3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(18):5573-5582. doi:10.7150/jca.56532 This issue Cite
Review
Advances in deubiquitinating enzymes in lung adenocarcinoma
1. Department of Pulmonary and Critical Care Medicine, Qilu Hospital, Cheeloo College of Medicine, Shandong University (Jinan 250012, China)
2. Department of Pulmonary and Critical Care Medicine, Qilu Hospital of Shandong University (Jinan 250012, China)
Received 2020-11-29; Accepted 2021-7-9; Published 2021-7-25
Abstract
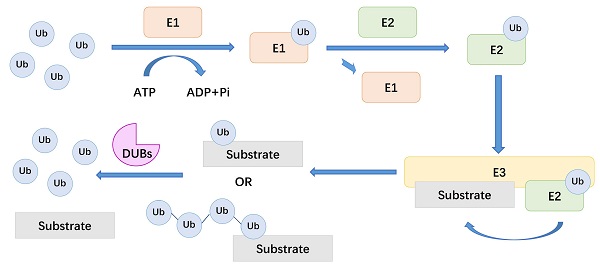
The process of ubiquitination and deubiquitination is widely present in the human body's protein reactions and plays versatile roles in multiple diseases. Deubiquitinating enzymes (DUBs) are significant regulators of this process, which cleave the ubiquitin (Ub) moiety from various substrates and maintain protein stability. Lung adenocarcinoma (LUAD) is the most common type of non-small cell lung cancer (NSCLC) and remains refractory to treatment. To elucidate the mechanism of LUAD and advance new therapeutic targets, we review the latest research progress on DUBs in LUAD. We summarize the biological capabilities of these DUBs and further highlight those DUBs that may serve as anticancer target candidates for precision treatment. We also discuss deubiquitinase inhibitors, which are expected to play a role in targeted LUAD therapy.
Keywords: deubiquitinating enzymes, lung adenocarcinoma, deubiquitinase inhibitor, targeted therapy
Introduction
Lung cancer is a disease with the highest incidence and mortality worldwide. Lung adenocarcinoma (LUAD) is the most common subtype of lung cancer, which has characteristics of complex mechanisms, aggressiveness, and poor prognosis[1-3]. Although surgical treatment, radiotherapy, chemotherapy, and targeted, comprehensive therapy have made significant progress over the past decade, the five-year survival rate remains bleak[4]. Therefore, it is compelling to investigate the mechanisms of LUAD in order to identify new therapeutic targets to improve future treatment decisions[5, 6].
Among existing achievements, a breakthrough in tumor research has been the discovery of many proto-oncogenes and tumor suppressor genes[7, 8]. Mutation, fusion, deletion, overexpression, and suppression of these genes affect the biological process of tumor cells[9-11]. However, studies have proven that expression level of mRNA do not truly reflect protein expression[12]. Post-translational modifications (PTMs) of proteins refer the covalent modification of individual amino acid residues on a protein after the translation process is complete. PTMs further promote an increase in complexity from the genomic level to the proteome. Primary forms of PTMs include phosphorylation, glycosylation, acetylation, and ubiquitination[13, 14].
Ubiquitination is the covalent binding of ubiquitin (Ub) to a target protein under the catalysis of a series of enzymes[15, 16]. To be precise, ubiquitination is a kind of intracellular protein labelling system, which attach different labels to target proteins for the purpose of explicitly degrading proteins[17]. The action of three types of enzymes achieves this protein degradation mechanism: ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin ligases (E3). Among them, E3 is the most abundant enzyme type and is primarily responsible for ubiquitination specificity. There are more than 600 types of E3 ligases in the human body, which recognize substrate proteins and combine with E2-Ub conjugates[18]. Deubiquitinating enzymes (DUBs) cleave the covalent bonds between Ub and substrate proteins. During the process of PTM, deubiquitination and ubiquitination reactions usually maintain a dynamic balance[19].
There are about 100 types of DUBs in the human body. These enzymes can be divided into seven families based on the active site of DUBs, including ubiquitin-specific proteases (USPs), ubiquitin COOH-terminal hydrolases (UCHs), ovarian tumor proteases (OTUs), Josephine, the JAB1 / MPN / MOV34 family (JAMMs) and motif interacting with Ub-containing novel DUB family (MINDYs)[20, 21]. DUBs are enzymes that act between ubiquitin molecules or between ubiquitin molecules and substrate proteins[22]. In the process of ubiquitination, Ub provides eight attachment sites for the formation of polymeric chains, including seven lysine residues, K6, K11, K27, K29, K33, K48, K63 and its amino terminus. For example, K48-linked chain is considered to be related to the proteasomal degradation of the substrate. And DUBs usually deubiquitinate substrate proteins by removing these ubiquitin chains[23]. Here, we summarize a mechanistic diagram of ubiquitination and deubiquitination (Figure 1). By reversing the ubiquitination process, DUBs modulate a variety of cellular processes, including protein degradation, DNA damage and repair, endocytosis, apoptosis, cell cycle, and receptor signal transduction[24-27]. Therefore, the regulatory effect of DUBs in LUAD, especially the potential therapeutic value of DUBs on LUAD treatment, has attracted increasing attention[21, 28].
A brief account of DUBs
As significant regulators, DUBs are involved in various diseases, including neurodegenerative disorders, inflammation, and cancer[23, 27, 29, 30]. For example, UCHL1 plays a significant role in promoting corticospinal motor neuron (CSMN) stability by maintaining protein homeostasis[31]. USP7 and USP47 regulate the ubiquitination state of the NLR family pyrin domain containing 3 (NLRP3) and influence inflammasome activation in macrophages[32].
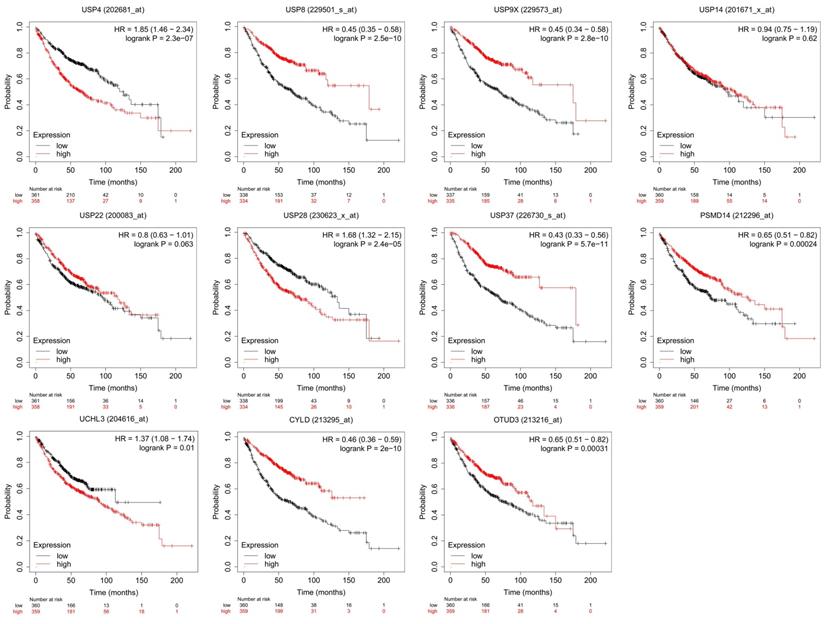
However, the most relevant research on DUBs is in the field of cancer[33, 34], including lung, liver[35], breast[36], stomach[37], colon[38], prostate[39] and ovarian cancers[40]. In this area, the mechanisms of DUBs in different tumors are described, and their potential as cancer therapeutics has attracted attention. For example, USP21-mediated deubiquitination of mitogen-activated protein kinase 2 (MEK2) promotes liver cancer cell proliferation in vitro and tumor growth in vivo[41]. BAP1 interacts with Krüppel-like factor 5 (KLF5) to stabilize KLF5 and promotes proliferation of breast cancer cells[42]. USP42 promotes the proliferation and invasion of gastric cancer cells by affecting matrix metalloproteinases (MMPs) and epithelial-mesenchymal transition (EMT)[43]. Further research focuses on the therapeutic effect of DUBs and their inhibitors on tumor therapy. For example, USP37 is overexpressed in breast cancer and promotes cell migration, invasion and EMT by regulating hedgehog signaling pathway. Downregulating of USP37 increases the sensitivity of breast cancer cells to cisplatin[44]. HBX19818 and P22077, two inhibitors that target USP10, induce anti-proliferative effects against FMS-like tyrosine kinase 3 (FLT3) mutant acute myeloid leukemia (AML)[45]. In this review, we concentrate on the particular role of DUBs in LUAD (Table 1 and Table 2). And the prognostic value of some essential DUBs was analyzed using Kaplan-Meier Plotter (http://kmplot.com/analysis/) (Figure 2).
The general mechanism of DUBs. Ub: ubiquitin, E1: ubiquitin-activating enzymes, E2: ubiquitin-conjugating enzymes, E3: ubiquitin ligases, DUBs: deubiquitinating enzymes.
The potential functions and mechanisms of DUBs in LUAD (USPs family)
| DUBs | Target | Potential functions | Potential mechanisms | Ref |
|---|---|---|---|---|
| USP4 | β-catenin | promote brain metastasis of LUAD cells | stabilize β-catenin protein | [74] |
| USP8 | PTK | promote the proliferative activity and inhibit apoptosis of tumor cells | the interaction between SFN and USP8 prevents the lysosomal degradation of RTK | [49] |
| EGFR | affect cell proliferation | regulate the expression of EGFR | [50] | |
| USP9X | PTGES | promote the metastasis of tumor cells | stabilize PTGES | [75, 76] |
| USP10 | PTEN | inhibit tumor growth and invasion | deubiquitinate and stabilize PTEN | [108] |
| p14ARF | prevent hyper-proliferation and transformation of cells | mediate the deubiquitination of p14ARF | [109] | |
| USP14 | β-catenin | promote proliferation | USP14 contributes to the accumulation of β-catenin protein level | [53, 56] |
| USP17 | Cyclin A | promote cell cycle from G0 / G1 to S phase | remove the polyubiquitin chains conjugated onto cyclin A and stabilize the cyclin A protein | [58] |
| USP18 | KRAS | regulating tumor cell proliferation | regulate KRAS protein stability | [110] |
| USP22 | EGFR | positively regulate cell proliferation and G1 / S phase transformation | stabilize the EGFR protein and increase the recovery and utilization of EGFR | [61] |
| USP28 | STAT3 | promote cell growth | induce STAT3 signaling | [111] |
| USP33 | PPM1A | suppress EMT | deubiquitinate PPM1A | [112] |
| USP37 | Snail protein | promote the invasion and migration of LUAD cells | deubiquitinate Snail protein, prevent the degradation of Snail protein | [81] |
| USP37 | c-Myc | affect the proliferation of cancer cells | deubiquitinate and stabilize c-Myc | [82] |
| USP44 | PTEN | inhibit cell growth | inhibit AKT signaling by stabilizing PTEN | [113] |
| USP49 | PTEN | inhibit cell growth | inhibit PI3K/AKT signaling by stabilizing PTEN | [114] |
Prognostic value of DUBs. We used Kaplan-Meier Plotter to analyze the prognostic value of DUBs. We searched the gene symbols, divided the LUAD patients into high-expression and low-expression groups based on the median and analyzed the difference in overall survival (OS) between the two groups. Some DUBs, for example, USP4, USP8, USP9X, are valuable in predicting the prognosis of LUAD patients.
The potential functions and mechanisms of DUBs in LUAD (other families)
| DUBs | Target | Potential functions | Potential mechanisms | Ref |
|---|---|---|---|---|
| BAP1 | inhibit lung cancer proliferation and promote apoptosis | deubiquitinate host cell factor-1 (HCF1) | [65] | |
| UCHL1 | inhibit cell proliferation | catalyze ubiquityl transfer to a lysine residue (presumably Lys63) on another ubiquitin molecule | [115] | |
| UCHL3 | LSH | influence the invasion and migration of LUAD cells | deubiquitinate LSH | [83] |
| OTUB2 | U2AF2 | promote cell growth, colony formation, migration, and invasive activities | stabilize U2AF2 and activate the AKT/mTOR pathway | [116] |
| OTUD3 | GRP78 | accelerate the growth of cancer cells | deubiquitinate and stabilize GRP78 | [68] |
| OTUD6B | regulate cell growth and proliferation | OTUD6B-1 represses DNA synthesis while OTUD6B-2 promotes it | [117] | |
| PSMD14 | promote the proliferative activity and inhibit apoptosis of tumor cells | p53-independent p21 down regulation | [64] |
DUBs affect proliferation and apoptosis in LUAD
The most fundamental feature of cancer cells is their infinite proliferation ability. Cancer cells control the release of growth-promoting signals and autonomously regulate their own growth[46, 47]. DUBs affect signaling pathways and regulatory factors related to LUAD cell proliferation and apoptosis.
USP8
USP8 is an essential cell growth-related DUB. USP8 maintains embryonic stem cell (ESC) stemness by deubiquitinating ectopic P-granules autophagy protein five homologs (EPG5). EPG5 is a specific autophagy protein, which protects pluripotency in ESCs by binding to LC3. USP8 directly deubiquitinates EPG5 by removing K63-linked ubiquitin and enhance the interaction between LC3 and EPG5[48].
In LUAD, USP8 stabilizes receptor tyrosine kinases (RTKs), contributes to proliferative activity, and inhibits apoptosis in A549 cells[49, 50]. Hyperactivation of AKT in LUAD promotes the combination of USP8 and stratifin (SFN). SFN-USP8 complex deubiquitinates RTKs and facilitates recycling of RTKs to the plasma membrane. The increase of RTKs leads to excessive activation of some signaling pathways, such as epidermal growth factor receptor (EGFR) signaling, and causes cell proliferation. Compared with normal lung tissue, USP8 is highly expressed in LUAD tissue, and is related to pathological subtype, lymphatic permeation, vascular invasion and EGFR overexpression of tumor tissue. Patients with high expression of USP8 have a significantly poorer prognosis, suggesting that USP8 has the potential as a prognostic marker. Furthermore, it has been reported that USP8 may represent a potential therapeutic target for gefitinib-resistant LUAD. Inhibition of USP8 activity significantly reduces the activity of gefitinib-resistant lung adenocarcinoma cell lines[51].
USP14
USP14 is a DUB associated with viral infection, neurodegenerative diseases and cancers[52]. USP14 has been reported to be overexpressed in LUAD and promote proliferation through the accumulation of β-catenin. β-catenin is a vital member of the Wnt-pathway that promotes proliferation in cancer cells. Silencing USP14 causes a sharp drop in β-catenin protein levels. Moreover, experiments confirmed that after knocking down USP14, the number of cells in the S phase significantly decreased (p<0.05), and the number of cells in the G0/G1 phase significantly increased (p<0.05). Meanwhile, expression of USP14 is significantly correlated with the prognosis of LUAD patients[53]. Therefore, USP14 may become a diagnostic marker and therapeutic target for LUAD patients. 1-[1-(4-fluorophenyl)-2,5-dimethylpyrrol-3-yl]-2-pyrrolidin-1-ylethanone, also known as IU1, is a kind of specific small-molecule inhibitor of USP14[54]. IU1 and its analogs can bound to USP14 and prevent the C-terminus of ubiquitin from approaching the catalytic center[55]. The results of in vitro experiments have confirmed that IU1-47 can inhibit the proliferation, invasion and migration of lung cancer cells[56].
USP17
USP17 regulates cell cycle progression by deubiquitinating cyclin A. Knockdown of USP17 decreased cyclin A levels. Cyclin A is a critical molecule that regulates the cell cycle[57]. USP17 interacts with cyclin A and removes the Ub chains conjugated onto cyclin A to stabilize it. Through this mechanism, USP17 drives cell cycle from G0/G1 to S phase and mediates the proliferation of LUAD cells[58]. Interestingly, USP17 has also been reported to drive a positive feedback loop between macrophages and LUAD cells. USP17 disrupts TRAF2/TRAF3 complex formation, enhancing inflammation and stemness in LUAD cells and promoting cell growth[59]. It is worth noting that USP17 is significantly highly expressed in NSCLC tissues and cells. In vivo experiments have also confirmed that knocking down USP17 inhibits the tumorigenesis of NSCLC[60]. Therefore, USP17 is a potential target for LUAD targeted therapy, which is worthy of further study.
USP22
USP22 is a DUB related to cell cycle regulation, tumor progression and EMT[61]. The expression of USP22 is significantly increased in LUAD tissues, suggesting that USP22 is a potential diagnostic marker of LUAD. In EGFR-mutated LUAD, USP22 positively regulates cell proliferation, G1/S phase transformation and metastasis. USP22 deubiquitinating EGFR localized on late endosomes, which stabilizes the EGFR protein, and increases the recovery and utilization of EGFR. Meanwhile, USP22 also promotes phosphorylation of EGFR downstream molecules. More importantly, USP22 promotes the resistance of EGFR-mutant lung ADC cells to EGFR-TKI, which provides new treatment strategies for patients with EGFR mutations[62].
PSMD14
Proteasome 26S subunit, non-ATPase 14 (PSMD14) is a DUB associated with the development of multiple tumors. For example, PSMD14 stabilizes ALK2 and contributes to tumor growth of colorectal cancers[63]. It has been reported that PSMD14 is upregulated in LUAD, and its high expression is related to a poor prognosis in LUAD patients. Knocking down PSMD14 inhibits proliferation of LUAD cells and induces cell cycle arrest and apoptosis. In H1299 and A549 cell lines, knocking down PSMD14 can induce G1 phase cell cycle arrest and cell senescence in both cell lines to inhibit cell survival. Among them, in the H1299 cell line, depletion of PSMD14 also leads to apoptosis in some cells[64]. In colorectal cancers, PSMD14 deubiquitinates K48-linked chains on ALK2 type I receptor to stabilize ALK2, resulting in the increases of cancer chemoresistance[63]. This provides a feasible direction for the study of PSMD14 in LUAD.
BAP1
BRCA1 related protein 1 (BAP1) is an essential nuclear localization DUB that inhibits lung cancer proliferation and promotes apoptosis. Experiments demonstrated that BAP1 is inhibited by the non-coding RNA miR-31. Combined with the prediction results of computer algorithms, such as TargetScan (http://www.targetscan.org/vert_72/), it was speculated that BAP1 represents a potential target of miR-31, and this conjecture was verified through Western blot and luciferase reporter assay[65]. In addition, some studies indicate that BAP1 mutation predisposes to LUAD[66]. Studies have shown that BAP1 inhibits the expression of solute carrier family 7 member 11 (SLC7A11) by decreasing H2Aub occupancy on it. And downregulation of SLC7A11 leads to ferroptosis, a non-apoptotic form of cell death, and inhibit tumor growth[67]. Unfortunately, the specific mechanism of BAP1 in LUAD has not been elucidated.
OTUD3
With respect to OTUs, it has been reported that OTUD3 accelerates the growth of human lung cancer. In LUAD, OTUD3 interacts with the glucose-regulated protein 78 (GRP78) and deubiquitinates it. GRP78 is a multifunctional protein and promotes tumor growth and metastasis. OTUD3 positively regulates GRP78, which in turn promotes the growth of LUAD cells[68]. Studies have shown that OTUD3 cleaves Lys6 and Lys11-linked diubiquitin[69], but the molecular mechanism of OTUD3 in LUAD is still unclear. It is worth mentioning that OTUD3 is decreased in breast cancer and suppresses tumorigenesis by deubiquitinating PTEN [70]. In contrast, OTUD3 is high-expressed in LUAD and indicates poorer overall survival for patients. Therefore, OTUD3 has the potential as a specific marker for diagnosing LUAD.
DUBs affect invasion and migration in LUAD
An important reason for the high rate of tumor recurrence and mortality is that tumor cells have a high incidence of invasion and migration. These tumor cells are different from normal cells with respect to cell metabolism, tumor microenvironment, and activation of signaling pathways[71]. In LUAD, some DUBs affect both tumor cell proliferation and migration. For example, USP22 mentioned above promotes LUAD cell invasion and migration[62]. It also promotes angiogenesis, proliferation, EMT, KRAS proto-oncogene (KRAS), and transcriptional regulator Myc-like (c-Myc) pathways in LUAD cells[72]. And OTUD3 also has specific functions to promote metastasis of LUAD cells[68].
USP4
USP4 is a member of the USPs family and affects the occurrence, invasion and migration of tumors. Through bioinformatics analysis in The Cancer Genome Atlas (TCGA) database, it was found that in LUAD tissues, expression of USP4 was significantly decreased. At the same time, USP4 expression was reported as a favorable independent prognostic factor for overall survival (OS) and recurrence-free survival (RFS) in LUAD patients[73]. USP4 influences brain metastasis of LUAD through β-catenin stabilization. β-catenin is a key molecule essential for acquisition of metastatic potential. Knockdown of USP4 reduces the level of β-catenin protein without affecting the level of mRNA expression. Therefore, it can be speculated that USP4 promotes brain metastasis of LUAD cells by stabilizing the β-catenin protein[74]. USP4 could be a potential therapeutic target for metastatic LUAD.
USP9X
USP9X is another DUB reported to be associated with LUAD invasion. Prostaglandin E synthase (PTGES) plays a key role in this function. PTGES, also known as mPGES-1, is an enzyme that promotes the conversion of prostaglandin H2 (PGH2) to prostaglandin E2 (PGE2)[75]. At the same time, PTGES is a key molecule related to the stemness of LUAD. Knocking down PTGES in LUAD cell lines inhibits cell invasion and migration. USP9X-mediated deubiquitinating reaction stabilized PTGES, and increased its expression in LUAD. Therefore, USP9X promotes the metastasis of tumor cells[76]. Another possible mechanism is that USP9X deubiquitinates dual specificity protein kinase TTK on K48-linked Ub chains. USP9X and TKK promote proliferation, invasion and migration in LUAD cells[77].
Interestingly, USP9X exerts different functions in different cancers. In colorectal cancer, USP9X stabilizes FBW7 and suppresses the cancer[78]. In another highly aggressive tumor, glioblastoma (GBM), USP9X interacts with aldehyde dehydrogenase one family member A3 (ALDH1A3) and stabilizes it to promote the tumorigenic capacity of mesenchymal stem cells[79]. These experiments suggest that USP9X could be a therapeutic target for various cancers. Inhibition of USP9X may play an important role in the treatment of metastatic LUAD.
USP37
EMT is the process in which epithelial cells acquire interstitial characteristics through specific procedures. EMT is related to tumor occurrence, invasion, migration, and resistance to treatment[80]. Snail protein is an essential positive regulator of EMT, which is upregulated by USP37. USP37 and Snail co-localize in the nucleus. USP37 deubiquitinates Snail protein, prevents the degradation of Snail, and stabilizes the initially unstable Snail protein. In this way, USP37 achieves the function of promoting the invasion and migration of LUAD cells[81]. It is worth mentioning that USP37 also deubiquitinates and stabilizes c-Myc to affect proliferation in cancer cells[82]. USP37 levels is significantly increased in LUAD tissues, which provides a clue that USP37 could be a specific molecule for LUAD diagnosis.
UCHL3
UCHL3 is a DUB belonging to the UCHs family. UCHL3 deubiquitinates chromatin modifier lymphoid-specific helicase (LSH). LSH is critical for the malignant progression of cancers by promoting cell proliferation and migration. Thus, UCHL3 contributes to the occurrence and transfer of LUAD. At the same time, the lncRNA GIAT4RA counteracts the deubiquitinating effects of LSH by interfering with interaction between LSH and UCH-L3, reducing levels of LSH protein, and playing the role in inhibiting the invasion and migration of LUAD cells[83]. Another article proposed that UCHL3 promotes the growth and stem-like properties of NSCLC cells through deubiquitinating and stabilizing Aryl hydrocarbon receptor (AhR)[84]. UCHL3 is significantly high-expressed in LUAD and associated with poor survival in LUAD. These data all indicate that UCHL3 could be a potential target for the treatment of metastatic LUAD.
DUBs and LUAD treatment
At present, there are many treatment methods for LUAD, including traditional surgical treatment, radiotherapy, and chemotherapy[85, 86]. Moreover, molecular targeted therapy is an essential part of precision treatment for LUAD[87, 88]. For example, in patients with EGFR mutations, gefitinib, erlotinib, afatinib, and osimertinib can be chosen clinically[89]. However, targets deficiency, drug resistance, and drug toxicity affect the clinical efficacy of molecular targeted therapies. Some astudies suggest that USP4[90], USP17[60], USP18[91], USP21[92], OTUD1, OTUD3, and OTUD4[93] may represent potential targets for targeted therapy of LUAD. DUBs are significant regulators in LUAD and have attracted much attention in the field of targeted drug development.
USP7
Macrophages (MΦs) mainly differentiate into two phenotypes, M1 MΦs is a tumor suppressor subtype, and M2 MΦs is a tumor promoting subtype. Studies have found that the inhibiting of USP17 plays an important role in reprogramming tumor associated macrophages (TAMs) to M1 MΦs through p38 MAPK pathway. Since this reprogramming of TAMs has the effect of eliminating tumor cells, the treatment targeting USP17 may play an important role in the treatment of lung cancer. P5091, a kind of USP7 inhibitor, can activate the anti-tumor immune responses. At the same time, P5091 and anti-pd-L1 have a synergistic anti-tumor effect in vivo. Therefore, P5091 may play an important role in LUAD immunotherapy[94].
DUBs inhibitor
| Name | Target DUB | Research stage | Ref |
|---|---|---|---|
| ML323 (70) | USP1/UAF1 | Preclinical | [118] |
| P5091 | USP7 | Preclinical | [119, 120] |
| P22077 | USP7 | Preclinical | [119, 120] |
| WP1130 | USP9X | Preclinical | [95] |
| Au (PPh3) PT | USP14; UCHL5 | Preclinical | [105] |
| AgDT | USP14; UCHL5 | Preclinical | [106] |
| AC17 | USP14; UCHL5 | Preclinical | [107] |
| IU1-47 | USP14 | Preclinical | [56] |
| [1,2,3]triazolo[4,5-d]pyrimidine derivatives | USP28 | Preclinical | [98] |
USP9X
USP9X inhibition was shown to increase tumor cell sensitivity to various chemotherapeutic agents, especially Bcl-2/Bcl-xL inhibitors. Bcl-xL and Mcl-1, members of the Bcl-2 family, convey resistance to drug therapy in many tumors. USP9X positively promotes the expression of Mcl-1. In contrast, the USP9X inhibitor WP1130 promotes the degradation of Mcl-1. Therefore, the combined use of WP1130 and Bcl-xL inhibitors may play a synergistic role in promoting apoptosis of LUAD cells[95].
USP28
In tumor cells, USP28 antagonizes the activity of the F-box and WD repeat domain containing 7 (FBXW7) to promoting the stability of c-Myc. As a target protein recognition component of the E3 complex, FBXW7 is involved in the degradation of various oncogenes, including cyclins and c-Myc. Moreover, USP28 is upregulated in LUAD and related to poor prognosis. Therefore, we suggest that USP28 can be used as a targeted therapy for LUAD[96, 97]. [1,2,3]triazolo[4,5-d]pyrimidine derivatives are highly effective and selective USP28 inhibitors and have the potential as targeted therapy drugs for LUAD patients[98].
CYLD
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is a member of the tumor necrosis factor (TNF) family. It has become a vital tumor biotherapeutic factor due to its ability to induce apoptosis of malignant cells. However, many tumor cells are resistant to TRAIL-induced apoptosis. Overexpression of CYLD lysine 63 deubiquitinase (CYLD) blocks TRAIL-induced nuclear factor-kappa B (NF-κB) activation, increases TRAIL-induced apoptosis of lung cancer cells and sensitizes lung cancer cells to TRAIL-induced apoptosis. Therefore, it has been suggested that CYLD is a therapeutic factor for lung cancer, especially when treating patients with high NF-κB activity[99-101].
Deubiquitinase inhibitors
Although they are still in preclinical stages, some deubiquitinase inhibitors have proven to have significant tumor-suppressing effects laboratory experiments[102-104]. According to a new report, a new gold(I) complex, Au(PPh3)PT, has been synthesized. Au (PPh3) PT selectively inhibits 19S proteasome-related subunits, such as UCHL5 and USP14, reducing hydrolysis of related proteins, and inducing the apoptosis in two lung cancer cell lines, A549 and H1299. In vivo experiments demonstrated that Au (PPh3) PT could also effectively inhibit the growth of transplanted tumors in LUAD in nude mice[105]. At present, research on deubiquitinase inhibitors is a hot topic in the field of anticancer drugs, and there are many related articles (Table 3). For example, ADT triggers the accumulation of ubiquitinated proteins in cells, effectively inhibiting proteasome subtypes USP14 and UCHL5 and inducing tumor cell apoptosis[106]. AC17 acts as an irreversible deubiquitinase inhibitor of 19S RP, leading to inhibition of the NF-κB pathway and reactivation of the pro-apoptotic protein p53[107].
Conclusions
This review focuses on research of DUBs in LUAD. On the one hand, we demonstrate the mechanisms of different DUBs in LUAD to understand the regulatory role of DUBs. In conclusion, the primary function of DUBs in LUAD is to inhibit ubiquitin degradation and stabilize proteins. In addition, DUBs also indirectly affect changes in multiple signaling pathways in the cell. On the other hand, this article primarily discusses whether DUB inhibitors may help treat tumors, especially respect to targeted therapy. Currently, only some small molecule inhibitors are included in the study, and all drug development is still in the preclinical stage. Meanwhile, many DUBs may eventually serve as therapeutic targets. Through these studies, new therapeutic targets have been provided for LUAD targeted therapy, conducive to further drug development.
However, insufficient research on the mechanism of DUBs is a defect of existing studies, resulting in related drug development still being limited. Compared to other types of tumors, such as breast and ovarian cancer, research on DUBs in LUAD is still scarce. Furthermore, substantial work is needed in pharmacology to apply DUBs in clinical therapy. Therefore, in future research on LUAD, DUBs are essential and should be extensively explored.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Inamura K. Clinicopathological Characteristics and Mutations Driving Development of Early Lung Adenocarcinoma: Tumor Initiation and Progression. International journal of molecular sciences. 2018;19(4):1259
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: a cancer journal for clinicians. 2019;69(1):7-34
3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA: a cancer journal for clinicians. 2020;70(1):7-30
4. Denisenko TV, Budkevich IN, Zhivotovsky B. Cell death-based treatment of lung adenocarcinoma. Cell death & disease. 2018;9(2):117
5. Calvayrac O, Pradines A, Pons E, Mazieres J, Guibert N. Molecular biomarkers for lung adenocarcinoma. The European respiratory journal. 2017;49(4):1601734
6. Kleczko EK, Kwak JW, Schenk EL, Nemenoff RA. Targeting the Complement Pathway as a Therapeutic Strategy in Lung Cancer. Frontiers in immunology. 2019;10:954
7. Lee JJ, Park S, Park H, Kim S, Lee J, Lee J. et al. Tracing Oncogene Rearrangements in the Mutational History of Lung Adenocarcinoma. Cell. 2019;177(7):1842-57 e21
8. Saito M, Shiraishi K, Kunitoh H, Takenoshita S, Yokota J, Kohno T. Gene aberrations for precision medicine against lung adenocarcinoma. Cancer science. 2016;107(6):713-20
9. Roman M, Baraibar I, Lopez I, Nadal E, Rolfo C, Vicent S. et al. KRAS oncogene in non-small cell lung cancer: clinical perspectives on the treatment of an old target. Molecular cancer. 2018;17(1):33
10. Gridelli C, Peters S, Sgambato A, Casaluce F, Adjei AA, Ciardiello F. ALK inhibitors in the treatment of advanced NSCLC. Cancer treatment reviews. 2014;40(2):300-6
11. Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M. et al. The Human Transcription Factors. Cell. 2018;172(4):650-65
12. Liu J, Qian C, Cao X. Post-Translational Modification Control of Innate Immunity. Immunity. 2016;45(1):15-30
13. Prus G, Hoegl A, Weinert BT, Choudhary C. Analysis and Interpretation of Protein Post-Translational Modification Site Stoichiometry. Trends in biochemical sciences. 2019;44(11):943-60
14. Nussinov R, Tsai CJ, Xin F, Radivojac P. Allosteric post-translational modification codes. Trends in biochemical sciences. 2012;37(10):447-55
15. Seo J, Kim MW, Bae KH, Lee SC, Song J, Lee EW. The roles of ubiquitination in extrinsic cell death pathways and its implications for therapeutics. Biochemical pharmacology. 2019;162:21-40
16. Mansour MA. Ubiquitination: Friend and foe in cancer. The international journal of biochemistry & cell biology. 2018;101:80-93
17. Chen Z, Zhou Y, Zhang Z, Song J. Towards more accurate prediction of ubiquitination sites: a comprehensive review of current methods, tools and features. Briefings in bioinformatics. 2015;16(4):640-57
18. Wang D, Bu F, Zhang W. The Role of Ubiquitination in Regulating Embryonic Stem Cell Maintenance and Cancer Development. International journal of molecular sciences. 2019;20(11):2667
19. Cai J, Culley MK, Zhao Y, Zhao J. The role of ubiquitination and deubiquitination in the regulation of cell junctions. Protein & Cell. 2017;9(9):754-69
20. He M, Zhou Z, Wu G, Chen Q, Wan Y. Emerging role of DUBs in tumor metastasis and apoptosis: Therapeutic implication. Pharmacology & therapeutics. 2017;177:96-107
21. Harrigan JA, Jacq X, Martin NM, Jackson SP. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nature reviews Drug discovery. 2018;17(1):57-78
22. Mevissen TET, Komander D. Mechanisms of Deubiquitinase Specificity and Regulation. Annu Rev Biochem. 2017;86:159-92
23. Mennerich D, Kubaichuk K, Kietzmann T. DUBs, Hypoxia, and Cancer. Trends in cancer. 2019;5(10):632-53
24. Heideker J, Wertz IE. DUBs, the regulation of cell identity and disease. The Biochemical journal. 2015;465(1):1-26
25. Kee Y, Huang TT. Role of Deubiquitinating Enzymes in DNA Repair. Molecular and cellular biology. 2016;36(4):524-44
26. Clague MJ, Urbe S, Komander D. Breaking the chains: deubiquitylating enzyme specificity begets function. Nature reviews Molecular cell biology. 2019;20(6):338-52
27. Kattah MG, Malynn BA, Ma A. Ubiquitin-Modifying Enzymes and Regulation of the Inflammasome. Journal of molecular biology. 2017;429(22):3471-85
28. Mofers A, Pellegrini P, Linder S, D'Arcy P. Proteasome-associated deubiquitinases and cancer. Cancer metastasis reviews. 2017;36(4):635-53
29. Hussain S, Zhang Y, Galardy PJ. DUBs and cancer: the role of deubiquitinating enzymes as oncogenes, non-oncogenes and tumor suppressors. Cell cycle. 2009;8(11):1688-97
30. Jacomin AC, Taillebourg E, Fauvarque MO. Deubiquitinating Enzymes Related to Autophagy: New Therapeutic Opportunities? Cells. 2018;7(8):112
31. Jara JH, Genc B, Cox GA, Bohn MC, Roos RP, Macklis JD. et al. Corticospinal Motor Neurons Are Susceptible to Increased ER Stress and Display Profound Degeneration in the Absence of UCHL1 Function. Cereb Cortex. 2015;25(11):4259-72
32. Palazon-Riquelme P, Worboys JD, Green J, Valera A, Martin-Sanchez F, Pellegrini C. et al. USP7 and USP47 deubiquitinases regulate NLRP3 inflammasome activation. EMBO reports. 2018;19(10):e44766
33. Gadhave K, Kumar P, Kapuganti SK, Uversky VN, Giri R. Unstructured Biology of Proteins from Ubiquitin-Proteasome System: Roles in Cancer and Neurodegenerative Diseases. Biomolecules. 2020;10(5):796
34. Pinto-Fernandez A, Davis S, Schofield AB, Scott HC, Zhang P, Salah E. et al. Comprehensive Landscape of Active Deubiquitinating Enzymes Profiled by Advanced Chemoproteomics. Front Chem. 2019;7:592
35. Sharma A, Liu H, Tobar-Tosse F, Chand Dakal T, Ludwig M, Holz FG. et al. Ubiquitin Carboxyl-Terminal Hydrolases (UCHs): Potential Mediators for Cancer and Neurodegeneration. International journal of molecular sciences. 2020;21(11):3910
36. Kim D, Hong A, Park HI, Shin WH, Yoo L, Jeon SJ. et al. Deubiquitinating enzyme USP22 positively regulates c-Myc stability and tumorigenic activity in mammalian and breast cancer cells. Journal of cellular physiology. 2017;232(12):3664-76
37. Saldana M, VanderVorst K, Berg AL, Lee H, Carraway KL. Otubain 1: a non-canonical deubiquitinase with an emerging role in cancer. Endocrine-related cancer. 2019;26(1):R1-R14
38. Zhang W, Sartori MA, Makhnevych T, Federowicz KE, Dong X, Liu L. et al. Generation and Validation of Intracellular Ubiquitin Variant Inhibitors for USP7 and USP10. Journal of molecular biology. 2017;429(22):3546-60
39. Liu Y, Xu X, Lin P, He Y, Zhang Y, Cao B. et al. Inhibition of the deubiquitinase USP9x induces pre-B cell homeobox 1 (PBX1) degradation and thereby stimulates prostate cancer cell apoptosis. The Journal of biological chemistry. 2019;294(12):4572-82
40. Zhang L, Wang H, Tian L, Li H. Expression of USP7 and MARCH7 Is Correlated with Poor Prognosis in Epithelial Ovarian Cancer. Tohoku J Exp Med. 2016;239(3):165-75
41. Li W, Cui K, Prochownik EV, Li Y. The deubiquitinase USP21 stabilizes MEK2 to promote tumor growth. Cell death & disease. 2018;9(5):482
42. Qin J, Zhou Z, Chen W, Wang C, Zhang H, Ge G. et al. BAP1 promotes breast cancer cell proliferation and metastasis by deubiquitinating KLF5. Nature communications. 2015;6:8471
43. Hou K, Zhu Z, Wang Y, Zhang C, Yu S, Zhu Q. et al. Overexpression and Biological Function of Ubiquitin-Specific Protease 42 in Gastric Cancer. PloS one. 2016;11(3):e0152997
44. Qin T, Li B, Feng X, Fan S, Liu L, Liu D. et al. Abnormally elevated USP37 expression in breast cancer stem cells regulates stemness, epithelial-mesenchymal transition and cisplatin sensitivity. Journal of experimental & clinical cancer research: CR. 2018;37(1):287
45. Weisberg EL, Schauer NJ, Yang J, Lamberto I, Doherty L, Bhatt S. et al. Inhibition of USP10 induces degradation of oncogenic FLT3. Nat Chem Biol. 2017;13(12):1207-15
46. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646-74
47. Shackleton M. Normal stem cells and cancer stem cells: similar and different. Semin Cancer Biol. 2010;20(2):85-92
48. Gu H, Shi X, Liu C, Wang C, Sui N, Zhao Y. et al. USP8 maintains embryonic stem cell stemness via deubiquitination of EPG5. Nature communications. 2019;10(1):1465
49. Kim Y, Shiba-Ishii A, Nakagawa T, Iemura SI, Natsume T, Nakano N. et al. Stratifin regulates stabilization of receptor tyrosine kinases via interaction with ubiquitin-specific protease 8 in lung adenocarcinoma. Oncogene. 2018;37(40):5387-402
50. Kim Y, Shiba-Ishii A, Nakagawa T, Husni RE, Sakashita S, Takeuchi T. et al. Ubiquitin-specific protease 8 is a novel prognostic marker in early-stage lung adenocarcinoma. Pathol Int. 2017;67(6):292-301
51. Byun S, Lee SY, Lee J, Jeong CH, Farrand L, Lim S. et al. USP8 is a novel target for overcoming gefitinib resistance in lung cancer. Clin Cancer Res. 2013;19(14):3894-904
52. Wang D, Ma H, Zhao Y, Zhao J. Ubiquitin-specific protease 14 is a new therapeutic target for the treatment of diseases. Journal of cellular physiology. 2021;236(5):3396-405
53. Wu N, Liu C, Bai C, Han YP, Cho WC, Li Q. Over-expression of deubiquitinating enzyme USP14 in lung adenocarcinoma promotes proliferation through the accumulation of beta-catenin. International journal of molecular sciences. 2013;14(6):10749-60
54. Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC. et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467(7312):179-84
55. Wang Y, Jiang Y, Ding S, Li J, Song N, Ren Y. et al. Small molecule inhibitors reveal allosteric regulation of USP14 via steric blockade. Cell Res. 2018;28(12):1186-94
56. Han KH, Kwak M, Lee TH, Park MS, Jeong IH, Kim MJ. et al. USP14 Inhibition Regulates Tumorigenesis by Inducing Autophagy in Lung Cancer In Vitro. International journal of molecular sciences. 2019;20(21):5300
57. Oakes V, Wang W, Harrington B, Lee WJ, Beamish H, Chia KM. et al. Cyclin A/Cdk2 regulates Cdh1 and claspin during late S/G2 phase of the cell cycle. Cell cycle. 2014;13(20):3302-11
58. Hu B, Deng T, Ma H, Liu Y, Feng P, Wei D. et al. Deubiquitinase DUB3 Regulates Cell Cycle Progression via Stabilizing Cyclin A for Proliferation of Non-Small Cell Lung Cancer Cells. Cells. 2019;8(4):297
59. Lu CH, Yeh DW, Lai CY, Liu YL, Huang LR, Lee AY. et al. USP17 mediates macrophage-promoted inflammation and stemness in lung cancer cells by regulating TRAF2/TRAF3 complex formation. Oncogene. 2018;37(49):6327-40
60. Zhang S, Yuan J, Zheng R. Suppression of Ubiquitin-Specific Peptidase 17 (USP17) Inhibits Tumorigenesis and Invasion in Non-Small Cell Lung Cancer Cells. Oncol Res. 2016;24(4):263-9
61. Hu J, Yang D, Zhang H, Liu W, Zhao Y, Lu H. et al. USP22 promotes tumor progression and induces epithelial-mesenchymal transition in lung adenocarcinoma. Lung Cancer. 2015;88(3):239-45
62. Zhang H, Han B, Lu H, Zhao Y, Chen X, Meng Q. et al. USP22 promotes resistance to EGFR-TKIs by preventing ubiquitination-mediated EGFR degradation in EGFR-mutant lung adenocarcinoma. Cancer Lett. 2018;433:186-98
63. Seo D, Jung SM, Park JS, Lee J, Ha J, Kim M. et al. The deubiquitinating enzyme PSMD14 facilitates tumor growth and chemoresistance through stabilizing the ALK2 receptor in the initiation of BMP6 signaling pathway. EBioMedicine. 2019;49:55-71
64. Zhang L, Xu H, Ma C, Zhang J, Zhao Y, Yang X. et al. Upregulation of deubiquitinase PSMD14 in lung adenocarcinoma (LUAD) and its prognostic significance. J Cancer. 2020;11(10):2962-71
65. Yu M, Liang H, Fu Z, Wang X, Liao Z, Zhou Y. et al. BAP1 suppresses lung cancer progression and is inhibited by miR-31. Oncotarget. 2016;7(12):13742-53
66. Abdel-Rahman MH, Pilarski R, Cebulla CM, Massengill JB, Christopher BN, Boru G. et al. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J Med Genet. 2011;48(12):856-9
67. Zhang Y, Shi J, Liu X, Feng L, Gong Z, Koppula P. et al. BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat Cell Biol. 2018;20(10):1181-92
68. Du T, Li H, Fan Y, Yuan L, Guo X, Zhu Q. et al. The deubiquitylase OTUD3 stabilizes GRP78 and promotes lung tumorigenesis. Nature communications. 2019;10(1):2914
69. Mevissen TE, Hospenthal MK, Geurink PP, Elliott PR, Akutsu M, Arnaudo N. et al. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell. 2013;154(1):169-84
70. Yuan L, Lv Y, Li H, Gao H, Song S, Zhang Y. et al. Deubiquitylase OTUD3 regulates PTEN stability and suppresses tumorigenesis. Nat Cell Biol. 2015;17(9):1169-81
71. Elia I, Doglioni G, Fendt SM. Metabolic Hallmarks of Metastasis Formation. Trends Cell Biol. 2018;28(8):673-84
72. Zhang K, Yang L, Wang J, Sun T, Guo Y, Nelson R. et al. Ubiquitin-specific protease 22 is critical to in vivo angiogenesis, growth and metastasis of non-small cell lung cancer. Cell Commun Signal. 2019;17(1):167
73. Zhong M, Jiang Q, Jin R. USP4 expression independently predicts favorable survival in lung adenocarcinoma. IUBMB Life. 2018;70(7):670-7
74. Hwang SJ, Lee HW, Kim HR, Lee H, Shin CH, Yun SI. et al. Ubiquitin-specific protease 4 controls metastatic potential through beta-catenin stabilization in brain metastatic lung adenocarcinoma. Sci Rep. 2016;6:21596
75. Fahmi H. mPGES-1 as a novel target for arthritis. Curr Opin Rheumatol. 2016;16(5):623-7
76. Wang T, Jing B, Sun B, Liao Y, Song H, Xu D. et al. Stabilization of PTGES by deubiquitinase USP9X promotes metastatic features of lung cancer via PGE2 signaling. Am J Cancer Res. 2019;9(6):1145-60
77. Chen X, Yu C, Gao J, Zhu H, Cui B, Zhang T. et al. A novel USP9X substrate TTK contributes to tumorigenesis in non-small-cell lung cancer. Theranostics. 2018;8(9):2348-60
78. Khan OM, Carvalho J, Spencer-Dene B, Mitter R, Frith D, Snijders AP. et al. The deubiquitinase USP9X regulates FBW7 stability and suppresses colorectal cancer. J Clin Invest. 2018;128(4):1326-37
79. Chen Z, Wang HW, Wang S, Fan L, Feng S, Cai X. et al. USP9X deubiquitinates ALDH1A3 and maintains mesenchymal identity in glioblastoma stem cells. J Clin Invest. 2019;129(5):2043-55
80. Pastushenko I, Blanpain C. EMT Transition States during Tumor Progression and Metastasis. Trends Cell Biol. 2019;29(3):212-26
81. Cai J, Li M, Wang X, Li L, Li Q, Hou Z. et al. USP37 Promotes Lung Cancer Cell Migration by Stabilizing Snail Protein via Deubiquitination. Front Genet. 2019;10:1324
82. Pan J, Deng Q, Jiang C, Wang X, Niu T, Li H. et al. USP37 directly deubiquitinates and stabilizes c-Myc in lung cancer. Oncogene. 2015;34(30):3957-67
83. Yang R, Liu N, Chen L, Jiang Y, Shi Y, Mao C. et al. GIAT4RA functions as a tumor suppressor in non-small cell lung cancer by counteracting Uchl3-mediated deubiquitination of LSH. Oncogene. 2019;38(46):7133-45
84. Ouyang L, Yan B, Liu Y, Mao C, Wang M, Liu N. et al. The deubiquitylase UCHL3 maintains cancer stem-like properties by stabilizing the aryl hydrocarbon receptor. Signal Transduct Target Ther. 2020;5(1):78
85. Dong ZY, Zhang C, Li YF, Su J, Xie Z, Liu SY. et al. Genetic and Immune Profiles of Solid Predominant Lung Adenocarcinoma Reveal Potential Immunotherapeutic Strategies. J Thorac Oncol. 2018;13(1):85-96
86. Roman M, Lopez I, Guruceaga E, Baraibar I, Ecay M, Collantes M. et al. Inhibitor of Differentiation-1 Sustains Mutant KRAS-Driven Progression, Maintenance, and Metastasis of Lung Adenocarcinoma via Regulation of a FOSL1 Network. Cancer Res. 2019;79(3):625-38
87. Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446-54
88. Nagasaka M, Gadgeel SM. Role of chemotherapy and targeted therapy in early-stage non-small cell lung cancer. Expert Rev Anticancer Ther. 2018;18(1):63-70
89. Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ, Wu Y-L. et al. Lung cancer: current therapies and new targeted treatments. The Lancet. 2017;389(10066):299-311
90. Lai CY, Yeh DW, Lu CH, Liu YL, Chuang YC, Ruan JW. et al. Epigenetic Silencing of Ubiquitin Specific Protease 4 by Snail1 Contributes to Macrophage-Dependent Inflammation and Therapeutic Resistance in Lung Cancer. Cancers (Basel). 2020;12(1):148
91. Mustachio LM, Lu Y, Kawakami M, Roszik J, Freemantle SJ, Liu X. et al. Evidence for the ISG15-Specific Deubiquitinase USP18 as an Antineoplastic Target. Cancer Res. 2018;78(3):587-92
92. Xu P, Xiao H, Yang Q, Hu R, Jiang L, Bi R. et al. The USP21/YY1/SNHG16 axis contributes to tumor proliferation, migration, and invasion of non-small-cell lung cancer. Exp Mol Med. 2020;52(1):41-55
93. Deng J, Hou G, Fang Z, Liu J, Lv XD. Distinct expression and prognostic value of OTU domain-containing proteins in non-small-cell lung cancer. Oncol Lett. 2019;18(5):5417-27
94. Dai X, Lu L, Deng S, Meng J, Wan C, Huang J. et al. USP7 targeting modulates anti-tumor immune response by reprogramming Tumor-associated Macrophages in Lung Cancer. Theranostics. 2020;10(20):9332-47
95. Peddaboina C, Jupiter D, Fletcher S, Yap JL, Rai A, Tobin RP. et al. The downregulation of Mcl-1 via USP9X inhibition sensitizes solid tumors to Bcl-xl inhibition. BMC Cancer. 2012;12:541
96. Gersch M, Wagstaff JL, Toms AV, Graves B, Freund SMV, Komander D. Distinct USP25 and USP28 Oligomerization States Regulate Deubiquitinating Activity. Mol Cell. 2019;74(3):436-51 e7
97. Wang X, Liu Z, Zhang L, Yang Z, Chen X, Luo J. et al. Targeting deubiquitinase USP28 for cancer therapy. Cell death & disease. 2018;9(2):186
98. Liu Z, Zhao T, Li Z, Sun K, Fu Y, Cheng T. et al. Discovery of [1,2,3]triazolo[4,5-d]pyrimidine derivatives as highly potent, selective, and cellularly active USP28 inhibitors. Acta Pharm Sin B. 2020;10(8):1476-91
99. Deng LL, Shao YX, Lv HF, Deng HB, Lv FZ. Over-expressing CYLD augments antitumor activity of TRAIL by inhibiting the NF-kappaB survival signaling in lung cancer cells. Neoplasma. 2012;59(1):18-29
100. Lin X, Chen Q, Huang C, Xu X. CYLD Promotes TNF-alpha-Induced Cell Necrosis Mediated by RIP-1 in Human Lung Cancer Cells. Mediators Inflamm. 2016;2016:1542786
101. Zhang J, Chen Y, Chen K, Huang Y, Xu X, Chen Q. et al. IL-33 drives the antitumour effects of dendritic cells via upregulating CYLD expression in pulmonary adenocarcinoma. Artif Cells Nanomed Biotechnol. 2019;47(1):1335-41
102. Yuan T, Yan F, Ying M, Cao J, He Q, Zhu H. et al. Inhibition of Ubiquitin-Specific Proteases as a Novel Anticancer Therapeutic Strategy. Front Pharmacol. 2018;9:1080
103. Poondla N, Chandrasekaran AP, Kim K-S, Ramakrishna S. Deubiquitinating enzymes as cancer biomarkers: new therapeutic opportunities? BMB Reports. 2019;52(3):181-9
104. Patel K, Ahmed ZS, Huang X, Yang Q, Ekinci E, Neslund-Dudas CM. et al. Discovering Proteasomal Deubiquitinating Enzyme Inhibitors for Cancer Therapy Lessons From Rational Design Nature and Old Drug Reposition. Future Med Chem. 2018;10(17):2087-108
105. Li X, Huang Q, Long H, Zhang P, Su H, Liu J. A new gold(I) complex-Au(PPh3)PT is a deubiquitinase inhibitor and inhibits tumor growth. EBioMedicine. 2019;39:159-72
106. Chen X, Yang Q, Chen J, Zhang P, Huang Q, Zhang X. et al. Inhibition of Proteasomal Deubiquitinase by Silver Complex Induces Apoptosis in Non-Small Cell Lung Cancer Cells. Cell Physiol Biochem. 2018;49(2):780-97
107. Zhou B, Zuo Y, Li B, Wang H, Liu H, Wang X. et al. Deubiquitinase Inhibition of 19S Regulatory Particles by 4-Arylidene Curcumin Analog AC17 Causes NF- B Inhibition and p53 Reactivation in Human Lung Cancer Cells. Molecular Cancer Therapeutics. 2013;12(8):1381-92
108. Sun J, Li T, Zhao Y, Huang L, Sun H, Wu H. et al. USP10 inhibits lung cancer cell growth and invasion by upregulating PTEN. Mol Cell Biochem. 2018;441(1-2):1-7
109. Ko A, Han SY, Choi CH, Cho H, Lee M-S, Kim S-Y. et al. Oncogene-induced Senescence Mediated by c-Myc Requires USP10 Dependen Deubiquitination and Stabilization of p14ARF. Cell Death Differ. 2018;25(6):1050-62
110. Mustachio LM, Lu Y, Tafe LJ, Memoli V, Rodriguez-Canales J, Mino B. et al. Deubiquitinase USP18 Loss Mislocalizes and Destabilizes KRAS in Lung Cancer. Mol Cancer Res. 2017;15(7):905-14
111. Li P, Huang Z, Wang J, Chen W, Huang J. Ubiquitin-specific peptidase 28 enhances STAT3 signaling and promotes cell growth in non-small-cell lung cancer. Onco Targets Ther. 2019;12:1603-11
112. Lu J, Zhong Y, Chen J, Lin X, Lin Z, Wang N. et al. Radiation Enhances the Epithelial- Mesenchymal Transition of A549 Cells via miR3591-5p/USP33/PPM1A. Cell Physiol Biochem. 2018;50(2):721-33
113. Zhang YK, Tian WZ, Zhang RS, Zhang YJ, Ma HT. Ubiquitin-specific protease 44 inhibits cell growth by suppressing AKT signaling in non-small cell lung cancer. Kaohsiung J Med Sci. 2019;35(9):535-41
114. Shen WM, Yin JN, Xu RJ, Xu DF, Zheng SY. Ubiquitin specific peptidase 49 inhibits non-small cell lung cancer cell growth by suppressing PI3K/AKT signaling. Kaohsiung J Med Sci. 2019;35(7):401-7
115. Liu Y, Lashuel HA, Choi S, Xing X, Case A, Ni J. et al. Discovery of Inhibitors That Elucidate the Role of UCH-L1 Activity in the H1299 Lung Cancer Cell Line. Chem Biol. 2003;10(9):837-46
116. Li J, Cheng D, Zhu M, Yu H, Pan Z, Liu L. et al. OTUB2 stabilizes U2AF2 to promote the Warburg effect and tumorigenesis via the AKT/mTOR signaling pathway in non-small cell lung cancer. Theranostics. 2019;9(1):179-95
117. Sobol A, Askonas C, Alani S, Weber MJ, Ananthanarayanan V, Osipo C. et al. Deubiquitinase OTUD6B Isoforms Are Important Regulators of Growth and Proliferation. Mol Cancer Res. 2017;15(2):117-27
118. Dexheimer TS, Rosenthal AS, Luci DK, Liang Q, Villamil MA, Chen J. et al. Synthesis and structure-activity relationship studies of N-benzyl-2-phenylpyrimidin-4-amine derivatives as potent USP1/UAF1 deubiquitinase inhibitors with anticancer activity against nonsmall cell lung cancer. J Med Chem. 2014;57(19):8099-110
119. Chen C, Song J, Wang J, Xu C, Chen C, Gu W. et al. Synthesis and biological evaluation of thiazole derivatives as novel USP7 inhibitors. Bioorg Med Chem Lett. 2017;27(4):845-9
120. Altun M, Kramer HB, Willems LI, McDermott JL, Leach CA, Goldenberg SJ. et al. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem Biol. 2011;18(11):1401-12
Author contact
![]() Corresponding author: Prof. Yi-Qing Qu, MD, PhD, Department of Pulmonary and Critical Care Medicine, Qilu Hospital of Shandong University, Wenhuaxi Road 107#, Jinan 250012, China. Tel: +86 531 8216 9335; Fax: +86 531 8296 7544; E-mail: quyiqingedu.cn
Corresponding author: Prof. Yi-Qing Qu, MD, PhD, Department of Pulmonary and Critical Care Medicine, Qilu Hospital of Shandong University, Wenhuaxi Road 107#, Jinan 250012, China. Tel: +86 531 8216 9335; Fax: +86 531 8296 7544; E-mail: quyiqingedu.cn

 Global reach, higher impact
Global reach, higher impact