3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(18):5506-5518. doi:10.7150/jca.51524 This issue Cite
Research Paper
Diagnosis and prognostic value of C-X-C motif chemokine ligand 1 in colon adenocarcinoma based on The Cancer Genome Atlas and Guangxi cohort
1. Department of Colorectal and Anal Surgery, The First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi Zhuang Autonomous Region, People's Republic of China.
2. Department of Immunology, School of Preclinical Medicine, Guangxi Medical University, Nanning, Guangxi Zhuang Autonomous Region, People's Republic of China.
3. Department of Hepatobiliary Surgery, The First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi Zhuang Autonomous Region, People's Republic of China.
*Equal contributors
Received 2020-8-4; Accepted 2021-3-6; Published 2021-7-25
Abstract
Objective: The objective was to identify and validate C-X-C motif chemokine ligand 1(CXCL1) for diagnosis and prognosis in colon adenocarcinoma (COAD).
Methods: Our current study had enrolled one The Cancer Genome Atlas (TCGA) cohort and two Guangxi cohorts to identify and verify the diagnostic and prognostic values of CXCL1 in COAD. Functional enrichment was performed by gene set enrichment analysis (GSEA).
Results: In TCGA cohort, the expression of CXCL1 was significantly up-regulated in tumor tissues and decreased as the tumor stage developed. The receiver operating characteristic (ROC) curve showed that CXCL1 had a high diagnostic value for COAD. The result of Kaplan-Meier survival analysis showed that CXCL1 gene expression (P=0.045) was significantly correlated with overall survival (OS) of COAD. Results of Guangxi cohort also verified the diagnostic value of CXCL1 in COAD, and sub-group survival analyses also suggested that patients with high CXCL1 expression were related to a favorable OS (Corrected P=0.005). GSEA revealed that CXCL1 high expression phenotype was related to cytokine activity, cell apoptosis, P53 regulation pathway, and regulation of autophagy in COAD.
Conclusions: In this study, we found that CXCL1 gene might be a potential diagnostic biomarker for COAD, and might serve as a prognostic biomarker for specific subgroup of COAD.
Keywords: CXCL1, COAD, Diagnosis, Prognosis, Biomarker, GSEA
Introduction
Colorectal cancer (CRC) ranks as one of those diseases of the highest morbidity and mortality in the world [1]. The early treatment of CRC had a good prognosis, and the survival rate of patients with early cancer was about five times higher than that of patients with advanced cancer [2]. Colonoscopy remained the gold standard for CRC diagnosis, but the procedure was invasive, expensive, and had low patients' acceptance. Serum carcinoembryonic antigen (CEA) is a tumor marker and is more meaningful for CRC diagnosis and postoperative monitoring. However, serum CEA positive rate in CRC patients was less than 50% in some clinical trials [3-5]. Therefore, it was necessary to identify better biomarker to improve the effectiveness of early diagnosis of CRC and prognosis prediction of the patients.
CXC motif chemokine ligand 1 (CXCL1), also known as the GRO1 oncogene, is a small cytokine of the CXC chemokine family [6]. It is expressed by macrophages, neutrophils, and epithelial cells and has neutrophil chemoattractant activity [6-8]. CXCL1 was taken part in the processes of angiogenesis, arteriogenesis, inflammation, wound healing, and tumorigenesis [9, 10]. This chemokine triggered its above actions by signaling from the chemokine receptor CXCR2 [10]. Previous researches had discovered that CXCL1 was markedly upregulated in CRC cancer tissues [11, 12], and the overexpress of CXCL1 was connected to the poor prognosis of CRC stage III [13].
More than 60% of CRC occurred in the colon. Global cancer statistics showed 1,096,601 new colon cancer cases and 551,269 deaths in 2018, accounting for about 6% of all tumors [1]. The cause of colon cancer is not the same as that of rectal cancer [1, 14], so their pathogenesis might also be different. Majority of the pathological type of colon cancer is colon adenocarcinoma (COAD). Previous studies had not systematically reported the diagnostic and prognostic value of CXCL1 in COAD. In this study, we first explored the diagnostic and prognostic values of CXCL1 gene mRNA expression in COAD applying the Cancer Genome Atlas (TCGA) database, and then validated the TCGA results with the cohort of the First Affiliated Hospital of Guangxi Medical University.
Materials and Methods
Expression of CXCL1 in COAD and normal tissues
Human Protein Atlas (HPA: https://www.proteinatlas.org, accessed December 27, 2018) is used to reveal the distribution of CXCL1 in normal human tissues [15]. Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/detail.php?gene=cxcl1, accessed February 17, 2019) was used to explore the distribution of CXCL1 between COAD and normal colon tissues [16].
RNA sequencing data in TCGA
RNA sequencing dataset and patient parameters of COAD was got from TCGA (https://cancergenome.nih.gov, Accessed time: November 27, 2018) [17, 18]. We compared the expression of CXCL1 in tumor and paracancerous tissues of COAD patients to evaluate its diagnostic value, high- and low-expression CXCL1 phenotypes of COAD for survival analysis were grouped according to median value.
Real-time RT-PCR (RT-qPCR) of CXCL1 expressions in COAD
Patient tissue samples
From April to June 2018, we continuously collected tumor and paracancerous tissues from the surgery of the patients with COAD in the Department of Colorectal and Anal Surgery, the First Affiliated Hospital of Guangxi Medical University (Nanning, Guangxi). The patient's tissue was soaked in RNA store reagent immediately after surgery and subsequently frozen in a -80 ° C refrigerator. These patients were those who had no radiation or chemotherapy before surgery and their postoperative pathological diagnosis was COAD. In the Guangxi cohort, we only collected tissues from patients who had not received preoperative chemoradiotherapy and who were pathologically confirmed to have COAD after surgery. All patients in this study signed informed consent, and the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University approved the experimental protocol [Ethics no.:2019(KY-E-001)].
RNA extraction and RT-qPCR
First, we extracted the total RNA from tissues via the TRIzol reagent (15596026, Invitrogen). Then, we applied the PrimeScript™ RT Reagent Kit with gDNA Eraser (RR047A, Takara) to synthesize the total RNA into first-strand cDNA. After that, the expression of CXCL1 was normalized to GAPDH expression. At the same time, quantitative real-time PCR (qPCR) was conducted via the FastStart Universal SYBR Green Master (ROX) (Roche) in the Applied Biosystems Quantsudio TM Real-PCR System (Q6). All the above experiments were carried out according to the instructions. The relative gene expression level was performed according to 2‑ ∆∆ Ct [19, 20].
The primer sequences were as follows:
CXCL1-forward: 5'-CAAACCGAAGTCATAGCCACA-3'
CXCL1-reverse: 5'-CTCCTAAGCGATGCTCAAACA-3'
GAPDH-forward: 5'-GTCAGCCGCATCTTCTTT-3'
GAPDH-reverse: 5'-CGCCCAATACGACCAAAT-3'
Immunohistochemistry (IHC) of CXCL1 expressions in COAD
Patient tissue samples
We retrospectively collected tumor and paracancerous tissues wax blocks from patients who underwent colonic tumor resection in the First Affiliated Hospital of Guangxi Medical University from 2012 to 2013. The patients did not have any other known tumors. No radiotherapy or chemotherapy had been performed before surgery. The pathological diagnosis was COAD, and the tumors were identified and categorized according to the American Joint Committee on Cancer (AJCC) tumor node metastasis (TNM) staging system (8th edition, 2017) [21]. We routinely collected clinical parameters and survival dataset for these patients. Inclusion criteria for COAD patients were described above. All patients in this study signed informed consent, and the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University approved the experimental protocol [Ethics no.:2019(KY-E-001)].
Evaluation of IHC
We used the CXCL1 antibody supplied by Signalway Antibody LLC, and the immunohistochemical staining reagents from Shanghai ChangDao Biotech Company, China. IHC procedure carried out in accordance with the manufacturer's instructions. Two pathologists respectively evaluated the percentage of positive cells according to the following norm: 0 (0%); 1 (1-25%); 2 (26-50%); 3 (51-75%); and 4 (76- 100%). According to the intensity of staining, the staining results were divided into four levels: negative, weak, moderate and strong, and give four corresponding scores of 0, 1, 2, and 3, respectively. We multiply the percentage and staining intensity score to get the final IHC score. The results of these two independent pathologists were calculated and got the average score. When the scores were over two, the positive staining results were confirmed [22].
Gene set enrichment analysis (GSEA)
We divided the TCGA patients into two groups, as high one and low on, based on the expression of CXCL1. Then we applied GSEA (http://software.broadinstitute.org/gsea/index.jsp, accessed December 24, 2018) v3.0 to investigate the prognosis molecular mechanism of CXCL1 in patients with COAD by enriching metabolic pathways and biological processes [23]. The reference gene sets of GSEA were obtained from Molecular Signatures Database (MSIGDB): c5 (GO gene sets: bp. V6.2.symbols.gmt, cc.v6.2.symbols.gmt, and mf. v6.2.symbols.gmt) and c2 (KEGG gene sets: c2.cp.kegg.v6.2.symbols.gmt), respectively. Nominal P less than 0.05 and false discovery rate (FDR) less than 0.25 were considered statistically significant.
Statistical Analysis
We conducted a t-test to assess the CXCL1 expression between tumor and paracancerous tissues. Kaplan-Meier method was performed for survival analysis. We applied the Cox regression model to evaluate the hazard ratio (HR) and 95% confidence interval (CI). The FDR in GSEA was carried out according to the Benjamini-Hochberg procedure [24, 25]. The drawing of the figures were performed by GraphPad Prism 7.0. P<0.05 was regarded as statistically significant. SPSS v.24.0 software (IBM, Chicago, IL, USA) was used for statistics.
Results
Expression of CXCL1 in COAD and normal tissues
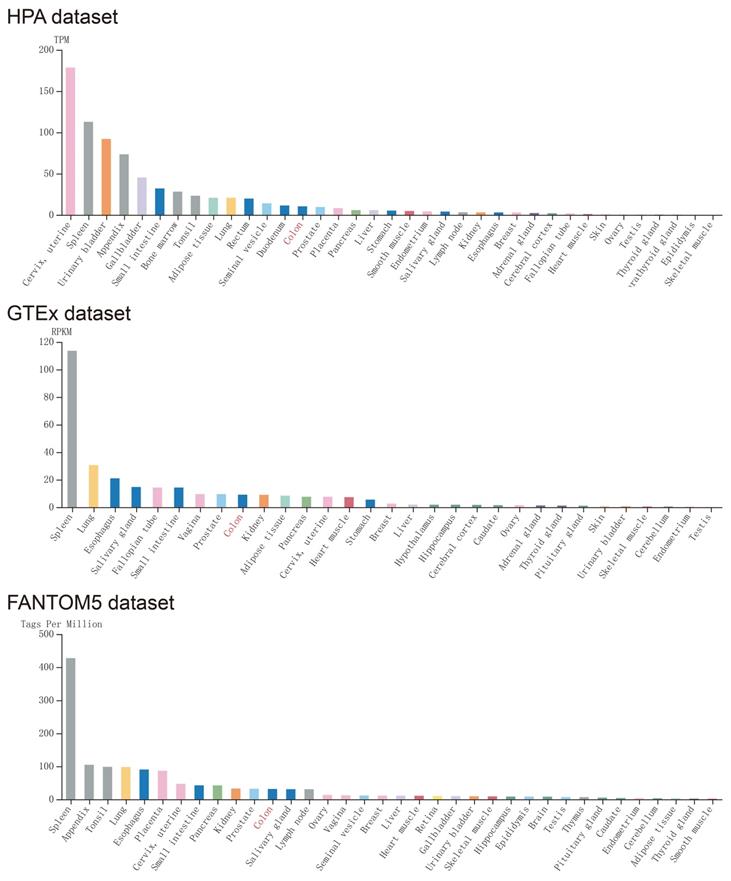
Expression of CXCL1 in normal human tissues was got from Human Protein Atlas, which was based on Functional Annotation of Mammalian Genomes 5 (FANTOM5), Genotype-Tissue Expression (GTEx), and HPA RNA-seq dataset (Fig. 1), CXCL1 gene was highly expressed in normal human colon tissue. The expression of CXCL1 gene in COAD tumor tissues was significantly higher than that in normal colon tissues (Fig. 2a).
COAD data analysis in TCGA database
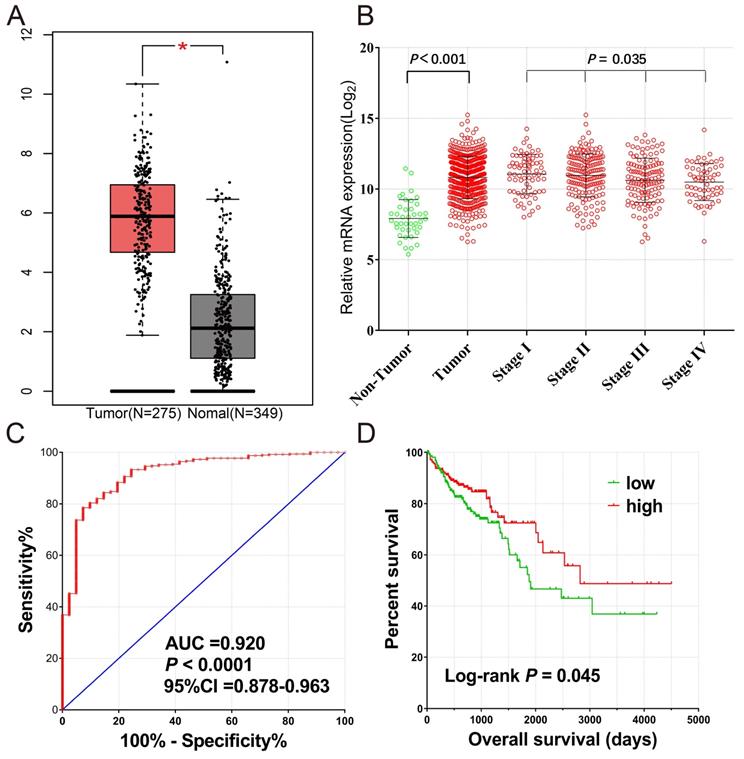
A total of 461 COAD patients were enrolled in the project. There were RNA sequencing data in 480 tumor and 41 paracancerous tissue samples from 456 patients. The expression of CXCL1 was markedly up-regulated in tumor tissues, and it decreased as the tumor stage developed (Fig. 2b). The ROC curve (Fig. 2c) showed that CXCL1 had a high accurately for COAD diagnosis [AUC(95% CI)=0.920(0.878-0.963)]. We excluded 5 patients without mRNA expression data, 2 patients without clinical data, 1 patient with postoperative survival time of “unknown”, and 15 patients with postoperative survival time of 0. Finally, 438 COAD patients with both survival data and genome-wide RNA sequencing data were included for survival analysis (Table 1). The results of the Kaplan-Meier survival analysis showed that the TNM stage (Log-rank P<0.0001) and CXCL1 gene expression (P=0.045) were significantly correlated with overall survival (OS) of COAD (Fig 2d). However, the results of multivariate analysis showed that the OS of the CXCL1 gene and COAD was not statistically significant in the correction of TNM staging of tumors (Corrected P=0.364, Corrected HR (95% CI) = 0.825 (0.544-1.250)).
The expression level of CXCL1 in normal human tissues. Notes: GTEx, Genotype-Tissue Expression; FANTOM5, the Functional Annotation of Mammalian Genomes 5.
Clinical factors sheet for COAD patients in TCGA cohort
| Variables | Patients (n=438) | No. of event (%) | MST (days) | HR (95%CI) | Log-rank P value |
|---|---|---|---|---|---|
| Sex | 0.545 | ||||
| Female | 204 | 44(21.6%) | NA | 1 | |
| Male | 234 | 54(23.1%) | 2475 | 1.131(0.759-1.686) | |
| Age& | 0.112 | ||||
| ≤65 | 168 | 29(17.3%) | NA | 1 | |
| >65 | 268 | 68(25.4%) | 2475 | 1.420(0.919-2.194) | |
| TNM stage† | <0.001 | ||||
| I | 73 | 4(5.5%) | NA | 1 | |
| II | 167 | 27(16.2%) | 2821 | 2.240(0.781-6.421) | |
| III | 126 | 31(24.6%) | NA | 4.068(1.434-11.538) | |
| IV | 61 | 31(50.8%) | 858 | 11.291(3.980-32.026) | |
| CXCL1 | 0.045 | ||||
| 219 | 58 | 1881 | 1 | ||
| 219 | 40 | 2821 | 0.665(0.444-0.995) |
Note: &Age information of 2 patients was not obtained; †Tumor TNM staging information was not obtained in 11 patients; MST: median survival time; HR: hazard ratio; 95% CI: 95% confidence interval; NA: Not available.
(A) The mRNA expression levels of CXCL1 gene comparison in normal colon tissue and primary colon tumors based on TCGA and GTEx database; (B) The Scatter plot of CXCL1 gene mRNA expression in tumor tissues and paracancerous non-tumor colon tissues of TCGA cohort, as well as those of different tumor stages; (C) The ROC curves of CXCL1 gene mRNA expression in tumor tissues and paracancerous non-tumor colon tissues of TCGA cohort; (D) Kaplan-Meier curves for CXCL1 gene in COAD of TCGA cohort.
The mRNA expression of CXCL1 in Guangxi COAD cohort
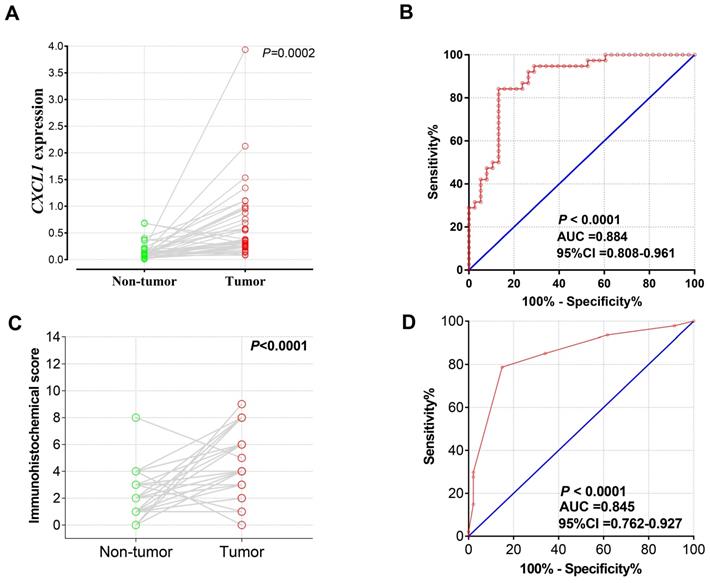
A total of 38 patients with COAD were recruited into current study, with a median age of 61 years (ranged 35 to 85 years), 25 men and 13 women. The result of the pair-t test showed that CXCL1 mRNA expression in COAD tumor tissues was markedly up-regulated than in paracancerous non-tumor colon tissues (Fig. 3a), and the diagnostic ROC curve (Fig. 3b) showed that CXCL1 had a high accurately for COAD diagnosis (P<0.0001, AUC (95% Cl)=0.884(0.808-0.961) ).
IHC expression of CXCL1 in Guangxi COAD cohort
Basic characteristics of the study population
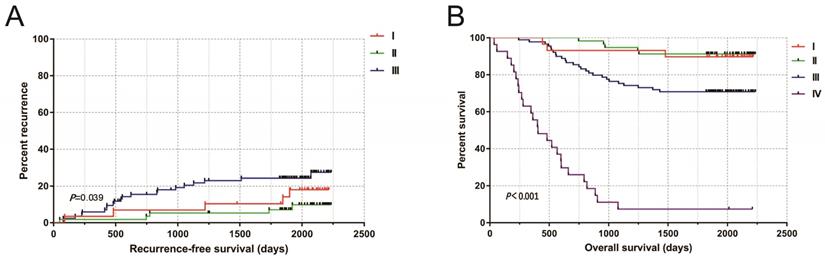
In this study, a total of 216 patients with COAD were retrospectively collected, 4 cases of tumor tissue wax mass could not be obtained, and 212 patients were included in the study (including 212 tumor tissues and 47 paracancerous non-tumor colon tissues). The median age was 59 years (ranged 17 to 87 years). The median follow-up time was 1934 days (ranged 36 to 2236 days). Ten people lost to follow-up. The tumor-free survival curves of COAD patients performed radical resection were shown in Fig 4a. The 5-year survival rate of TNM stage I and II patients was 90.7% that of stage III patients was 70.8%, and that of stage IV patients was 7.41% (Fig. 4b).
IHC results and clinicopathological factors
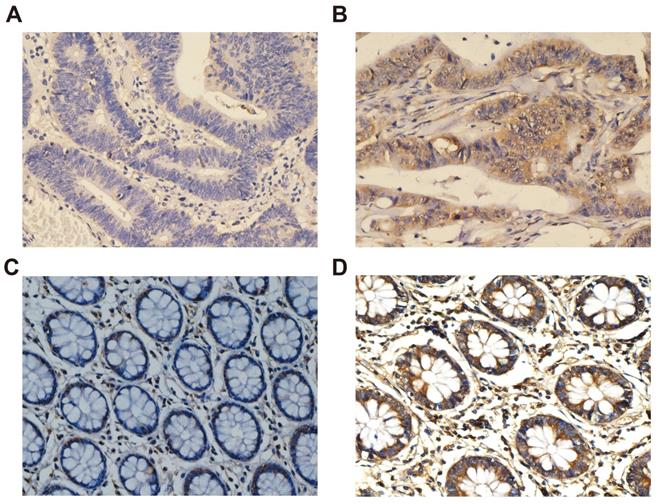
The positive signal of CXCL1 was the formation of diffuse brownish yellow or dark brown in the cytoplasm of the target cells (Fig. 5). The positive rate of CXCL1 staining was 81.6% (173 / 212) in COAD patients and 34.0% (16 / 47) in paracancerous non-tumor colon tissues. We collected clinicopathological factors that might be relevant to prognosis to perform correlation analysis with CXCL1, the results showed that the expression of CXCL1 protein in COAD patients was correlated with preoperative carcinoembryonic antigen (CEA) (Table 2).
CXCL1 mRNA expression in Guangxi Medical University COAD cohorts. Notes: The Scatter plot (A) and the ROC curves (B) of 38 pairs samples with RT-qPCR. The Scatter plot (C) and the ROC curves (D) of 47 pairs of samples with IHC.
(A) Kaplan-Meier curves of TNM stage in COAD RFS; (B) Kaplan-Meier curves of TNM stage in COAD OS.
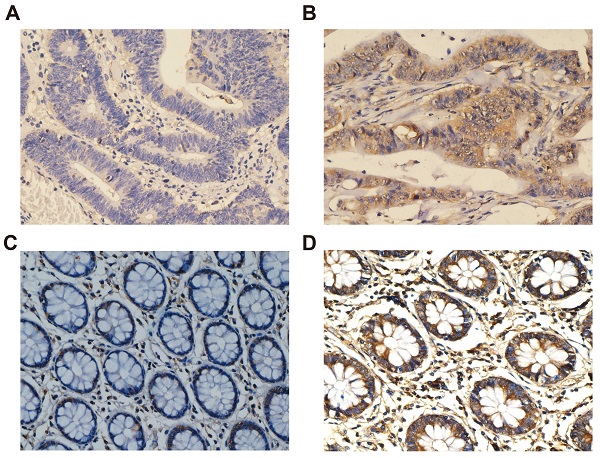
Immunohistochemical staining of CXCL1 expression in COAD. Notes: CXCL1 signaling was predominantly under the observation in the cytoplasm of colon cancer cells. (A) negative staining in tumor tissues; (B) positive expression in tumor tissues; (C) negative staining in paracancerous non-tumor colon tissues; (D) positive expression in paracancerous non-tumor colon tissues. Magnification, 400×.
Correlation analysis between CXCL1 expression and clinical characteristics in 212 patients with COAD
| Variable | Negative | positive | χ2 | P-value | ||
|---|---|---|---|---|---|---|
| Gender | 3.720 | 0.054 | ||||
| Male | 20 | 117 | ||||
| Female | 19 | 56 | ||||
| Age (years) | 0.087 | 0.768 | ||||
| ≤65 | 26 | 111 | ||||
| >65 | 13 | 62 | ||||
| CEAa (ng/ml) | 6.671 | 0.010 | ||||
| 1-5 | 28 | 85 | ||||
| >5 | 10 | 83 | ||||
| TNM stage | 5.058 | 0.168 | ||||
| I | 9 | 21 | ||||
| II | 8 | 50 | ||||
| III | 14 | 78 | ||||
| IV | 8 | 24 | ||||
| Location | 0.962 | 0.327 | ||||
| Right | 16 | 86 | ||||
| Left | 23 | 87 | ||||
| Tumor typeb | 0.214 | 0.898 | ||||
| Invasive | 2 | 9 | ||||
| ulcerative | 28 | 125 | ||||
| mass | 9 | 33 | ||||
| Tumor differentiation | 1.401 | 0.496 | ||||
| Well | 3 | 7 | ||||
| Moderately | 27 | 133 | ||||
| poor | 9 | 33 | ||||
| Tumor trombusc | 0.415 | 0.519 | ||||
| No | 33 | 152 | ||||
| Yes | 6 | 20 | ||||
| Tumor sized (cm) | 0.754 | 0.385 | ||||
| <5 | 19 | 71 | ||||
| ≥5 | 19 | 97 | ||||
| Tumor number | <0.001 | 1.000 | ||||
| Single | 38 | 167 | ||||
| Two | 1 | 6 | ||||
| Lymph nodee | 0.086 | 0.769 | ||||
| Negative | 23 | 97 | ||||
| Positive | 16 | 75 | ||||
| Tumor transfer | 0.890 | 0.346 | ||||
| No | 31 | 148 | ||||
| Yes | 8 | 25 | ||||
| Nerve infiltrationf | 0.561 | |||||
| No | 38 | 169 | ||||
| Yes | 1 | 3 | ||||
Notes: a, 6 patients without CEA information; b, 6 patients without general classification information; c, 1 patient without tumor thrombosis information; d, 6 patients without tumor size information; e, 1 patient without lymph node information; f, 1 patient without nerve infiltration information; NA: Not available.
Analysis of the diagnostic value of CXCL1 Immunohistochemical staining
Paired t-test analysis showed that the immunohistochemical score of CXCL1 in COAD carcinoma was considerably higher than that in paracancerous non-tumor colon tissues (Fig. 3c). At the same time, the results of the diagnostic ROC curve (Fig. 3d) revealed that CXCL1 has a high accurately for COAD diagnosis (P < 0.0001, AUC= 0.845, 95% Cl ( 0.762 - 0.927).
Prognostic value of CXCL1 immunohistochemical staining in COAD
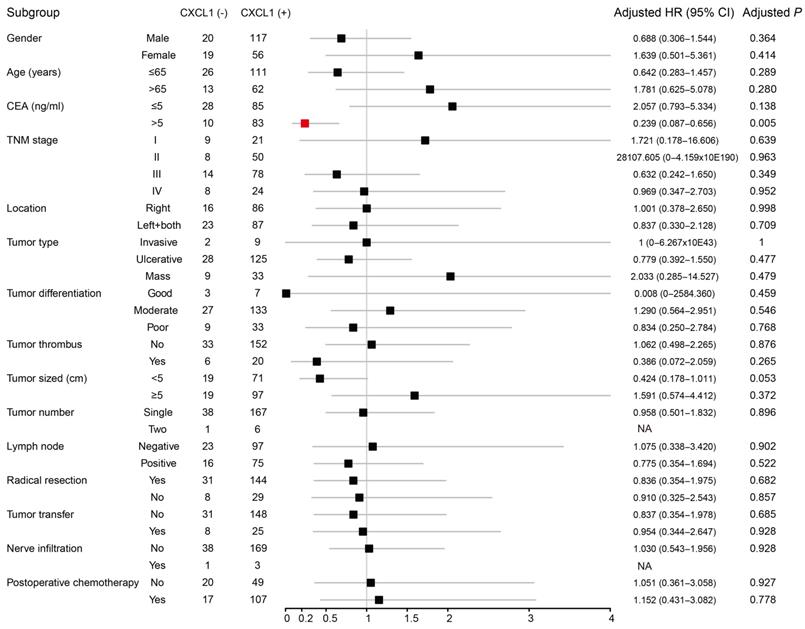
We performed Kaplan-Meier analysis to compare clinicopathological factors and prognosis of COAD patients (Table 3), the results showed that the recurrence-free survival time (RFS) was relatively short for patients with tumor TNM stage III and lymph node positive after radical resection. After adjusting for TNM staging, the expression of CXCL1 (corrected P ≥ 0.925, corrected HR (95% CI) = 0.957 (0.38 - 2.409) was not significantly correlated with tumor-free survival in COAD patients. The patients with early TNM stage, good tumor differentiation, no tumor thrombus, lymph nodes (-), radical resection, and no tumor metastasis had a relatively long OS. After correcting factors as the TNM stage, the tumor differentiation, with or without tumor thrombus and performed radical resection or palliative operation, Multivariate COX regression model showed that the expression of CXCL1 (corrected P ≥ 0.737, corrected HR (95% CI) = 0.898 (0.478 - 1.685) was not significantly correlated with OS. To further understand the relationship between the expression of CXCL1 protein and prognosis in COAD patients, we carried out the stratified analysis. There was no perceivable correlation between the expression of CXCL1 protein and RFS in the subgroup of clinicopathological factors. OS of CEA positive patients before operation was longer than that of CXCL1 positive patients. (Corrected P = 0.005 corrected HR (95% CI) = 0.239 (0.087 - 0.656) (Fig 6).
Gene set enrichment analysis
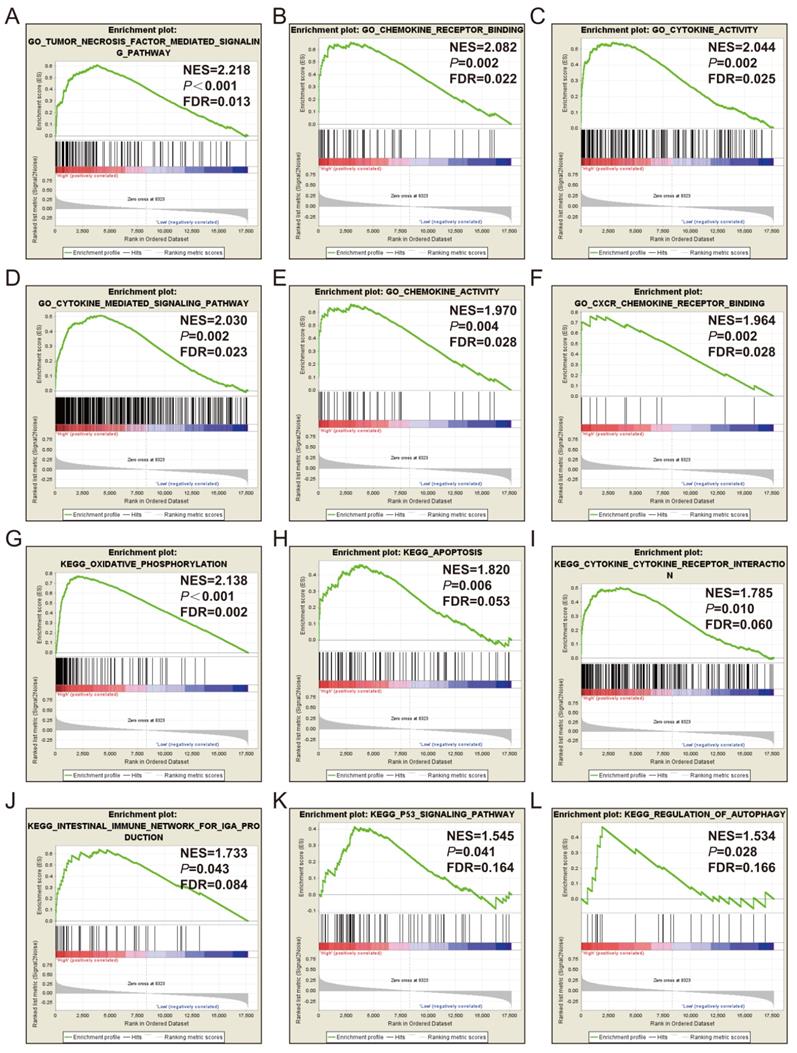
GSEA of CXCL1 was also performed by TCGA cohort. The RNA sequencing dataset of COAD patients was divided into 2 phenotypes through the median value of CXCL1 expressions in tumor tissues. The results of GSEA were displayed in Fig. 7 and Table S, which indicated that the high expression of CXCL1 was appreciably relevant to cytokine activity, cell apoptosis, P53 regulation pathway and regulation of autophagy.
Forest plot of the stratified analysis of CXCL1 with OS in different strata of characteristics in Guangxi cohort. Note: 6 patients without CEA information; 6 patients without general classification information; 1 patient without tumor thrombosis information; 6 patients without tumor size information; 1 patient without lymph node information; 1 patient without nerve infiltration information; 19 patients without postoperative chemotherapy information; NA, not obtained
Associations between clinical factors with OS and RFS in 212 COAD patients
| Variable | OS* | RFS# | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients | MST (days) | HR (95% CI) | Log-rank P | Patients | MRT (days) | HR (95% CI) | Log-rank P | ||||
| Gender | 0.801 | 0.467 | |||||||||
| Male | 137 | NA | 1 | 111 | NA | 1 | |||||
| Female | 75 | NA | 0.934 (0.552-1.582) | 64 | NA | 1.302 (0.637-2.661) | |||||
| Age (years) | 0.536 | 0.915 | |||||||||
| ≤65 | 137 | NA | 1 | 109 | NA | 1 | |||||
| >65 | 75 | NA | 1.174 (0.707-1.950) | 66 | NA | 1.040 (0.505-2.143) | |||||
| CEA (ng/ml) | 0.169 | 0.462 | |||||||||
| 1-5 | 113 | NA | 1 | 101 | NA | 1 | |||||
| >5 | 93 | NA | 1.424 (0.858-2.363) | 70 | NA | 0.759 (0.364-1.585) | |||||
| Missing | 6 | 4 | |||||||||
| TNM stage | <0.0001 | 0.039 | |||||||||
| I | 29 | NA | 1 | 30 | 1 | ||||||
| II | 57 | NA | 0.742 (0.209-2.629) | 58 | 0.499 (0.144-1.725) | ||||||
| III | 89 | NA | 2.469 (0.864-7.060) | 87 | 1.625 (0.612-4.311) | ||||||
| IV | 27 | 405 | 22.253 (7.607-65.098) | 0 | |||||||
| Location | 0.806 | 0.627 | |||||||||
| Right | 102 | NA | 1 | 86 | NA | 1 | |||||
| Left+both | 110 | NA | 0.929 (0.565-1.529) | 89 | NA | 1.191 (0.587-2.418) | |||||
| Tumor type | 0.691 | 0.358 | |||||||||
| Invasive | 11 | NA | 1 | 7 | NA | 1 | |||||
| ulcerative | 153 | NA | 1.511 (0.367-6.221) | 126 | NA | 25897.646 (0-2.4428763792086E+116) | |||||
| mass | 42 | NA | 1.203 (0.267-5.428) | 36 | NA | 17555.202 (0-1.657925218057E+116) | |||||
| Missing | 6 | 6 | |||||||||
| Tumor differentiation | 0.019 | 0.639 | |||||||||
| Well | 10 | NA | 1 | 10 | NA | 1 | |||||
| Moderate | 160 | NA | 1.451 (0.352-5.993) | 138 | NA | 0.582 (0.175-1.941) | |||||
| Poor | 42 | NA | 3.076 (0.710-13.318) | 27 | NA | 0.724 (0.173-3.031) | |||||
| Tumor thrombus | <0.0001 | 0.095 | |||||||||
| No | 185 | NA | 1 | 163 | NA | 1 | |||||
| Yes | 26 | 660 | 4.571 (2.568-8.134) | 12 | NA | 2.389 (0.833-6.858) | |||||
| Missing | 1 | ||||||||||
| Tumor size (cm) | 0.236 | 0.573 | |||||||||
| <5 | 90 | NA | 1 | 75 | NA | 1 | |||||
| ≥5 | 116 | NA | 0.739 (0.447-1.221) | 95 | NA | 0.817 (0.404-1.654) | |||||
| Missing | 6 | 5 | |||||||||
| Tumor number | 0.138 | 0.160 | |||||||||
| Single | 205 | NA | 1 | 170 | NA | 1 | |||||
| Two | 7 | 1917 | 2.119 (0.768-5.844) | 5 | 1900 | 2.687 (0.64-11.291) | |||||
| Lymph node | <0.0001 | 0.001 | |||||||||
| Negative | 120 | NA | 1 | 107 | NA | 1 | |||||
| Positive | 91 | NA | 3.546 (2.075-6.061) | 68 | NA | 3.411 (1.633-7.126) | |||||
| Missing | 1 | ||||||||||
| Radical resection | <0.0001 | ||||||||||
| Yes | 175 | NA | 1 | ||||||||
| No | 37 | 481 | 11.536 (6.836-19.469) | ||||||||
| Tumor transfer | <0.0001 | ||||||||||
| No | 179 | NA | 1 | ||||||||
| Yes | 33 | 401 | 14.344 (8.376-24.565) | ||||||||
| Nerve infiltration | 0.173 | 0.152 | |||||||||
| No | 207 | NA | 1 | 173 | NA | 1 | |||||
| Yes | 4 | 1079 | 2.572 (0.628-10.540) | 2 | 412 | 3.864 (0.525-28.419) | |||||
| Missing | 1 | ||||||||||
| Postopetative chemotherapy | 0.833 | 0.744 | |||||||||
| No | 69 | NA | 1 | 59 | NA | 1 | |||||
| Yes | 124 | NA | 1.061 (0.610-1.846) | 100 | NA | 0.887 (0.43-1.828) | |||||
| Missing | 19 | 16 | |||||||||
| CXCL1 | 0.444 | 0.738 | |||||||||
| Negative | 36 | NA | 1 | 31 | NA | 1 | |||||
| Positive | 166 | NA | 0.788 (0.427-1.452) | 144 | NA | 0.859 (0.352-2.094) | |||||
Notes: * 202 patients had OS information; # 175 patients underwent radical surgery, of which 171 had RFS information; MST: median total survival; MRT: median recurrence time, NA: Not available.
GSEA results of CXCL1 expressed in COAD tumor tissues. Notes: (A-F) GSEA results of c5 reference gene sets for high CXCL1 expression groups; (G-L) GSEA results of c2 reference gene sets for high CXCL1 expression groups; ES, enrichment score.
Discussion
Cancer metastasis was still the main cause of death in CRC patients. The 5-year overall survival rate of CRC patients could be as high as 80-90%, but it would decrease to 5-10% after tumor metastasis [26, 27]. Therefore, early detection of CRC are particularly important for patients' clinical outcome. Tumor markers with high sensitivity and specificity contributed to the early detection of tumors, and previous studies of CRC biomarkers had not yielded ideal results [28-33]. In the prognostic study of CRC, some prognostic markers had been found to be used to screen the risk of recurrence or metastasis, however, their performance in clinical application was not perfect due to the limitation of technology, cost, and their complicated testing methods [34, 35].
In this study, by comparing the expression distribution of CXCL1 in normal human organs and tissues, we observed that expression of CXCL1 in intestinal tissues was higher than that in most other organs, indicating that CXCL1 played an indispensable role in normal physiological process of intestinal tissues. At the same time, by comparing the expression of CXCL1 between tumor and paracancerous tissues in COAD patients from TCGA cohort, we also observed that the expression of CXCL1 was dysfunctional between tumor and paracancerous tissues, and CXCL1 was significantly up-regulated in tumor tissues. We verified this result through the cohort of the first affiliated Hospital of Guangxi Medical University from the perspectives of genetic and protein levels. The diagnostic ROC curves also suggested that CXCL1 had a high diagnostic value for COAD. These results were accord with Wen Y et al [11] and Zhuo C et al. [36].
In previous studies, multiple studies reported the prognostic value of CXCL1 in colorectal cancer [36-38], and there were reports verifying the molecular mechanism of CXCL1 in colorectal cancer through in vivo and in vitro experiments [39-43]. In the TCGA cohort, Kaplan-Meier analysis showed that the OS of patients with high expression of CXCL1 was longer than that of patients with low expression of CXCL1, and multivariate analysis showed a similar trend. In the Guangxi Medical University cohort, we found that the expression of CXCL1 in tumor tissues was significantly correlated with preoperative CEA. In the sub-group of CEA positive, the OS of patients with high expression of CXCL1 was longer than that of patients with low expression of CXCL1. This result was different from previous studies [36, 41, 44]. Interleukin-8, CXCL1, and other chemokines had a strong chemotactic effect on a series of inflammatory cells, such as T cells, neutrophils, and basophils, but their entire functions had not been fully elucidated [45]. Our study provided new evidence for the significance of CXCL1 expression. The good prognostic effect of infiltrated CXCL1 positive was most likely to indicate the immune function of this chemokine and the anti-tumor effect of inflammatory cells.
Through GSEA analysis, we enriched some meaningful biological functions and metabolic pathways. The research of Cabrero-de et al showed that chemokine CXC subfamily genes were widely related to the occurrence and development of CRC [38]. Soreide K et al reported cell apoptosis was associated with the prognosis of CRC [46]. There were also many studies reporting the correlation between P53 and CRC [47-49]. Zhou H et al.'s study suggested that autophagy was related to tumorigenesis and the protection of cancer [50]. However, the role of autophagy in CRC remained unclear. The advantage of the present study compared with previous studies was that we used TCGA whole-genome RNA sequencing data and GSEA method to further investigate the molecular mechanism of CXCL1 in COAD.
Although we first found the diagnostic and prognostic value of CXCL1 in COAD (rather than colorectal cancer), there were still some shortcomings in this study: a) There was imperfectness in the clinical information of COAD patients from TCGA database, and some important information such as tumor size, histological classification, degree of differentiation had not been provided. b) The sample size of this study was limited. c) Functional tests were needed to further verify the mechanism of the CXCL1 gene in the occurrence and development of COAD.
Conclusion
In this study, we found that the CXCL1 gene might function as a potential biomarker for the diagnosis of COAD, and might serve as a prognostic biomarker for a specific subgroup of COAD. Investigation of the molecular mechanism of CXCL1 in COAD, GSEA revealed that CXCL1 high expression phenotype was related to cytokine activity, cell apoptosis, P53 regulation pathway, and regulation of autophagy. However, further research and verification were still needed in the future.
Supplementary Material
Supplementary data.
Acknowledgements
The authors thank the contributors of The Cancer Genome Atlas (https://portal.gdc.cancer.gov/), GTEx website, and the website of https://www.proteinatlas.org for their contribution to sharing the COAD dataset and CXCL1 expression data on open access. The authors also thank the pathologist (Dr. Jia Li and Chuan-Li Su) for helping us interpret the results of immunohistochemistry.
Funding
The present study was supported by the Innovation Project of Guangxi Graduate Education (YCBZ2018036), The Basic Ability Improvement Project for Middle-aged and Young Teachers in Colleges and Universities in Guangxi (2020KY12026) and Innovation Project of Guangxi Graduate Education (YCBZ2020048).
Availability of data and materials
The datasets generated during the current study are available in The Cancer Genome Atlas (https://portal.gdc.cancer.gov/) and GTEx website.
Ethics approval
The study was conducted in accordance with the Declaration of Helsinki. The research program of the First Affiliated Hospital of Guangxi Medical University was approved by the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University (Ethical number: 2019(KY-E-001)).
Informed consent
All enrolled cases were signed informed consent.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
2. Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B. et al. Colorectal cancer. Lancet. 2010;375:1030-47
3. Huang EY, Chang JC, Chen HH, Hsu CY, Hsu HC, Wu KL. Carcinoembryonic antigen as a marker of radioresistance in colorectal cancer: a potential role of macrophages. BMC cancer. 2018;18:321
4. Shinkins B, Nicholson BD, Primrose J, Perera R, James T, Pugh S. et al. The diagnostic accuracy of a single CEA blood test in detecting colorectal cancer recurrence: Results from the FACS trial. PLoS One. 2017;12:e0171810
5. Huang SC, Huang SF, Chen YT, Chang Y, Chiu YT, Chang IC. et al. Overexpression of MutL homolog 1 and MutS homolog 2 proteins have reversed prognostic implications for stage I-II colon cancer patients. Biomedical journal. 2017;40:39-48
6. Haskill S, Peace A, Morris J, Sporn SA, Anisowicz A, Lee SW. et al. Identification of three related human GRO genes encoding cytokine functions. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:7732-6
7. Moser B, Clark-Lewis I, Zwahlen R, Baggiolini M. Neutrophil-activating properties of the melanoma growth-stimulatory activity. The Journal of experimental medicine. 1990;171:1797-802
8. Becker S, Quay J, Koren HS, Haskill JS. Constitutive and stimulated MCP-1, GRO alpha, beta, and gamma expression in human airway epithelium and bronchoalveolar macrophages. The American journal of physiology. 1994;266:L278-86
9. Vries MH, Wagenaar A, Verbruggen SE, Molin DG, Dijkgraaf I, Hackeng TH. et al. CXCL1 promotes arteriogenesis through enhanced monocyte recruitment into the peri-collateral space. Angiogenesis. 2015;18:163-71
10. Tsai HH, Frost E, To V, Robinson S, Ffrench-Constant C, Geertman R. et al. The chemokine receptor CXCR2 controls positioning of oligodendrocyte precursors in developing spinal cord by arresting their migration. Cell. 2002;110:373-83
11. Wen Y, Giardina SF, Hamming D, Greenman J, Zachariah E, Bacolod MD. et al. GROalpha is highly expressed in adenocarcinoma of the colon and down-regulates fibulin-1. Clinical cancer research: an official journal of the American Association for Cancer Research. 2006;12:5951-9
12. Miyake M, Lawton A, Goodison S, Urquidi V, Gomes-Giacoia E, Zhang G. et al. Chemokine (C-X-C) ligand 1 (CXCL1) protein expression is increased in aggressive bladder cancers. BMC cancer. 2013;13:322
13. Oladipo O, Conlon S, O'Grady A, Purcell C, Wilson C, Maxwell PJ. et al. The expression and prognostic impact of CXC-chemokines in stage II and III colorectal cancer epithelial and stromal tissue. British journal of cancer. 2011;104:480-7
14. Garcia-Larsen V, Morton V, Norat T, Moreira A, Potts JF, Reeves T. et al. Dietary patterns derived from principal component analysis (PCA) and risk of colorectal cancer: a systematic review and meta-analysis. Eur J Clin Nutr. 2019;73:366-86
15. Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A. et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419
16. Tang Z, Li C, Kang B, Gao G, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic acids research. 2017;45:W98-W102
17. Zheng S, Cherniack AD, Dewal N, Moffitt RA, Danilova L, Murray BA. et al. Comprehensive Pan-Genomic Characterization of Adrenocortical Carcinoma. Cancer cell. 2016;29:723-36
18. Network TCGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330-7
19. Rong M, He R, Dang Y, Chen G. Expression and clinicopathological significance of miR-146a in hepatocellular carcinoma tissues. Ups J Med Sci. 2014;119:19-24
20. Dai J, Wu H, Zhang Y, Gao K, Hu G, Guo Y. et al. Negative feedback between TAp63 and Mir-133b mediates colorectal cancer suppression. Oncotarget. 2016;7:87147-60
21. Amin MB, Edge S, Greene F, Byrd DR, Brookland RK, Washington MK. et al. AJCC Cancer Staging Manual (8th Edition). Chicago. 2017 Springer: 20
22. Zhang Y, Luo J, He R, Huang W, Li Z, Li P. et al. Expression and clinicopathological implication of DcR3 in lung cancer tissues: a tissue microarray study with 365 cases. OncoTargets and therapy. 2016;9:4959-68
23. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545-50
24. Y B, Y H. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological). 1995;57:289-300
25. Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368-75
26. Pita-Fernandez S, Gonzalez-Saez L, Lopez-Calvino B, Seoane-Pillado T, Rodriguez-Camacho E, Pazos-Sierra A. et al. Effect of diagnostic delay on survival in patients with colorectal cancer: a retrospective cohort study. BMC cancer. 2016;16:664
27. Garborg K. Colorectal Cancer Screening. The Surgical clinics of North America. 2015;95:979-89
28. Uchiyama K, Naito Y, Yagi N, Mizushima K, Higashimura Y, Hirai Y. et al. Selected reaction monitoring for colorectal cancer diagnosis using a set of five serum peptides identified by BLOTCHIP((R))-MS analysis. Journal of gastroenterology. 2018;53:1179-85
29. Chen JJ, Wang AQ, Chen QQ. DNA methylation assay for colorectal carcinoma. Cancer biology & medicine. 2017;14:42-9
30. Abdulla MH, Valli-Mohammed MA, Al-Khayal K, Al Shkieh A, Zubaidi A, Ahmad R. et al. Cathepsin B expression in colorectal cancer in a Middle East population: Potential value as a tumor biomarker for late disease stages. Oncology reports. 2017;37:3175-80
31. Hashimoto Y, Zumwalt TJ, Goel A. DNA methylation patterns as noninvasive biomarkers and targets of epigenetic therapies in colorectal cancer. Epigenomics. 2016;8:685-703
32. Zhang A, Sun H, Yan G, Wang P, Han Y, Wang X. Metabolomics in diagnosis and biomarker discovery of colorectal cancer. Cancer letters. 2014;345:17-20
33. Rosenberg EE. et al. D-glucuronyl C5-epimerase cell type specifically affects angiogenesis pathway in different prostate cancer cells. Tumour Biol. 2014;35:3237-45
34. Nakamura Y, Yoshino T. Clinical Utility of Analyzing Circulating Tumor DNA in Patients with Metastatic Colorectal Cancer. The oncologist. 2018;23:1310-8
35. Feng X, Liu J, Gong Y, Gou K, Yang H, Yuan Y. et al. DNA repair protein XPA is differentially expressed in colorectal cancer and predicts better prognosis. Cancer medicine. 2018;7:2339-49
36. Zhuo C, Wu X, Li J, Hu D, Jian J, Chen C. et al. Chemokine (C-X-C motif) ligand 1 is associated with tumor progression and poor prognosis in patients with colorectal cancer. Bioscience reports. 2018;38:BSR20180580
37. le Rolle AF, Chiu TK, Fara M, Shia J, Zeng Z, Weiser MR. et al. The prognostic significance of CXCL1 hypersecretion by human colorectal cancer epithelia and myofibroblasts. Journal of translational medicine. 2015;13:199
38. Cabrero-de Las Heras S, Martinez-Balibrea E. CXC family of chemokines as prognostic or predictive biomarkers and possible drug targets in colorectal cancer. World journal of gastroenterology. 2018;24:4738-49
39. Liu ZY, Zheng M, Li YM, Fan XY, Wang JC, Li ZC. et al. RIP3 promotes colitis-associated colorectal cancer by controlling tumor cell proliferation and CXCL1-induced immune suppression. Theranostics. 2019;9:3659-73
40. Ogawa R, Yamamoto T, Hirai H, Hanada K, Kiyasu Y, Nishikawa G. et al. Loss of SMAD4 Promotes Colorectal Cancer Progression by Recruiting Tumor-Associated Neutrophils via the CXCL1/8-CXCR2 Axis. Clinical cancer research: an official journal of the American Association for Cancer Research. 2019;25:2887-99
41. Wang D, Sun H, Wei J, Cen B, DuBois RN. CXCL1 Is Critical for Premetastatic Niche Formation and Metastasis in Colorectal Cancer. Cancer research. 2017;77:3655-65
42. Wang D, Wang H, Brown J, Daikoku T, Ning W, Shi Q. et al. CXCL1 induced by prostaglandin E2 promotes angiogenesis in colorectal cancer. The Journal of experimental medicine. 2006;203:941-51
43. Hsu YL, Chen YJ, Chang WA, Jian SF, Fan HL, Wang JY. et al. Interaction between Tumor-Associated Dendritic Cells and Colon Cancer Cells Contributes to Tumor Progression via CXCL1. International journal of molecular sciences. 2018;19:2427
44. Divella R, Daniele A, R DEL, Simone M, Naglieri E, Savino E. et al. Circulating Levels of VEGF and CXCL1 Are Predictive of Metastatic Organotropismin in Patients with Colorectal Cancer. Anticancer research. 2017;37:4867-71
45. Oppenheim JJ, Zachariae CO, Mukaida N, Matsushima K. Properties of the novel proinflammatory supergene "intercrine" cytokine family. Annual review of immunology. 1991;9:617-48
46. Soreide K, Buter TC, Janssen EA, Gudlaugsson E, Skaland I, Korner H. et al. Cell-cycle and apoptosis regulators (p16INK4A, p21CIP1, beta-catenin, survivin, and hTERT) and morphometry-defined MPECs predict metachronous cancer development in colorectal adenoma patients. Cellular oncology: the official journal of the International Society for Cellular Oncology. 2007;29:301-13
47. Noda M, Okayama H, Kofunato Y, Chida S, Saito K, Tada T. et al. Prognostic role of FUT8 expression in relation to p53 status in stage II and III colorectal cancer. PloS one. 2018;13:e0200315
48. Wu Y, Li Y, Zhao X, Dong D, Tang C, Li E. et al. Combined detection of the expression of Nm23-H1 and p53 is correlated with survival rates of patients with stage II and III colorectal cancer. Oncology letters. 2017;13:129-36
49. Katkoori VR, Manne U, Chaturvedi LS, Basson MD, Haan P, Coffey D. et al. Functional consequence of the p53 codon 72 polymorphism in colorectal cancer. Oncotarget. 2017;8:76574-86
50. Zhou H, Yuan M, Yu Q, Zhou X, Min W, Gao D. Autophagy regulation and its role in gastric cancer and colorectal cancer. Cancer biomarkers: section A of Disease markers. 2016;17:1-10
Author contact
![]() Corresponding author: Prof. Feng Gao, Department of Colorectal and Anal Surgery, The First Affiliated Hospital of Guangxi Medical University, 6 ShuangYong Road, Nanning 530021, Guangxi Zhuang Autonomous Region, People's Republic of China. Tel: (+86)-771-5356529. Fax: (+86)-771-5356585. Email: gaofengedu.cn.
Corresponding author: Prof. Feng Gao, Department of Colorectal and Anal Surgery, The First Affiliated Hospital of Guangxi Medical University, 6 ShuangYong Road, Nanning 530021, Guangxi Zhuang Autonomous Region, People's Republic of China. Tel: (+86)-771-5356529. Fax: (+86)-771-5356585. Email: gaofengedu.cn.

 Global reach, higher impact
Global reach, higher impact