38].
Gastric cancer
Gastric cancer (GC), as one of the most frequently occurring malignancy, has become the third major cause of cancer deaths in China [65]. Although rapid advances in diagnosis and treatment had partly improved patient outcomes, the 5-year survival rate is still quite poor, as shown in global cancer statistics [65]. Therefore, it is imperative to seek novel biomarkers and significative therapeutic target for diagnosis and treatment for GC.
Mounting data suggests that circRNAs are related to gastric cancer (GC) development, but only a few studies uncover their mechanism of action in GC development, especially the circRNAs-RBPs interaction.
As a significant cancer-related circRNA, CircAGO2 is up-regulated in human cancer tissues, such as gastric cancer, colon cancer, prostate cancer, and is closely relevant to poor prognosis of tumor sufferers. The physically binding CircAGO2 to HuR was confirmed by RNA pull-down and western blot assays, which played an important role in promoting gastric cancer progression. Further studies discovered that CircAGO2-HuR complex enhanced the translocation of HuR from nucleus to cytoplasm, where HuR directionally tethered to the 3'-UTR regions of target oncogenes and followingly prevented AGO2 from binding and blocked downstream AGO2/miRNA signal pathway [66].
According to functional assays, Fang et al. indicated a negative correlation between the expression of circFAT1 (e2) and GC cell proliferation, migration and invasion. Besides, Y-box binding protein-1 (YBX1), a RBP known as a potential biomarker in GC diagnosis, has been found to positively regulate gastric cancer cells migration [67, 68]. Moreover, it was demonstrated that circFAT1 (e2) may directly interact with YBX1 in the nucleus and initiate its tumor suppressive effect by performing the online catRAPID analysis and circRIP assay [69].
Another pathway associated with gastric cancer progression is the CircMRPS35 interacting with histone acetyltransferase KAT7.CircMRPS35, a noval circRNA, was first found in gastric cancer tissues with a low expression level, selected from differential expression profiles of circRNAs in human gastric cancer and adjacent normal tissues by RNA-seq. It was proposed that overexpression CircMRPS35 could specifically bind to FOXO1/3a promoter regions and hence activate the transcription of FOXO1/3a via H4K5 acetylation after recruitment of KAT7.In turn, the activation of FOXO1 positively stimulates the expression of its downstream genes p21, p27, PUMA and Bim, while activated FOXO3a upregulates E-cadherin and downregulated Twist1, eventually restraining cell invasion and metastasis in GC [70, 71]. Therefore, we could hypothesize that CircMRPS35 is becoming a promising druggable target for antitumor treatment under the regulation of histone modification [72].
In addition, another new circRNA called Circ-HuR (hsa_circ_0049027) was shown to be remarkably down-regulated in gastric cancer tissue, whose high expression was related to inhibiting aggressive features such as the growth, invasion, and metastasis of gastric cancer cells. CCHC-type zinc finger nucleic acid binding protein (CNBP), known as a transcription factor, was previously proven its meaningful impact on malignant behavior of tumor cells [73, 74]. Particularly, a proteomic analysis was performed to suggest the binding of circ-HuR to CNBP arrested the function of CNBP on facilitating HuR expression and suppress tumor progression [75].
Lung cancer
Lung cancer (LC) is the most dominant cause of cancer-related death worldwide, mainly owing to highly reoccurrence and metastasis after therapy [76, 77]. Non-small-cell lung cancer (NSCLC), representing 85% of the total lung cancer cases, has been considered as the most common histological kind of lung cancer, while small-cell lung cancer (SCLC) is known as the most lethal lung cancer with very low survival rates [78]. To cut down LC deaths, scientists are trying to understand the detailed molecular mechanisms in LC in order to find more potential therapeutic strategies for LC.
Studies have suggested that circRNAs and related RBPs could serve as diagnostic or predictive biomarkers for lung cancer, and could provide new insights especially in NSCLC.
It was well recognized that circNOL10 was low-expressed in lung cancer tissues, as well as lung cancer cells, whereas circNOL10 upregulation contributed to repression of tumor development. Most notably, circNOL10 was shown to physically bind to the transcription factor SCML1, and subsequently increased the SCML1 expression, instead of inhibiting binding protein functions. Moreover, the circNOL10-SCML1 complex was able to restrain from SCML1 degradation induced by ubiquitination, thus up-regulated SCML1 further activated transcription of the HN polypeptide family associated with apoptosis, proliferation, and cell cycle progression in lung cancer [79] (Figure 2E).
A newly study verified that circ-SOX4 is positively correlation with CD133 expression and highly expressed in CD133+ lung cancer cells. Results from functional assays proved that downregulated circ-SOX4 restrained lung tumor-initiating cells (TICs) proliferation, self-renewal, migration and invasion. c-MYC, a pivotal RNA-binding protein, has been previously delineated involvement in NSCLC progression [80]. In this study, circ-SOX4 upregulation was illustrated to enhance c-MYC expression via triggering the Wnt/β-catenin axis in NSCLC, while up-regulated c-MYC enabled circ-SOX4 to accelerate transcription by binding to circ-SOX4 promoter, thus forming a positive feedback [81].
G protein-coupled oestrogen receptor (GPER), a component of the G protein-coupled receptor (GPCR) family, has been regarded as a oncogenic driver to promote tumor initiation and development through YAP1/TEAD signalling in breast cancer [82, 83]. Similarly, Shen et al. suggested that GPER enhanced cell growth of non-small cell lung cancer cells by circNOTCH1/m6A methylated NOTCH1 axis, whereby the physical interaction between circNOTCH1 and METTL14 played a compelling role in GPER downstream regulatory network. As a RBP, METTL14 is capable of undergoing target mRNAs m6A modifications and affecting its stability. Besides, circNOTCH1 served as endogenous competitive circRNA to compete for METTL14 binding. Due to the lack of METTL14, NOTCH1 mRNA appeared to be more stable and tended to be transcribed to high expression of NOTCH1, ultimately leading to initiate NOTCH1-mediated signal pathway [84].
Glioma
Glioma is the most prevailing tumor of the central nervous system (CNS), accounting for 30% of all CNS tumors, of which glioblastoma multiforme (GBM) is regarded as most aggressive glioma [85]. In spite of tremendous efforts and progress made in surgical operation, radiotherapies to prolong survival time, the prognosis of patients with GBM is still poor, with a median overall survival at approximately 15 months [86]. Hence, elucidating the underlying mechanisms of glioma progression is warrant to screen more proper biomarkers and effective therapeutic measures for glioma.
Numerous circRNAs, along with its binding proteins, have been widely detected at aberrant expressions in normal tissue of central nervous system, including glioblastoma, emphasizing their potential roles as promising prognostic, diagnostic, and therapeutic molecules.
It was observed that circSMARCA5 expressed at low expression in glioblastoma multiforme GBM biopsies, and was negatively correlated with glioma's histological grade in affected subjects. Furthermore, circSMARCA5 could not only suppress the migration of glioblastoma cells, but also involve in VEGFA mRNA splicing and angiogenesis in glioblastoma multiforme [87, 88]. To further explore the underlying molecular mechanism, a bioinformatics analysis revealed that circSMARCA5 harbored abundant binding sites for RNA binding proteins, strongly suggesting that the circRNA-RBP interaction was essential for performing the action of circSMARCA5. Among the RBPs, erine and arginine rich splicing factor 1 (SRSF1), a splicing factor, was predicted to bind circSMARCA5 and then disrupt splicing within GBM cells, participating in GBM pathogenesis. SRSF1 is an upregulated protein in several cancer and exhibits many biological function depending on its various downstream target genes of splicing pattern. On the one hand, overexpressed SRSF1 was able to promote the expression of PTBP1 and SRSF3, which were likely the positive regulators of GBM cells migration [87, 89]. On the another hand, SRSF1 had an influence on VEGFA-mediated angiogenesis through affecting alternative splicing of VEGFA pre-mRNA [88]. Therefore, further studies revealed that circSMARCA5 exerted its function in GBM by specially binding to SRSF1, makes it possible that circSMARCA5 works as GBM biomarker into clinical practice [87].
As one of the potential circRNA in glioma, CDR1as expression is greatly decreased in glioma as comparing to adjacent normal brain, which is positively correlation with patients outcomes. A latest article was firstly reported that CDR1as exerted its suppression effect on tumorigenesis by binding firmly to tumor suppressor p53 protein at core DNA-binding domain, rather than sponging miRNA, where is exactly essential for MDM2 interaction. Consequently, binding tightly to p53, CDR1as acted as a protein defender to protect p53 from ubiquitination and degradation in MDM2-depent manner, eventually repressing tumorigenesis of glioma [90].
Clinical implication of the circRNAs-RBPs interactions
Biomarker
In line with different circRNA-RBP interactions in different types of tumors, we find that the abnormally expressed circRNAs or RBPs in some tumors are closely related to the patients prognosis respectively, highlighting their potential for tumor biomarkers.
circRNA is a kind of promising candidates for predicting living status of tumor patients. In a study published in 2017, a cohort of 116 HCC patients with survival data and corresponding circRNA-MTO1 expression were collectively analyzed by Kaplan-Meier survival curve, revealing the positive correlation between circRNA-MTO1 expression and prognosis of HCC patients and thus suggesting its potential as a prognosis biomarker [91]. Microarray analysis of HCC tumor tissues of 112 patients in another study indicated that downregulated circZKSCAN1 was associated with various HCC clinic pathologic features. Based on Kaplan-Meier survival analysis, HCC patients with high circZKSCAN1 expression level were likely to own a better overall and recurrence-free survival. It was demonstrated that circZKSCAN1 expression was an independent factor relating with overall survival and relapse-free survival rate of HCC patients, supported by univariate and multivariate analysis [57]. In addition to the stability of the looping structure of circRNAs, its widespread distribution in body fluids makes it more possible to be used as a simple clinical indicator.
The other protagonist in our article, RBPs, have clinical significances as well, depending on differential expression of RBPs in tumor and normal tissues. One of the representatives for RBPs is YBX1, which has been recognized as a marker for tumor aggressiveness and poor prognosis in various cancers such as breast, ovary, and liver cancer [92-94]. The researchers indicated ectopic YBX1 expression associated with shorter DFS and verified its role as a biomarker, shown as analysis of 94 patients who underwent surgery in advanced GC [95]. More recently, deregulated ALKBH5 may be capable of predicting an unfavorable clinical outcome in NSLC, as patients with low ALKBH5 expression tended to survive shorter than those with low ALKBH5 expression [96].
However, in light of the detection of circRNA in fluid, it seems more applicable to clinical using than RBPs. In fact, more datas and experimental proof are needed to support their role of biomarker before being put into clinical setting.
Therapeutic strategy
We can propose an effective therapeutic envision for cancer treatment through distinctively targeting circRNA-RBP axes we summarized in several tumors above.
When it comes to circRNAs, it can be classified as tumor suppressors and oncogenes. Some deregulated circRNAs in tumors are considered as tumor suppressors. Therefore, it is a promising targeted therapy that artificially synthesizing tumor suppressor circRNAs. Previously, the efficiency of synthetic circRNAs was proved to be significant for gastric and esophageal cancers therapy [97, 98], thus similar thoughts may also be broadly applicable to other type of cancers. For another, oncogenic circRNAs are overexpressed in tumors, inhibiting its expression levels may work as another therapeutic strategy. Recently, shRNA-based knockdown of FECR1 was confirmed to diminish recruitment for TET1 demethylase and inactivate transcription of FLI1, resulting in repression of breast tumor metastasis [46]. Similar to significant circRNAs, some circRNA-binding proteins can become a therapeutic target by regulating the expression or activity of RBPs or modulating connection of the circRNA-RBP comlex. For example, MS-444 is an inhibitor of the oncogenic RNA-binding protein HuR, leading to tumor cell apoptosis in malignant glioma via affecting HuR ability in the RNA binding and trafficking [99]. Besides, CMLD-2, another HuR inhibitor, was found to reduce cell viability and promote apoptosis in thyroid cancer by obstructing interaction between HuR and mRNA targets [100]. Above results suggest that potential HuR-targeted therapy for cancers may be put into clinical practice. Additionally, TET1, which belongs to ten eleven translocation (Tet) family dioxygenases, was shown to be essential for DNA demethylation and enhanced target FLI1 gene transcription in BC [46]. Interestingly, it was previously reported that Vitamin C served as a cofactor and increased TET activity in HCC cells through directly binding to the catalytic domain of TET proteins [101]. Notably, the circRNAs -RBPs interaction, along with their downstream regulatory networks, is likely to be specific targets for treatment of cancers, providing us with more ideas for developing novel anti-tumor therapies. However, whether these findings exert a meaningful role in clinical setting still need further investigation.
Conclusions and perspectives
Over the years, the number of studies on the function of circRNAs, as shown in cancer cell lines and models, dramatically increases to the peak, most of which outlines the role of circRNAs as miRNA sponge, proteins sponge, gene transcriptional regulator, a template for proteins coding. Among them, the ability of circRNAs to bind to proteins gained a growing attention, which was predicted by valid bioinformatics algorithms and confirmed by RIP, RNA pulldown and other binding assays. As previous studies suggested, circRNAs-RBPs axes made difference to tumorigenesis and aggressiveness of tumors. However, the understanding of circRNAs-RBPs interaction in diverse tumor types have not been fully disclosed. Thus, we not only intended to expound biogenesis and properties of circRNAs, elaborate the reciprocity of circRNAs and RBPs, but also summarize manifold circRNAs-RBPs relationships in several common tumors, such as breast cancer, hepatocellular carcinoma, gastric cancer, lung cancer and glioma.
A number of studies have revealed the relationship between circRNAs and RNA-related proteins, including RBP. In brief, a few proteins were involved in circRNAs biogenesis and inversely some circRNAs were associated with expression of proteins. On the other hand, a number of RNA-related proteins were competent in influencing the regulation of target genes expression medicated by circRNAs, directly or indirectly. Moreover, it was discovered that many proteins, like PES1 and METTL3/14, were capable of promoting specific circRNAs translated to proteins. Besides, circRNA-RBPs interactions also vary under different pathophysiological conditions. Although uncountable circRNAs in human tissues or cells have been recognized, there are many unsolved puzzles about more detailed connections between circRNAs and RBPs, possibly due to the lack of detection methods for their relationships in living cells. Surprisingly, it is understood that many emerging technologies for RNA-proteins interactions, such as a psoralen probe (PP)-based method [102], have been developed and expected to be extended to this field to solve this problem, aiming to make great progress in the field of circRNAs.
Here, we firstly roundly discussed the roles of circRNAs-RBPs connection in cancer, enumerating studies that might deepen our comprehension of how it modulates cancer malignant behaviors. In several universal tumors we mentioned, circRNAs were able to interact with relevant proteins by acting as protein scaffold, protein recruiter, protein translocation, protein decoy and protein defender, exerting a vital role in tumor cell proliferation, apoptosis, autophagy, migration, invasion, metastasis. Without a doubt, the pathways we reviewed underlined clinical application of circRNAs and RBPs as the prognosis biomarkers for cancer patients and provided a wealth of targets for oncotherapy, including circRNAs-RBPs complex and downstream regulators. M.Puttaraju and his colleague have shown the intron-exon array (PIE) method, which could be used to generate circRNA drugs [103]. This synthetic circRNA inhibitor may be an important member of future targeted therapies for cancer. In addition, our results attempted to lay a firm foundation for researchers to explore more precise regulatory networks in tumors.
It is worth noting that one of the hot spots in the field of circRNA today is the discovery of circRNA enrichment in various body fluids, including plasma [104], serum [105] and saliva [106], as well as exosomes [107]. This facilitates the detection of circRNAs and thus makes them strong candidates for biomarkers in early diagnosis of cancer. Additionally, Xinyi Wang et.al found that exosomes were involved in intercellular signal delivery and might provide a promising therapeutic target for cancer [108]. But nowadays more researches on exosomes continue to deepen.
In conclusion, some circRNA-RBPs interactions widely exist in a variety of cells, not just in specific cells, but others only under certain pathophysiological situations. Therefore, wider and more comprehensive researches are urged to perform to understand this relationship, which is expected to become a novel research hotspot with clinical promise to benefit tumor patients.
Abbreviations
AUF1: AU-rich RNA-binding protein; BC: breast cancer; CCAR1: protein-cell cycle and apoptosis regulator 1; ChIP: chromatin immunoprecipitation; circRNAs: circular RNAs; ciRNAs: intronic circRNAs; CNBP: CCHC-type zinc finger nucleic acid binding protein; CNS: central nervous system; DHX9: DExH-box helicase 9; EcircRNAs: exonic circRNAs; EIciRNAs: exon-intron circRNAs; EMSA: electrophoretic mobility shift assay; FISH: fluorescence in situ hybridization; GBM: glioblastoma multiforme; GC: gastric cancer; GPCR: G protein-coupled receptor; HCC: hepatocellular carcinoma; hnRNPs: heterogeneous nuclear ribonucleoprotein; HuR: human antigen R; Itgb1: integrin b1; LC: lung cancer; MBL: muscleblind; MDM2: murine double minute 2; miRNAs: microRNAs; NF90/NF110: double-stranded RNA-binding domain containing immune factors; NSCLC: non-small-cell lung cancer; pre-mRNA: precursor-mRNA; QKI : Quaking; RBM3: RNA-binding protein 3; RBP: RNA binding protein; RIP: RNA immunoprecipitation; RNA-seq: RNA sequencing; SCLC: small-cell lung cancer; SF: sorafenib; snRNA: small nuclear RNA; snRNP: mall nuclear ribonucleoprotein; SR proteins: serine-arginine proteins; SRSF1: erine and arginine rich splicing factor 1; Tip60: 60 kDa Tat-interactive protein; TRAF4: tumor necrosis factor receptor associated factor 4; YBX1: Y-box binding protein-1.
Acknowledgements
This review was supported by the National Key Research and Development Program of China (No. 2016YFC0905900), National Natural Science Foundation of China (No. 81872365) and Jiangsu Provincial Key Research Development Program (No. BE2019731).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73:3852-6
2. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733
3. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-8
4. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK. et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-8
5. Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B. et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215
6. Peng L, Chen G, Zhu Z, Shen Z, Du C, Zang R. et al. Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung's disease. Oncotarget. 2017;8:808-18
7. Zang J, Lu D, Xu A. The interaction of circRNAs and RNA binding proteins: An important part of circRNA maintenance and function. J Neurosci Res. 2020;98:87-97
8. Gebauer F, Schwarzl T, Valcarcel J, Hentze MW. RNA-binding proteins in human genetic disease. Nature reviews Genetics. 2021;22:185-98
9. Zhou H, Mangelsdorf M, Liu J, Zhu L, Wu JY. RNA-binding proteins in neurological diseases. Sci China Life Sci. 2014;57:432-44
10. Huang A, Zheng H, Wu Z, Chen M, Huang Y. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics. 2020;10:3503-17
11. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141-57
12. Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829-42
13. Cui X, Wang J, Guo Z, Li M, Li M, Liu S. et al. Emerging function and potential diagnostic value of circular RNAs in cancer. Mol Cancer. 2018;17:123
14. Wilusz JE. Repetitive elements regulate circular RNA biogenesis. Mob Genet Elements. 2015;5:1-7
15. Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife. 2015;4:e07540
16. Kramer MC, Liang D, Tatomer DC, Gold B, March ZM, Cherry S. et al. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015;29:2168-82
17. Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic acids research. 2006;34:e63
18. Li HM, Ma XL, Li HG. Intriguing circles: Conflicts and controversies in circular RNA research. Wiley Interdiscip Rev RNA. 2019;10:e1538
19. Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S. et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell. 2015;58:870-85
20. Dang Y, Yan L, Hu B, Fan X, Ren Y, Li R. et al. Tracing the expression of circular RNAs in human pre-implantation embryos. Genome Biol. 2016;17:130
21. Xia S, Feng J, Lei L, Hu J, Xia L, Wang J. et al. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinform. 2017;18:984-92
22. Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN. et al. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9:e90859
23. Hall MP, Nagel RJ, Fagg WS, Shiue L, Cline MS, Perriman RJ. et al. Quaking and PTB control overlapping splicing regulatory networks during muscle cell differentiation. RNA. 2013;19:627-38
24. Wu JI, Reed RB, Grabowski PJ, Artzt K. Function of quaking in myelination: regulation of alternative splicing. Proc Natl Acad Sci U S A. 2002;99:4233-8
25. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA. et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125-34
26. Xiao Y, Liu G, Sun Y, Gao Y, Ouyang X, Chang C. et al. Targeting the estrogen receptor alpha (ERalpha)-mediated circ-SMG1.72/miR-141-3p/Gelsolin signaling to better suppress the HCC cell invasion. Oncogene. 2020;39:2493-508
27. Aktas T, Avsar Ilik I, Maticzka D, Bhardwaj V, Pessoa Rodrigues C, Mittler G. et al. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017;544:115-9
28. Dong W, Dai ZH, Liu FC, Guo XG, Ge CM, Ding J. et al. The RNA-binding protein RBM3 promotes cell proliferation in hepatocellular carcinoma by regulating circular RNA SCD-circRNA 2 production. EBioMedicine. 2019;45:155-67
29. Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF. et al. Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Mol Cell. 2017;67:214-27.e7
30. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M. et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55-66
31. Chen S, Thorne RF, Zhang XD, Wu M, Liu L. Non-coding RNAs, guardians of the p53 galaxy. Semin Cancer Biol. 2020
32. Du WW, Yang W, Li X, Awan FM, Yang Z, Fang L. et al. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene. 2018;37:5829-42
33. Fang L, Du WW, Lyu J, Dong J, Zhang C, Yang W. et al. Enhanced breast cancer progression by mutant p53 is inhibited by the circular RNA circ-Ccnb1. Cell Death Differ. 2018;25:2195-208
34. Chaudhary R, Muys BR, Grammatikakis I, De S, Abdelmohsen K, Li XL. et al. A Circular RNA from the MDM2 Locus Controls Cell Cycle Progression by Suppressing p53 Levels. Molecular and cellular biology. 2020;40:e00473-19
35. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X. et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256-64
36. Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S. et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361-9
37. Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560-4
38. Sun S, Wang W, Luo X, Li Y, Liu B, Li X. et al. Circular RNA circ-ADD3 inhibits hepatocellular carcinoma metastasis through facilitating EZH2 degradation via CDK1-mediated ubiquitination. Am J Cancer Res. 2019;9:1695-707
39. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
40. Malmgren JA, Mayer M, Atwood MK, Kaplan HG. Differential presentation and survival of de novo and recurrent metastatic breast cancer over time: 1990-2010. Breast Cancer Res Treat. 2018;167:579-90
41. Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296-9
42. Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z. et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357-70
43. Du WW, Yang W, Li X, Fang L, Wu N, Li F. et al. The Circular RNA circSKA3 Binds Integrin beta1 to Induce Invadopodium Formation Enhancing Breast Cancer Invasion. Mol Ther. 2020;28:1287-98
44. Mueller SC, Chen WT. Cellular invasion into matrix beads: localization of beta 1 integrins and fibronectin to the invadopodia. J Cell Sci. 1991;99( Pt 2):213-25
45. Burger KL, Learman BS, Boucherle AK, Sirintrapun SJ, Isom S, Diaz B. et al. Src-dependent Tks5 phosphorylation regulates invadopodia-associated invasion in prostate cancer cells. Prostate. 2014;74:134-48
46. Chen N, Zhao G, Yan X, Lv Z, Yin H, Zhang S. et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018;19:218
47. Zhang L, Zhou F, García de Vinuesa A, de Kruijf EM, Mesker WE, Hui L. et al. TRAF4 promotes TGF-β receptor signaling and drives breast cancer metastasis. Mol Cell. 2013;51:559-72
48. Zhang X, Wen Z, Sun L, Wang J, Song M, Wang E. et al. TRAF2 regulates the cytoplasmic/nuclear distribution of TRAF4 and its biological function in breast cancer cells. Biochem Biophys Res Commun. 2013;436:344-8
49. Liu Y, Dong Y, Zhao L, Su L, Luo J. Circular RNAMTO1 suppresses breast cancer cell viability and reverses monastrol resistance through regulating the TRAF4/Eg5 axis. Int J Oncol. 2018;53:1752-62
50. Bassi C, Li YT, Khu K, Mateo F, Baniasadi PS, Elia A. et al. The acetyltransferase Tip60 contributes to mammary tumorigenesis by modulating DNA repair. Cell Death Differ. 2016;23:1198-208
51. Cregan S, McDonagh L, Gao Y, Barr MP, O'Byrne KJ, Finn SP. et al. KAT5 (Tip60) is a potential therapeutic target in malignant pleural mesothelioma. Int J Oncol. 2016;48:1290-6
52. Shiota M, Yokomizo A, Masubuchi D, Tada Y, Inokuchi J, Eto M. et al. Tip60 promotes prostate cancer cell proliferation by translocation of androgen receptor into the nucleus. Prostate. 2010;70:540-54
53. Xu P, Li C, Chen Z, Jiang S, Fan S, Wang J. et al. The NuA4 Core Complex Acetylates Nucleosomal Histone H4 through a Double Recognition Mechanism. Mol Cell. 2016;63:965-75
54. Wang L, Long H, Zheng Q, Bo X, Xiao X, Li B. Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol Cancer. 2019;18:119
55. Alpatov R, Lesch BJ, Nakamoto-Kinoshita M, Blanco A, Chen S, Stützer A. et al. A chromatin-dependent role of the fragile X mental retardation protein FMRP in the DNA damage response. Cell. 2014;157:869-81
56. Li Y, Tang Y, Ye L, Liu B, Liu K, Chen J. et al. Establishment of a hepatocellular carcinoma cell line with unique metastatic characteristics through in vivo selection and screening for metastasis-related genes through cDNA microarray. J Cancer Res Clin Oncol. 2003;129:43-51
57. Zhu YJ, Zheng B, Luo GJ, Ma XK, Lu XY, Lin XM. et al. Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma. Theranostics. 2019;9:3526-40
58. Guo J, Wu Y, Du J, Yang L, Chen W, Gong K. et al. Deregulation of UBE2C-mediated autophagy repression aggravates NSCLC progression. Oncogenesis. 2018;7:49
59. Zhu H, Gan X, Jiang X, Diao S, Wu H, Hu J. ALKBH5 inhibited autophagy of epithelial ovarian cancer through miR-7 and BCL-2. J Exp Clin Cancer Res. 2019;38:163
60. Liu Z, Wang Q, Wang X, Xu Z, Wei X, Li J. Circular RNA cIARS regulates ferroptosis in HCC cells through interacting with RNA binding protein ALKBH5. Cell Death Discov. 2020;6:72
61. Kullmann M, Gopfert U, Siewe B, Hengst L. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5'UTR. Genes Dev. 2002;16:3087-99
62. Bencivenga D, Caldarelli I, Stampone E, Mancini FP, Balestrieri ML, Della Ragione F. et al. p27(Kip1) and human cancers: A reappraisal of a still enigmatic protein. Cancer Lett. 2017;403:354-65
63. Liu B, Yang G, Wang X, Liu J, Lu Z, Wang Q. et al. CircBACH1 (hsa_circ_0061395) promotes hepatocellular carcinoma growth by regulating p27 repression via HuR. J Cell Physiol. 2020;235:6929-41
64. Sun S, Gao J, Zhou S, Li Y, Wang Y, Jin L. et al. A novel circular RNA circ-LRIG3 facilitates the malignant progression of hepatocellular carcinoma by modulating the EZH2/STAT3 signaling. J Exp Clin Cancer Res. 2020;39:252
65. Chen W, Sun K, Zheng R, Zeng H, Zhang S, Xia C. et al. Cancer incidence and mortality in China, 2014. Chin J Cancer Res. 2018;30:1-12
66. Chen Y, Yang F, Fang E, Xiao W, Mei H, Li H. et al. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019;26:1346-64
67. Zhang Z, Dou M, Yao X, Tang H, Li Z, Zhao X. Potential Biomarkers in Diagnosis of Human Gastric Cancer. Cancer Invest. 2016;34:115-22
68. Guo TT, Yu YN, Yip GW, Matsumoto K, Bay BH. Silencing the YB-1 gene inhibits cell migration in gastric cancer in vitro. Anat Rec (Hoboken). 2013;296:891-8
69. Fang J, Hong H, Xue X, Zhu X, Jiang L, Qin M. et al. A novel circular RNA, circFAT1(e2), inhibits gastric cancer progression by targeting miR-548g in the cytoplasm and interacting with YBX1 in the nucleus. Cancer Lett. 2019;442:222-32
70. Evans-Anderson HJ, Alfieri CM, Yutzey KE. Regulation of cardiomyocyte proliferation and myocardial growth during development by FOXO transcription factors. Circ Res. 2008;102:686-94
71. Shiota M, Song Y, Yokomizo A, Kiyoshima K, Tada Y, Uchino H. et al. Foxo3a suppression of urothelial cancer invasiveness through Twist1, Y-box-binding protein 1, and E-cadherin regulation. Clin Cancer Res. 2010;16:5654-63
72. Jie M, Wu Y, Gao M, Li X, Liu C, Ouyang Q. et al. CircMRPS35 suppresses gastric cancer progression via recruiting KAT7 to govern histone modification. Mol Cancer. 2020;19:56
73. Lee E, Lee TA, Yoo HJ, Lee S, Park B. CNBP controls tumor cell biology by regulating tumor-promoting gene expression. Mol Carcinog. 2019;58:1492-501
74. D'Amico D, Antonucci L, Di Magno L, Coni S, Sdruscia G, Macone A. et al. Non-canonical Hedgehog/AMPK-Mediated Control of Polyamine Metabolism Supports Neuronal and Medulloblastoma Cell Growth. Dev Cell. 2015;35:21-35
75. Yang F, Hu A, Li D, Wang J, Guo Y, Liu Y. et al. Circ-HuR suppresses HuR expression and gastric cancer progression by inhibiting CNBP transactivation. Mol Cancer. 2019;18:158
76. Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Nikšić M. et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. The Lancet. 2018;391:1023-75
77. Zhou M, Wang H, Zeng X, Yin P, Zhu J, Chen W. et al. Mortality, morbidity, and risk factors in China and its provinces, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. The Lancet. 2019;394:1145-58
78. Sharma P, Allison James P. Immune Checkpoint Targeting in Cancer Therapy: Toward Combination Strategies with Curative Potential. Cell. 2015;161:205-14
79. Nan A, Chen L, Zhang N, Jia Y, Li X, Zhou H. et al. Circular RNA circNOL10 Inhibits Lung Cancer Development by Promoting SCLM1-Mediated Transcriptional Regulation of the Humanin Polypeptide Family. Adv Sci (Weinh). 2019;6:1800654
80. Wang J, Jia Y, Zhao S, Zhang X, Wang X, Han X. et al. BIN1 reverses PD-L1-mediated immune escape by inactivating the c-MYC and EGFR/MAPK signaling pathways in non-small cell lung cancer. Oncogene. 2017;36:6235-43
81. Liang Wang, Chao Zheng, Xiaoying Wu, Yuefeng Zhang, Sunshun Yan, Luya Ruan. et al. Circ-SOX4 promotes non-small cell lung cancer progression by activating the Wnt/beta-catenin pathway. Mol Oncol. 2020;14:3253
82. Shibata M, Ham K, Hoque MO. A time for YAP1: Tumorigenesis, immunosuppression and targeted therapy. Int J Cancer. 2018;143:2133-44
83. Zhao L, Guan H, Song C, Wang Y, Liu C, Cai C. et al. YAP1 is essential for osteoclastogenesis through a TEADs-dependent mechanism. Bone. 2018;110:177-86
84. Shen Y, Li C, Zhou L, Huang JA. G protein-coupled oestrogen receptor promotes cell growth of non-small cell lung cancer cells via YAP1/QKI/circNOTCH1/m6A methylated NOTCH1 signalling. Journal of cellular and molecular medicine. 2020;25:284-96
85. Ostrom QT, Cioffi G, Gittleman H, Patil N, Waite K, Kruchko C. et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2012-2016. Neuro Oncol. 2019;21:v1-v100
86. Khasraw M, Lassman AB. Advances in the treatment of malignant gliomas. Curr Oncol Rep. 2010;12:26-33
87. Barbagallo D, Caponnetto A, Cirnigliaro M, Brex D, Barbagallo C, D'Angeli F. et al. CircSMARCA5 Inhibits Migration of Glioblastoma Multiforme Cells by Regulating a Molecular Axis Involving Splicing Factors SRSF1/SRSF3/PTB. Int J Mol Sci. 2018;19:480
88. Barbagallo D, Caponnetto A, Brex D, Mirabella F, Barbagallo C, Lauretta G. et al. CircSMARCA5 Regulates VEGFA mRNA Splicing and Angiogenesis in Glioblastoma Multiforme Through the Binding of SRSF1. Cancers (Basel). 2019;11:194
89. Cheung HC, Hai T, Zhu W, Baggerly KA, Tsavachidis S, Krahe R. et al. Splicing factors PTBP1 and PTBP2 promote proliferation and migration of glioma cell lines. Brain. 2009;132:2277-88
90. Lou J, Hao Y, Lin K, Lyu Y, Chen M, Wang H. et al. Circular RNA CDR1as disrupts the p53/MDM2 complex to inhibit Gliomagenesis. Mol Cancer. 2020;19:138
91. Han D, Li J, Wang H, Su X, Hou J, Gu Y. et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151-64
92. Janz M, Harbeck N, Dettmar P, Berger U, Schmidt A, Jurchott K. et al. Y-box factor YB-1 predicts drug resistance and patient outcome in breast cancer independent of clinically relevant tumor biologic factors HER2, uPA and PAI-1. Int J Cancer. 2002;97:278-82
93. Yasen M, Kajino K, Kano S, Tobita H, Yamamoto J, Uchiumi T. et al. The up-regulation of Y-box binding proteins (DNA binding protein A and Y-box binding protein-1) as prognostic markers of hepatocellular carcinoma. Clin Cancer Res. 2005;11:7354-61
94. Kamura T, Yahata H, Amada S, Ogawa S, Sonoda T, Kobayashi H. et al. Is nuclear expression of Y box-binding protein-1 a new prognostic factor in ovarian serous adenocarcinoma? Cancer. 1999;85:2450-4
95. Wu Y, Yamada S, Izumi H, Li Z, Shimajiri S, Wang KY. et al. Strong YB-1 expression is associated with liver metastasis progression and predicts shorter disease-free survival in advanced gastric cancer. J Surg Oncol. 2012;105:724-30
96. Jin D, Guo J, Wu Y, Yang L, Wang X, Du J. et al. m(6)A demethylase ALKBH5 inhibits tumor growth and metastasis by reducing YTHDFs-mediated YAP expression and inhibiting miR-107/LATS2-mediated YAP activity in NSCLC. Mol Cancer. 2020;19:40
97. Liu X, Abraham JM, Cheng Y, Wang Z, Wang Z, Zhang G. et al. Synthetic Circular RNA Functions as a miR-21 Sponge to Suppress Gastric Carcinoma Cell Proliferation. Mol Ther Nucleic Acids. 2018;13:312-21
98. Wang Z, Ma K, Cheng Y, Abraham JM, Liu X, Ke X. et al. Synthetic circular multi-miR sponge simultaneously inhibits miR-21 and miR-93 in esophageal carcinoma. Lab Invest. 2019;99:1442-53
99. Wang J, Hjelmeland AB, Nabors LB, King PH. Anti-cancer effects of the HuR inhibitor, MS-444, in malignant glioma cells. Cancer Biol Ther. 2019;20:979-88
100. Allegri L, Baldan F, Roy S, Aubé J, Russo D, Filetti S. et al. The HuR CMLD-2 inhibitor exhibits antitumor effects via MAD2 downregulation in thyroid cancer cells. Sci Rep. 2019;9:7374
101. Yin R, Mao SQ, Zhao B, Chong Z, Yang Y, Zhao C. et al. Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J Am Chem Soc. 2013;135:10396-403
102. Zhang Z, Liu T, Dong H, Li J, Sun H, Qian X. et al. An RNA tagging approach for system-wide RNA-binding proteome profiling and dynamics investigation upon transcription inhibition. Nucleic acids research. 2021 [Epub ahead of print]
103. Puttaraju M, Been MD. Group I permuted intron-exon (PIE) sequences self-splice to produce circular exons. Nucleic acids research. 1992;20:5357-64
104. Ouyang Q, Huang Q, Jiang Z, Zhao J, Shi GP, Yang M. Using plasma circRNA_002453 as a novel biomarker in the diagnosis of lupus nephritis. Mol Immunol. 2018;101:531-8
105. Wu J, Zhou Q, Niu Y, Chen J, Zhu Y, Ye S. et al. Aberrant expression of serum circANRIL and hsa_circ_0123996 in children with Kawasaki disease. J Clin Lab Anal. 2019;33:e22874
106. Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y. et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61:221-30
107. Wang Y, Zhao R, Liu W, Wang Z, Rong J, Long X. et al. Exosomal circHIPK3 Released from Hypoxia-Pretreated Cardiomyocytes Regulates Oxidative Damage in Cardiac Microvascular Endothelial Cells via the miR-29a/IGF-1 Pathway. Oxid Med Cell Longev. 2019;2019:7954657
108. Wang X, Zhang H, Yang H, Bai M, Ning T, Deng T. et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol Oncol. 2020;14:539-55
Author contact
![]() Corresponding author: Jin-hai Tang, MD, Ph.D, Department of General Surgery, the First Affiliated Hospital of Nanjing Medical University, Guangzhou Road 300, Nanjing, Jiangsu, 210029, China. E-mail: jhtangedu.cn.
Corresponding author: Jin-hai Tang, MD, Ph.D, Department of General Surgery, the First Affiliated Hospital of Nanjing Medical University, Guangzhou Road 300, Nanjing, Jiangsu, 210029, China. E-mail: jhtangedu.cn.

 Global reach, higher impact
Global reach, higher impact