3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(24):7266-7276. doi:10.7150/jca.63919 This issue Cite
Research Paper
ZNF750 exerted its Antitumor Action in Oral Squamous Cell Carcinoma by regulating E2F2
1. Central laboratory of Liaocheng People's Hospital, Liaocheng, 252000, P.R. China.
2. School of Chemistry and Chemical Engineering, Liaocheng University, Liaocheng 252059, P. R. China.
#These authors contributed equally to this work.
Received 2021-6-16; Accepted 2021-10-7; Published 2021-10-25
Abstract
Cell cycle activator E2F transcription factor 2 (E2F2) play a key role in tumor development and metastasis. Previous RNA sequence analysis (GSE134835) revealed E2F2 was significantly reduced by Zinc-finger protein 750 (ZNF750) in oral squamous cell carcinoma (OSCC). This study was aimed to determine the involvement of E2F2 in antitumor action of ZNF750. The nude mouse xenograft model was established by subcutaneously injection of stable cell line CAL-27oeZNF750 or CAL-27shZNF750. Xenograft tumor volume and tumor weight was measured. The expression of E2F2, transcriptional repressors such as enhancer of zeste 2 (Ezh2), PHD finger protein 19 (PHF19), and the genes related to cell proliferation or metastasis was studied in vivo or in vitro. Luciferase assay was performed to investigate regulation effect of ZNF750 on E2F2 luciferase activity. The involvement of E2F2 in the antitumor action of ZNF750 was studied by cotransduced ZNF750 with E2F2 lentivirus. The tumor growth and metastasis was repressed by ZNF750 manifested by reduced tumor size, tumor weight and the genes related to cell proliferation and metastasis. However, all of these were reversed by knockdown of the ZNF750 gene. Furthermore, E2F2 luciferase activity was inhibited by ZNF750. E2F2 partly blocked the antitumor action of ZNF750 manifested by increased self-renewal, invasion, migration, elevated Ezh2 and MMP13 protein expression in ZNF750 + E2F2 groups. However, silenced E2F2 further enhanced the antitumor action of ZNF750. ZNF750 depressed E2F2 activity and played a critical role in regulating transcriptional repressors for inhibiting the OSCC growth and metastasis in OSCC.
Keywords: Oral squamous cell carcinoma (OSCC), Zinc-finger protein 750, E2F transcription factor 2 (E2F2), Enhancer of zeste 2 (Ezh2), PHD Finger protein 19 (PHF19)
Introduction
Oral squamous cell carcinoma (OSCC) is one of the most common malignant tumors worldwide, and approximately 90% of the total oral malignancies are squamous cell carcinomas [1]. OSCC poses significant mortality and morbidity in the patients [2], therefore, it is urgently to develop new targeted therapy strategies.
Transcription factor Zinc-finger protein 750 (ZNF750) is a remarkable lineage-specific tumor suppressor gene shown to have a relationship with squamous cell carcinoma [3]. Previous study had testified that ZNF750 was reduced or deleted in OSCC tissues, as a potential tumor suppressor, ZNF750 inhibited cell proliferation and cell metastasis in CAL-27 cells [4, 5]. RNA sequence (NCBI/GEO/GSE134835) analysis showed that PHD finger protein 19 (PHF19), enhancer of zeste 2 (Ezh2) and E2F transcription factor 2 (E2F2) was downregulated by ZNF750 [4].
Deregulations of PHD finger protein 19 (PHF19) and enhancer of zeste 2 (Ezh2) implied in epigenetic regulation have been frequently associated with cancers [6]. PHF19, also known as PCL3 (polycomb-like 3), is a member of the polycomb-like (PCL) family with PHD finger domain. The increased PHF19 expression correlates with advanced cancers [7]. PHF19 is polycomb repressive complexes 2 (PRC2) associated factor, which form sub-complexes with PRC2 three essential core components, including suppressor of zeste 12 homolog (SUZ12), embryonic ectoderm development (EED), and enhancer of zeste 1/2 (Ezh1/2), to modulate the activity of PRC2 [8].
PRC2 is one of the multimeric polycomb group (PcG) protein complex. PcG proteins are essential epigenetic repressors. They form two major protein complexes, called polycomb repressive complexes 1 and 2 (PRC1 and PRC2) whose function is to maintain transcriptional repression [9]. PRC2 is mainly trimethylate lysine 27 of histone H3 (H3K27me3). Trimethylation of H3K27 correlates with transcriptionally repressed chromatin. Ezh2 functions as a H3K27 methyltransferase when comprising the PRC2 [10]. Over-expression of Ezh2 has been reported in many cancers. It was testified that Ezh2 promoted cell proliferation, migration, invasion and metastasis of cancer cells [11].
Ezh2 are E2F transcription factors-regulated genes [12]. E2F is a family of transcription factor proteins that have been implicated in multiple biological functions in human cancer [13]. E2F are divided into two subfamilies: E2F transcription factors are divided into two subfamilies: transcription activators (E2F1, E2F2, E2F3a) and repressors (E2F3b, E2F4, E2F5, E2F6, E2F7 and E2F8). E2F2 is a member of E2F family of transcription activators [14]. High E2F2 expression was correlated to worsen overall survival [15]. Our previous RNA sequence analysis has found that ZNF750 has the ability to reduce E2F2 expression [4].
Currently, the transcriptional regulation of ZNF750 in OSCC is still unclear, and it remains to be fully determined the antitumor action of ZNF750 was relevance to its regulation on E2F2. The present study was to explore the mechanism of ZNF750 for inhibiting the cell proliferation or metastasis in OSCC cell line CAL-27 cells and xenograft model in nude mice.
Materials and methods
Animals, cell lines and plasmids
The five-weeks-old male BALB/c-nu mice weighing 16-17 g (certificate number SCXK Beijing 20160006) were obtained from the Beijing Vital River Laboratory Animal Technology Co., Ltd (Beijing, China). The mice were kept under conventional housing conditions (22 ± 1 °C, 40-70% humidity) with food and water supply ad libitum and 12 h day/dark cycle. The mice were acclimated for one week before the beginning of the experiment. The experimental protocol was approved by Liaocheng People's Hospital Research Ethics Committee. All studies with animals were treated in accordance with the “Guide for the Care and Use of Laboratory Animals”.
The 293T cells and oral squamous cell carcinoma (OSCC) cell line CAL-27 cells were purchased from Shanghai Zhong Qiao Xin Zhou Biotechnology Co., Ltd (Shanghai, China) and Procell Life Science & Technology Co., Ltd (Wuhan, China) respectively. The lentiviral packaging plasmids psPax2, VSV-G and pRSV-Rev were given as a kind gift by Dr Padraig Strappe (Central Queensland University, Australian). The plvx-PGK-Puro lentiviral vector backbone (oe-Con), plvx-ZNF750-PGK-Puro (oe-ZNF750) and plvx-E2F2-PGK-Puro (oe-E2F2) was purchased from Biowit Technologies (Shenzhen, China). The hU6-MCS-CMV-Puromycin-negative control (sh-Con), hU6-MCS-CMV-Puromycin-shZNF750 (sh-ZNF750) silenced the ZNF750 gene, hU6-MCS-CMV-Puromycin-shE2F2 (sh-E2F2) silenced the E2F2 gene and the renilla reniformis plasmids were purchased from Shanghai GeneChem Co., LTD. (Shanghai, China). The plasmids pcDNA3.1A-ZNF750 and PGL3-E2F2-promoter (-2500 to +100) for luciferase assay was purchased from Sangon Biotech Co., Ltd. (Shanghai, China).
Cell cultures and treatment
The 293T cells and CAL-27 cells were grown in DMEM media supplemented with 10% FBS, streptomycin (100 mg/ml) and penicillin (100 IU/ml) at 37 °C in a humidified incubator with 5% CO2. All experiments were performed with mycoplasma-free cells. The CAL-27 cells growing at an exponential phase were randomly divided into four groups: Control (oe-Con and sh-Con) groups were transduced with oe-Con or sh-Con lentivirus respectively. Oe-ZNF750 (over-expression ZNF750 gene) and sh-ZNF750 (knockdown of the ZNF750 gene) groups were transduced with oe-ZNF750 or sh-ZNF750 lentivirus respectivly. For investigating the antitumor action of ZNF750 in CAL-27 cells was involved by E2F2, additional group Z+E (ZNF750+E2F2, over-expression of ZNF750 and E2F2 gene), Z+shE (over-expression of ZNF750 and silenced E2F2 gene) and oe-sh-Con (co-transduced with oe-Con and sh-Con lentivirus) was included.
Lentiviral packaging and cell infection
The lentiviral packaging and infection was performed as we described previously [16]. Briefly, lentiviral particles were produced in 293T cells by transfection with Lipofectamine 2000 (Thermo Fisher Scientific). Lipofectamine 2000/DNA complexes were added into 293T cells with the addition of caffeine (final concentration of 4 mM) to achieve higher titer lentivirus [17]. Lentivirus-containing supernatant was collected at 48 and 72 h post-transfection, filtered and concentrated using SBI's one-step virus concentration solution, PEG-it TM (SBI, USA). The CAL-27 cells grown at 30-50% confluence were infected with the oe-Con, oe-ZNF750, sh-Con, sh-ZNF750 lentivirus respectively, or cotransduced oe-ZNF750 with oe-E2F2 or sh-E2F2, oe-Con with sh-Con respectively in the presence of Polybrene (5 μg/ml, Sigma). Cells were allowed to recover for 48 h before being subjected to puromycin selection to obtain a stable cell line CAL-27oeZNF750, CAL-27shZNF750, CAL-27oeZNF750+oeE2F2, and CAL-27oeZNF750+shE2F2. The stable cell line CAL-27oeCon, CAL-27shCon and CAL-27oe-sh-Con using as a control.
Animal models and tumor xenograft growth
To evaluate the antitumor effect of ZNF750 in the nude mice, 20 nude mice were randomly divided into four groups (n=5 of each), oe-Con, oe-ZNF750, sh-Con and sh-ZNF750 groups. The stable cell line CAL-27oeCon and CAL-27oeZNF750, CAL-27shCon and CAL-27shZNF750 cells (3×106) were inoculated right armpit of nude mice to construct xenografts tumor model. The needle was stopped internally for 5 sec, rotated and pulled out to avoid leakage of the cell suspension. The activity, diet and mental state of nude mice were observed daily. The tumor volume was measured by a caliper every 6-7 days and calculated using the formla length × width 2 × 1/2. At the end of the experiment, the mice were euthanized 40 days later following inoculation. Tumors were excised and weighed to evaluate tumor growth, and then were fixed or frozen under liquid nitrogen for quantitative real-time PCR (qPCR) and western blot analysis.
Immunohistochemistry
The antigen identified by monoclonal antibody Ki67 and proliferating cell nuclear antigen (PCNA) protein expression in xenograft tumor samples was investigated by immunohistochemistry to evaluate the proliferation of cancer cells. Briefly, xenograft tumor samples were isolated from surrounding normal tissues. Fresh tissue samples were fixed in 10% formaldehyde and embedded in paraffin, Serial 5 µm sections were cut, deparaffinized in xylene, and hydrated through an ethanol series. Endogenous peroxidase was quenched in 3% hydrogen peroxide for 15 min incubation at room temperature. The sectioned slides were incubated with anti-Ki67 (1:16,000) and anti-PCNA (1:1000) antibody antibodies (all from Proteintech) at 4 °C overnight. Then the sections were incubated with KIT-5010 MaxVision TM HRP-Polymer anti-Mouse/Rabbit IHC Kit (Maxin-Bio, Co., Fuzhou, China) for 15 min. The slides were stained using a DAB Chromogen Substrate Kit (Maxin-Bio, Co., Fuzhou, China) for 3-5 min, and sections were counterstained with hematoxylin to identify nuclei. Images were acquired using a digital camera under the microscope (CKX71, Olympus).
Quantitative real-time PCR (qPCR)
Extraction of total RNA was performed using TRIzol® reagent (Thermo Fisher Scientific). RNA purity was detected by NanoDrop 2000 (Thermo Fisher, America). RNA (1 μg) was reverse-transcribed to cDNA using a PrimeScript® RT Kit in 20 μl reactions. The cDNA product was diluted 2-folds, aliquoted, and stored at -80 °C. QPCR was performed on an ABI 7500 Sequence Detection System (Applied Biosystems) using SYBR® Premix Ex Taq™ II Kit (all from Takara, Dalian, China). Amplification parameters for qPCR was 30 sec pre-incubation at 95 °C for one cycle, followed by 40 cycles of 95 °C for 5 sec and 60 °C for 34 sec. Table 1 summarizes the sequences of primers (Sangon Biotech) used in this study. The fold changes of genes were normalized to the housekeeping gene GAPDH by the 2-ΔΔCT method. Samples were analyzed in triplicate and each experiment was repeated for at least three times.
The sequences of primers
| Gene | Refseq Accession# | Direction | Sequence |
|---|---|---|---|
| E2F2 | NM_004091.3 | Forward | GGGCCCATCGAAGTCTACCT |
| E2F2 | Reverse | ATGATGCTAGGGTCGGTGCT | |
| EED | NM_003797 | Forward | GTCCTGTGGTATGGATCATTCT |
| EED | Reverse | GTATCAAATCGCCTAACCATCG | |
| Ezh2 | NM_004456.4 | Forward | AAATCAGAGTACATGCGACTGA |
| Ezh2 | Reverse | GTATCCTTCGCTGTTTCCATTC | |
| MMP1 | NM_002421.4 | Forward | TGGGCTGAAAGTGACTGGGA |
| MMP1 | Reverse | ATGGCATGGTCCACATCTGC | |
| MMP3 | NM_002422.5 | Forward | GGCCAGGGATTAATGGAGAT |
| MMP3 | Reverse | TGAAAGAGACCCAGGGAGTG | |
| MMP7 | NM_002423.5 | Forward | ACCGTGCTGTGTGCTGTGTG |
| MMP7 | Reverse | AGTCCTGAGCCTGTTCCCACTG | |
| MMP13 | NM_002427.3 | Forward | CACTTTATGCTTCCTGATGACG |
| MMP13 | Reverse | TCTGGCGTTTTTGGATGTTTAG | |
| MMP17 | NM_016155.7 | Forward | CACTCATGTACTACGCCCTCA |
| MMP17 | Reverse | TGGAGAAGTCGATCTGGATGTC | |
| PHF19 | NM_015651.3 | Forward | ACCCCAGTGACAGATCGAGG |
| PHF19 | Reverse | AGAGGAGGCGCTGTTCTCAT | |
| TIMP1 | NM_003254.3 | Forward | CATCACTACCTGCAGTTTTGTG |
| TIMP1 | Reverse | TGGATAAACAGGGAAACACTGT | |
| UBE2C | NM_007019.4 | Forward | ACCCTCATGATGTCTGGCGA |
| UBE2C | Reverse | ATAGCAGGGCGTGAGGAACT | |
| ZNF750 | NM_024702.3 | Forward | GCCACCATCTACTCGCCTTA |
| ZNF750 | Reverse | GCAGGAAGTGTCTCGGGTCT | |
| SUZ12 | NM_015355 | Forward | CAAACTGAAGCAAGAGATGACC |
| SUZ12 | Reverse | GCTATGGCAGAGTTTAAGATGC |
Western blot
Total protein was extracted from culture cells or tumor tissue from xenograft model of OSCC using 100 μl of ice cold lysis buffer including PMSF. The cell lysates were centrifuge for 5 min at 12000 × g at 4 °C and supernatant was collected as total protein. Protein concentrations were determined using the BCA protein assay kit (all from Beyotime, Jiangsu, China). Equal amount of proteins (15 µg) was used for Western blot. The proteins were denatured in 1× SDS - PAGE sample buffer (Beyotime, Jiangsu, China) for 5 min at 95 °C. Then, the denatured protein was electrophoresed on a 10% sodium dodecyl sulfate-polyacrylamide electophoresis (SDS-PAGE) and was transferred to polyvinylidene difluoride membranes (Millipore, Bedford, MA) after electrophoresis. Nonspecific bindings to the membranes were blocked with 5% (w/v) skimmed milk in TBST (Tris-buffered saline-Tween 20) at room temperature for 1 hour, and then incubated with the appropriate primary antibodies, anti-ZNF750, anti-E2F2 (1:1000, all from Abcam), anti-PHF19 (1:1000), anti-Ezh2 (1:2000), anti-cyclin D1 (1:5000 dilution), anti-matrix metalloproteinase (MMP) 9 (1:500, all from Proteintech), and mouse monoclonal anti-β-actin antibody (1:10000, Proteintech) overnight at 4 °C. After washing with TBST for three times, the membranes were incubated with species specific horseradish peroxidase coupled secondary antibodies for one hour. After washing, the membranes were visualized by ECL western blot detection reagents (Beyotime, Jiangsu, China). Quantification of the protein bands was visualized and analyzed with AlphaView analysis system (ProteinSimle, USA). The β-actin antibody was used as protein loading control. The values of ZNF750, PHF19, Ezh2, E2F2, cyclin D1 and MMP9 proteins expression were normalized against β-actin.
Luciferase assay
To investigate the regulation effect of ZNF750 on E2F2, the Dual Luciferase Reporter Assay Kit (E1910; Promega, Mannheim, Germany) was used to check luciferase and renilla signals according to a protocol provided by the manufacturer. Cells were divided into two groups: control groups (co-transfected with pcDNA3.1A, renilla reniformis plasmids and PGL3-E2F2-promoter) and ZNF750wt group (co-transfected with pcDNA3.1A-ZNF750, renilla reniformis plasmids and PGL3-E2F2-promoter). Luciferase activity of cell lysates were analyzed at 72 h after transfection using the GloMax® Navigator (Promega), and the relative luciferase activity was determined as the quotient of firefly luciferase and renilla luciferase activity. The folds change of E2F2 luciferase activity was calculated against control groups. The analysis was performed in triplicate.
Evaluation of E2F2 involvement in the antitumor action of ZNF750
For investigating the antitumor action of ZNF750 in CAL-27 cells were involved by E2F2, cell propagation, invasion, migration, Ezh2 and MMP13 protein expression were investigated by cell counting kit-8 (CCK-8), tumor sphere, colony formation assay, transwell assay, western blot and flow cytometry assay respectively. The CAL-27 cells were divided into seven groups: oe-Con, oe-ZNF750, sh-Con, sh-ZNF750, Z+E (co-transduced with ZNF750 and E2F2 virus), oe-sh-Con, and Z+shE (co-transduced with ZNF750 and sh-E2F2 virus) groups.
To evaluate the tumor propagation potency and self-renewal of cells, cells viability, tumor sphere and colony forming assays was performed. For cells viability, it was evaluated by CCK-8 assay (Beyotime, Jiangsu, China) according to the manufacturer's instructions. Briefly, 10 μl of CCK-8 solution was added to each well during the 2 h of culture at 37 °C, and the absorbance in each well was measured at 450 nm using a 96-well Multiskan MK3 microplate reader and experiments were repeated three times.
For tumor sphere formation, each group of cells (1 cells/μl) was trypsinized, counted, and cultured in ultra-low adherent six well plate (Corning Costar) with serum free DMEM medium supplemented with B27 serum free supplements (1:50; Invitrogen, Thermo Fisher Scientific, Inc.), 20 ng/ml human recombinant epidermal growth factor and 20 ng/ml basic fibroblast growth factor (bFGF) (all from PeproTech, Inc., Rocky Hill, NJ, USA). The spheroid formation was imaged and counted after 7 days of culture.
For colony formation, the cells were seeded in six well plates at low density (500 cells/well) in triplicate, and cultured for 7 days. The plates were then washed with PBS and fixed with 4% paraformaldehyde for 30 min followed by staining with 0.5% crystal violet for 1 min. after washed with PBS, and the images of each well were captured and counted by AlphaView (ProteinSimple, Santa Clara, CA, USA). The test was repeated for three times.
The cell invasion and migration assay was performed as we described previous [5, 16]. For cell invasion assay, Corning transwell chambers with polycarbonate membrane (8 μm pore size) were used to evaluate the cell invasion, and matrigel (BD Biosciences, Franklin Lakes, NJ, USA) was used as the substrate for invasion. Cells that invaded to the lower surfaces of the membrane were fixed with 4% formaldehyde, stained with 0.1% crystal, and visualized under light microscopy (CKX71, Olympus). The average number of invasion cells per field was assessed by Image-Pro Plus 6 software. Cell migration assay use a similar approach without matrigel coating. Three independent experiments were performed.
The protein expression of ZNF750, E2F2 and Ezh2 was investigated by western blot as we described above. MMP13 positive cells were assayed by flow cytometry. Each group of stable cell line cells (1×106) was washed twice with ice cold PBS, resuspended and incubated cells in BD Cytofix/Cytoperm™ solution at a concentration of 1×106 cells/0.5 ml for 20 min on ice. After washed twice with BD Perm/Wash™ buffer (1×), the cells were stained with 5 μl primary rabbit anti-MMP13 (Abcam) at 37 °C for 20 min, and then incubated with 5 μl secondary antibody Alexa 488 goat anti-rabbit IgG (H+L) for 20 min. Samples were analyzed by the BD FACSDiva software.
Statistical analysis
The values were expressed as means ± SD. Data were analyzed by one-way ANOVA, followed by a post hoc SNK-q test using the SPSS 23.0 statistical package. When comparing two conditions, the data were analyzed by Student's t-test between two groups. P < 0.05 was considered a significant difference.
Results
E2F2 and transcriptional repressors were repressed by ZNF750 in vivo and in vitro
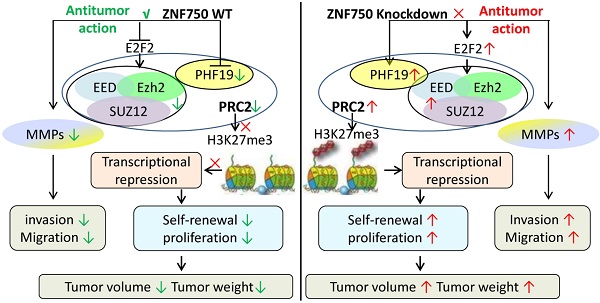
In this study, the expression of ZNF750 was detected by qPCR and western blot in vivo or in vitro to confirm stable cell lines were obtained. The ZNF750 mRNA expression was significantly increased 4505.9-folds and 55.8-folds in the CAL-27oeZNF750 cells and xenograft tumor samples respectively, while knockdown of the ZNF750 gene leaded to 77.8% and 60% down-regulation of ZNF750 in the CAL-27shZNF750 cells and xenografts tumor samples. Furthermore, the protein expression of ZNF750 was increased in oe-ZNF750 groups and decreased in sh-ZNF750 groups respectively in vivo (Figure 1A-C). The present study showed that the transcriptional repressors (PHF19, Ezh2, EED, SUZ12) and UBE2C mRNA or protein expression was all downregulated by ZNF750, whereas, it was all upregulated in sh-ZNF750 groups in vivo or in vitro (Figure 1D-G). Moreover, the cell cycle activator E2F2 and cell cycle regulator cyclinD1 expression was repressed in oe-ZNF750 groups but it was increased in sh-ZNF750 groups compared to their matched control groups (Figure 1E, 1H).
ZNF750 inhibited the cell growth in vivo
To confirm the inhibitory effect of ZNF750 on tumor growth in vivo, we subcutaneously injected the stable cell lines CAL-27oeCon, CAL-27oeZNF750, CAL-27shCon and CAL-27shZNF750 into nude mice, and then the tumor volume was measured about weekly for 40 days. The tumor formation rate in the nude mice was 100% (20/20). The present study showed that the tumor volumes were smaller in oe-ZNF750 groups than in oe-con groups at check point, but it was bigger in sh-ZNF750 groups than in sh-Con groups (Figure 2A, 2B). Furthermore, the tumor weight was reduced in oe-ZNF750 groups but it was increased in sh-ZNF750 groups than in their matched control groups, which indicated that ZNF750 had significant inhibitory effect on tumor growth (Figure 2C, 2D). Moreover, the proliferation markers Ki67 and PCNA protein expression in xenograft tumor samples was lower in the oe-ZNF750 groups than in oe-Con groups, whereas, it was higher expressed in sh-ZNF750 groups than in sh-Con groups (Figure 2E, 2F). The expression level of Ki67 and PCNA in oe-ZNF750 and sh-ZNF750 groups was in consistent with the observation results from the tumor growth.
ZNF750 repressed the genes related to metastasis in vivo
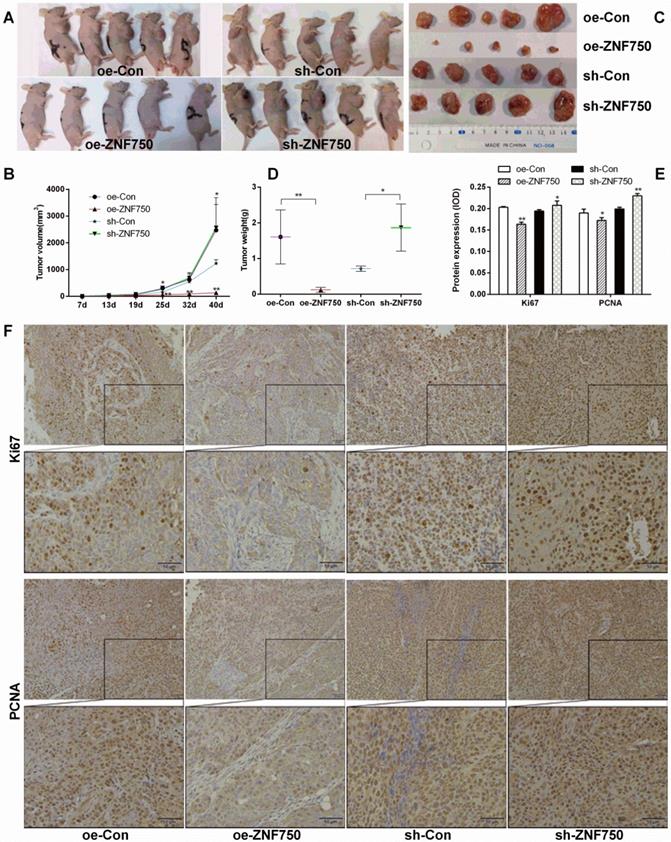
The expression of metastasis related genes MMP1, MMP3, MMP7, MMP9, MMP13, MMP17 and their endogenous inhibitor tissue inhibitor of metalloproteinase-1 (TIMP1) was studied to evaluate the inhibitory effect of ZNF750 on cell metastasis in vivo. The results showed that the above mentioned matrix metalloproteinases (MMPs) expression were all reduced by ZNF750, whereas, it was all increased in sh-ZNF750 groups compared to their matched control groups. However, the MMP inhibitor TIMP1 was downregulated in sh-ZNF750 groups but was upregulated in oe-ZNF750 groups (Figure 3A-D).
ZNF750 inhibited E2F2 luciferase activity
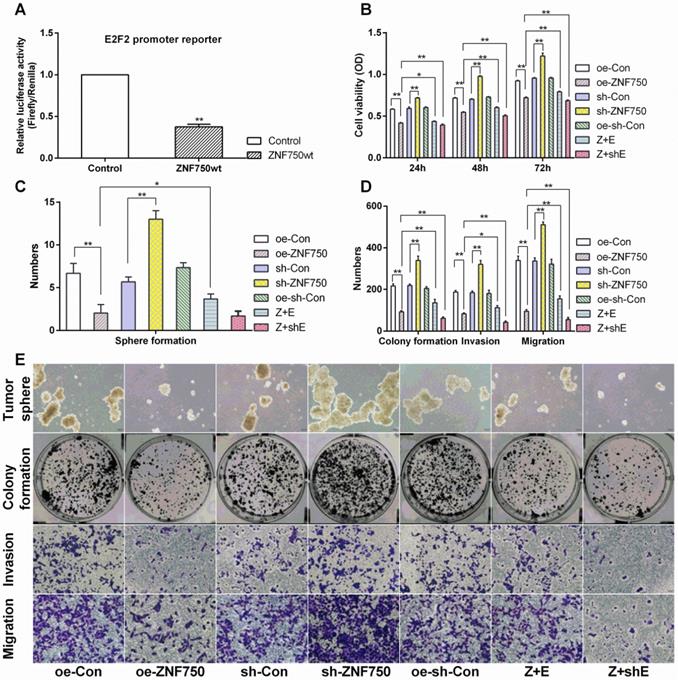
Luciferase reporter assay indicated that the E2F2 gene relative luciferase activity was significantly decreased in ZNF750wt groups compared to control groups (p<0.01), which indicated that ZNF750 negatively regulated the E2F2 expression (Figure 4A).
E2F2 involved in the inhibitory action of ZNF750 on cell proliferation, invasion and migration
The present results indicated that ZNF750 could abolish the cell proliferation, invasion and migration ability, manifested by reduced cell viability (reduced about 1.40-folds), number of tumor sphere (3.34-folds), colony formation (2.33-folds), cell invasion (2.27-folds) and migration (3.58-folds) in oe-ZNF750 groups compared to oe-con groups, but it was all increased in sh-ZNF750 groups compared to sh-con groups. Furthermore, E2F2 partly blocked the antitumor function of ZNF750 manifested by increased cell proliferation, invasion and migration in Z+E groups compared to oe-ZNF750 groups. However, cotransduced sh-E2F2 with ZNF750 lentivirus further enhanced the antitumor function of ZNF750 in Z+shE groups (Figure 4B-E).
ZNF750 inhibited the transcriptional repressors and cell cycle related genes in vivo and in vitro. A-C: ZNF750 mRNA and protein expression in CAL-27 cells or xenograft tumor samples from nude mice. D-F: The PHF19, Ezh2, UBE2C, E2F2, EED and SUZ12 mRNA expression in CAL-27 cells or in xenograft tumor samples. G-H: Effect of ZNF750 on transcriptional repressors (PHF19, Ezh2) and cell cycle activator (E2F2, cyclin D1) protein expression. * p<0.05, ** p<0.01. 1: oe-control groups, 2: oe-ZNF750 groups, 3: sh-control groups, 4: sh-ZNF750 groups. The experiment was repeated at least three times. E2F2, E2F transcription factor 2; EED, Embryonic ectoderm development; Ezh2, Enhancer of zeste 2; PHF19, PHD Finger protein 19; SUZ12, Suppressor of zeste 12 homolog; UBE2C, ubiquitin-conjugating enzyme E2C; ZNF750, Zinc finger protein 750.
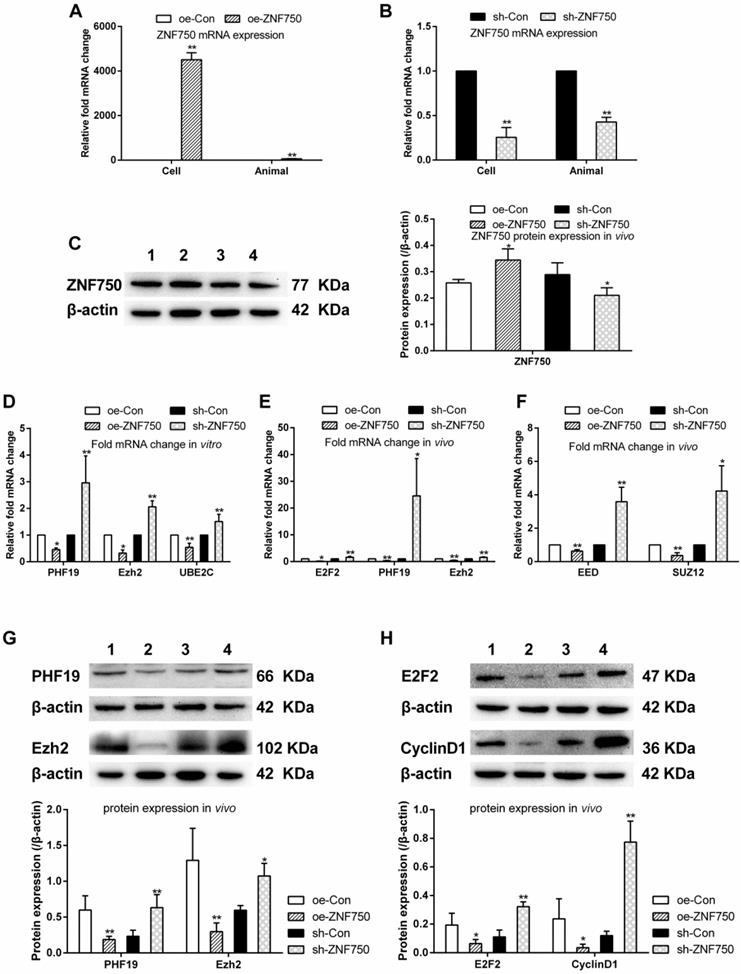
Effect of ZNF750 on tumor volume and tumor weight. A-B: The changes of tumor volume in each group (n=5 of each). The mice were euthanized 40 days later following inoculation. C-D: The tumor weight was reduced in oe-ZNF750 groups but elevated in sh-ZNF750 groups. E-F: The proliferation marker Ki67 and PCNA (Proliferating cell nuclear antigen) protein expression in xenograft tumor samples. Bar=20×. * p<0.05, ** p<0.01.
E2F2 blocked the depressed expression of Ezh2 and MMP13 induced by ZNF750
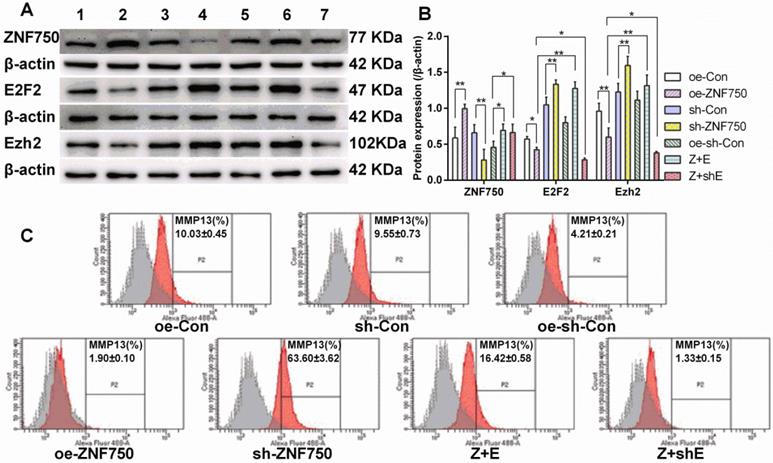
Western blot showed that E2F2 partly reversed the inhibitory effect of ZNF750 on transcriptional repressor Ezh2 expression. The Ezh2 protein expression was reduced in ZNF750 groups but it was elevated in sh-ZNF750 groups compared to their matched groups (Figure 5A, 5B). Moreover, compared to ZNF750 groups, Ezh2 protein expression were elevated in Z+E groups but slightly reduced in Z+shE groups (Figure 5A, 5B). The flow cytometry analysis further testified that MMP13 protein expression in CAL-27 cells was repressed (from 10.03% to 1.90%) by overexpression of ZNF750 but it was elevated (from 9.55% to 63.60%) by knockdown of the ZNF750 gene compared to their matched groups, and more importantly, cotransduced E2F2 with ZNF750 lentivirus partly blocked the inhibitory effect of ZNF750 on MMP13 protein expression (from 1.90% to 16.42%) compared to oe-ZNF750 groups (Figure 5C).
Effect of ZNF750 on metastasis related genes expression in vivo. A-B: The mRNA expression of MMPs (MMP1, MMP3, MMP7, MMP13, MMP17) and their endogenous inhibitor TIMP1 (Tissue inhibitor of metalloproteinase-1). C-D: The protein expression of MMP9. * p<0.05, ** p<0.01. 1: oe-control groups, 2: oe-ZNF750 groups, 3: sh-control groups, 4: sh-ZNF750 groups. MMP, Matrix metalloproteinase.
The involvement of E2F2 on antitumor action of ZNF750. A: ZNF750 negative regulated the E2F2 luciferase activity. Control groups: co-transfected with pcDNA3.1A, renilla reniformis and PGL3-E2F2-promoter plasmids, ZNF750wt groups: co-transfected with pcDNA3.1A-ZNF750, renilla reniformis and PGL3-E2F2-promoter plasmids. B: Cell viability, C-E: ZNF750 led to reduced self-renewal, cell invasion and migration in CAL-27 cells.
E2F2 partly blocked the inhibitory function of ZNF750 on Ezh2 and MMP13 protein expression. A-B: ZNF750, E2F2 and Ezh2 protein expression. C: MMP13 protein expression was detected by flow cytometry. ZNF750 reduced the MMP13 protein expression from 10.03% to 1.90%, however, E2F2 partly blocked the inhibitory function of ZNF750 on MMP13 expression (from 1.90% to 16.42%). * p<0.05, ** p<0.01. 1: oe-control groups, 2: oe-ZNF750 groups, 3: sh-control groups, 4: sh-ZNF750 groups. 5: oe-sh-control groups, 6: Z+E groups, 7: Z+shE groups. E2F2, E2F transcription factor 2; Ezh2, Enhancer of zeste 2; ZNF750, Zinc finger protein 750.
Discussion
Our previous study had found that the potential tumor suppressor ZNF750 inhibited cell cycle activator E2F2 and transcriptional repressors (PHF19, Ezh2) expression analyzed by RNA sequence (NCBI/GEO/GSE134835) [4]. In this study, we elucidated the antitumor mechanism of ZNF750 on OSCC in vivo or in vitro. The present study revealed that knockdown of the ZNF750 gene enhanced the expression of E2F2, transcriptional repressors and UBE2C in CAL-27 cells or xenograft model in nude mice. On the contrary, increased ZNF750 expression leaded to decreased expression of it in vivo or in vitro. These observations are consistent with our previous RNA sequence analysis (NCBI/GEO/GSE134835) that transcriptional repressor PHF19 and Ezh2 expression was repressed by ZNF750 [4].
PHF19 (PHD finger protein 19) is PRC2 associated factors that form sub-complexes with PRC2 core components to modulate the enzymatic activity of PRC2 [8]. PRC2 is the major H3K27 methyltransferase and is responsible for maintaining repressed gene expression patterns throughout development [18]. Thus, PHF19 functions as a transcriptional repressor, and play an important role in regulating transcription and histone demethylation [19]. Decreasing level of PHF19 resulted in reduced H3K27me3 while overexpressing PHF19 led to increased H3K27me3 level [20]. The PRC2 complex is composed of a trimeric core of Ezh1/2, EED and SUZ12, catalyzes the trimethylation of histone H3 at lysine 27 (H3K27me3) [21]. PHF19 could form the PRC2 with Ezh2, EED, and SUZ12, knockdown of the PHF19 gene suppressed Ezh2 phosphorylation and proliferation in glioma cells [22]. Therefore, the increased PHF19 with Ezh2, EED, and SUZ12 which form the PRC2 in sh-ZNF750 groups may result in abnormal repressed gene expression.
Ezh2 is downstream of the pRB-E2F pathway, amplified in cancer and strongly associated with tumor proliferation and aggressiveness [23, 24]. Targeting Ezh2 markedly suppressed OSCC invasion [25]. It was found that Ezh2 could interact with ubiquitin-conjugating enzyme E2C (UBE2C) [26], and UBE2C involved in head and neck tumorigenesis through cell cycle [27]. Cell cycle inhibitor p21 caused growth arrest by inhibition of cyclin D1 [28]. Ezh2 suppressed the expression of p21 through histone methylation (H3K27me3) on the p21 promoter resulted in cell proliferation. E2F2 is a member of the E2F family of transcription factors. In addition to their well-characterized roles in cell cycle control, E2F2 play key roles in mediating tumor development and metastasis [29]. Our previous study had revealed that cell cycle was involved in the antitumor effect of ZNF750 [4]. The current study were consistent with above studies manifested by overexpressed ZNF750 accompany with reduced E2F2, PHF19, Ezh2, EED, SUZ12, UBE2C and cyclinD1, and the vise verse for knockdown of the ZNF750 gene. Thus, the increased E2F2, transcriptional repressor PHF19, Ezh2, EED and SUZ12 in sh-ZNF750 groups could regulate the repressive transcriptional activity resulted in enhanced cyclinD1expression and cell growth.
In line with above studies, we proofed that the changes of ZNF750 expression led to the changes of tumor growth, which manifested by decreased tumor volume and tumor weight, reduced Ki67 and PCNA expression in oe-ZNF750 groups, but all of these was increased in sh-ZNF750 groups. It is well known that Ki67 and PCNA are proliferation markers. The elevated expression level of Ki67 and PCNA in oe-ZNF750 and diminished level in sh-ZNF750 groups was in consistent with the observation results from the tumor growth in vivo.
Metastasis has been identified as the main cause of high mortality in OSCC [30]. MMPs and their endogenous inhibitors TIMPs regulate extracellular matrix degradation and synthesis critical for cancer cell metastasis. Imbalances of MMPs and TIMPs lead to progression of various diseases including cancer [31]. Activated MMPs mediated matrix degradation and leading to tumor cell invasion [32]. It has been reported that TIMP1 has strong inhibiting properties against MMP9 [31]. In the current study, a group of MMPs (including MMP1, MMP3, MMP7, MMP9, MMP13 and MMP17) were all decreased but TIMP1 was increased in ZNF750 groups. However, MMPs were elevated but TIMP1 was diminished in sh-ZNF750 groups. According to the above mentioned results, our observations proved the inhibitory effect of ZNF750 on cell metastasis.
The present study elucidated that E2F2 was downregulated by ZNF750 and involved in the antitumor action of ZNF750. Luciferase reporter assay indicated that ZNF750 regulated the E2F2 lead to reduced E2F2 relative luciferase activity, which testified the negative regulation effect of ZNF750 on E2F2. Overexpression of E2F2 partly blocked the inhibited effect of ZNF750 on cell proliferation, invasion, migration, Ezh2 and MMP13 protein expression. More recent work has found that knockdown of the ZNF750 gene significantly promoted cell proliferation, colony formation, migration and invasion in esophageal squamous cell carcinoma cells [33]. Parallel in other cell type support a potential tumor suppressor role of ZNF750, the present study testified that ZNF750 attenuated cell invasion, migration and MMP13 positive population. Moreover, the cell viability, colony forming activity, and the self-renewal capacity of each group in CAL-27 cells was paralleled with the cancer cell proliferation in the nude mice. Furthermore, compared to ZNF750 groups, the cell proliferation, invasion and migration were augmented in Z+E groups. More importantly, E2F2 could block the inhibitory function of ZNF750 on transcriptional repressor, which manifested by increased Ezh2 protein expression in Z+E groups than in oe-ZNF750 groups, but the Ezh2 was further reduced in Z+shE groups. Therefore, we postulated that ZNF750 may be a crucial mediator on Ezh2, leading to reduced tumor growth, metastasis, and the E2F2 was involved in its regulation.
Taken together, the present study revealed a possible novel mechanism underlying malignant biological behavior caused by loss function of ZNF750 in OSCC in vivo or in vitro, and indicated that ZNF750 as a tumor suppressor plays a vital role in regulating transcriptional repressors and this function was related to depressed expression of E2F2 by ZNF750. The detail mechanism of ZNF750 on tumor suppressing in OSCC involved by E2F2 will be further investigated in the future.
Abbreviations
E2F2: E2F transcription factor 2; EED: Embryonic ectoderm development; Ezh2: Enhancer of zeste 2; H3K27me3: Trimethylate lysine 27 of histone H3; MMP: Matrix metalloproteinase; OSCC: Oral squamous cell carcinoma; PCL: Polycomb-like; PCNA: Proliferating cell nuclear antigen; PHF19: PHD Finger protein 19; PRC2: Polycomb repressive complexes 2; SUZ12: Suppressor of zeste 12 homolog; TIMP1: Tissue inhibitor of metalloproteinase-1; UBE2C: ubiquitin-conjugating enzyme E2C; ZNF750: Zinc finger protein 750.
Acknowledgements
This work was supported by a grant from the National Natural Science Foundation of China [Grant Number 81773759], and Science Foundation of Liaocheng People's Hospital [Grant Number LYQN201906].
Author Contributions
Hai-Ying Chen and Li Pan contributed to the study conception and design. Data analysis and interpretation were performed by all authors. The first draft of the manuscript was written by Hong-Li Yang Hai-Ying Chen and Cong Xu. Hai-Ying Chen, Min Hong and Li Pan revised the manuscript for intellectual content. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. D'Souza S, Addepalli V. Preventive measures in oral cancer: An overview. Biomed Pharmacother. 2018;107:72-80
2. Dhanuthai K, Rojanawatsirivej S, Thosaporn W, Kintarak S, Subarnbhesaj A, Darling M. et al. Oral cancer: A multicenter study. Med Oral Patol Oral Cir Bucal. 2018;23:e23-e9
3. Hazawa M, Lin DC, Handral H, Xu L, Chen Y, Jiang YY. et al. ZNF750 is a lineage-specific tumour suppressor in squamous cell carcinoma. Oncogene. 2017;36:2243-54
4. Liu X, Yang Y, Xu C, Yang H, Chen S, Chen H. RNA sequencing analysis of the CAL-27 cell response to over-expressed ZNF750 gene revealed an extensive regulation on cell cycle. Biomed Pharmacother. 2019;118:109377
5. Yang H, Pan L, Xu C, Zhang Y, Li K, Chen S. et al. Overexpression of tumor suppressor gene ZNF750 inhibits oral squamous cell carcinoma metastasis. Oncol Lett. 2017;14:5591-6
6. Eich ML, Athar M, Ferguson JE 3rd, Varambally S. EZH2-Targeted Therapies in Cancer: Hype or a Reality. Cancer Res. 2020;80:5449-58
7. Ren Z, Ahn JH, Liu H, Tsai YH, Bhanu NV, Koss B. et al. PHF19 promotes multiple myeloma tumorigenicity through PRC2 activation and broad H3K27me3 domain formation. Blood. 2019;134:1176-89
8. Li H, Liefke R, Jiang J, Kurland JV, Tian W, Deng P. et al. Polycomb-like proteins link the PRC2 complex to CpG islands. Nature. 2017;549:287-91
9. Cameron SR, Nandi S, Kahn TG, Barrasa JI, Stenberg P, Schwartz YB. PTE, a novel module to target Polycomb Repressive Complex 1 to the human cyclin D2 (CCND2) oncogene. J Biol Chem. 2018;293:14342-58
10. Gorodetska I, Lukiyanchuk V, Peitzsch C, Kozeretska I, Dubrovska A. BRCA1 and EZH2 cooperate in regulation of prostate cancer stem cell phenotype. Int J Cancer. 2019;145:2974-85
11. Honma D, Kanno O, Watanabe J, Kinoshita J, Hirasawa M, Nosaka E. et al. Novel orally bioavailable EZH1/2 dual inhibitors with greater antitumor efficacy than an EZH2 selective inhibitor. Cancer Sci. 2017;108:2069-78
12. Mu W, Starmer J, Yee D, Magnuson T. EZH2 variants differentially regulate polycomb repressive complex 2 in histone methylation and cell differentiation. Epigenet Chromatin. 2018;11:71
13. Johnson ME, Cantalupo PG, Pipas JM. Identification of Head and Neck Cancer Subtypes Based on Human Papillomavirus Presence and E2F-Regulated Gene Expression. mSphere. 2018;3:e00580-17
14. Manicum T, Ni F, Ye Y, Fan X, Chen BC. Prognostic values of E2F mRNA expression in human gastric cancer. Biosci Rep. 2018;38:BSR20181264
15. Liang B, Zhao J, Wang X. Clinical performance of E2Fs 1-3 in kidney clear cell renal cancer, evidence from bioinformatics analysis. Genes Cancer. 2017;8:600-7
16. Pan L, Yang H, Xu C, Chen S, Meng Z, Li K. et al. ZNF750 inhibited the malignant progression of oral squamous cell carcinoma by regulating tumor vascular microenvironment. Biomed Pharmacother. 2018;105:566-72
17. Ellis BL, Potts PR, Porteus MH. Creating higher titer lentivirus with caffeine. Hum Gene Ther. 2011;22:93-100
18. Qin S, Guo Y, Xu C, Bian C, Fu M, Gong S. et al. Tudor domains of the PRC2 components PHF1 and PHF19 selectively bind to histone H3K36me3. Biochem Biophys Res Commun. 2013;430:547-53
19. Deng Q, Hou J, Feng L, Lv A, Ke X, Liang H. et al. PHF19 promotes the proliferation, migration, and chemosensitivity of glioblastoma to doxorubicin through modulation of the SIAH1/beta-catenin axis. Cell Death Dis. 2018;9:1049
20. Hunkapiller J, Shen Y, Diaz A, Cagney G, McCleary D, Ramalho-Santos M. et al. Polycomb-like 3 promotes polycomb repressive complex 2 binding to CpG islands and embryonic stem cell self-renewal. PLoS Genet. 2012;8:e1002576
21. Brien GL, Healy E, Jerman E, Conway E, Fadda E, O'Donovan D. et al. A chromatin-independent role of Polycomb-like 1 to stabilize p53 and promote cellular quiescence. Genes Dev. 2015;29:2231-43
22. Lu J, Ji H, Tang H, Xu Z. microRNA-124a suppresses PHF19 over-expression, EZH2 hyper-activation, and aberrant cell proliferation in human glioma. Biochem Biophys Res Commun. 2018;503:1610-7
23. Idris S, Lindsay C, Kostiuk M, Andrews C, Cote DW, O'Connell DA. et al. Investigation of EZH2 pathways for novel epigenetic treatment strategies in oropharyngeal cancer. J Otolaryngol Head Neck Surg. 2016;45:54
24. Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323-35
25. Wang Y, Guo W, Li Z, Wu Y, Jing C, Ren Y. et al. Role of the EZH2/miR-200 axis in STAT3-mediated OSCC invasion. Int J Oncol. 2018;52:1149-64
26. Martinez-Canales S, Lopez de Rodas M, Nuncia-Cantarero M, Paez R, Amir E, Gyorffy B. et al. Functional transcriptomic annotation and protein-protein interaction analysis identify EZH2 and UBE2C as key upregulated proteins in ovarian cancer. Cancer Med. 2018;7:1896-907
27. Jin Z, Zhao X, Cui L, Xu X, Zhao Y, Younai F. et al. UBE2C promotes the progression of head and neck squamous cell carcinoma. Biochem Biophys Res Commun. 2020;523:389-97
28. Li Z, Yu D, Li H, Lv Y, Li S. Long noncoding RNA UCA1 confers tamoxifen resistance in breast cancer endocrinotherapy through regulation of the EZH2/p21 axis and the PI3K/AKT signaling pathway. Int J Oncol. 2019;54:1033-42
29. Hollern DP, Honeysett J, Cardiff RD, Andrechek ER. The E2F transcription factors regulate tumor development and metastasis in a mouse model of metastatic breast cancer. Mol Cell Biol. 2014;34:3229-43
30. Bugshan A, Farooq I. Oral squamous cell carcinoma: metastasis, potentially associated malignant disorders, etiology and recent advancements in diagnosis. F1000Res. 2020;9:229
31. Kaczorowska A, Miekus N, Stefanowicz J, Adamkiewicz-Drozynska E. Selected Matrix Metalloproteinases (MMP-2, MMP-7) and Their Inhibitor (TIMP-2) in Adult and Pediatric Cancer. Diagnostics (Basel). 2020;10:547
32. Bagheri V, Memar B, Behzadi R, Aliakbarian M, Jangjoo A, Bahar MM. et al. Isolation and identification of chemotherapy-enriched sphere-forming cells from a patient with gastric cancer. J Cell Physiol. 2018;233:7036-46
33. Kong P, Xu E, Bi Y, Xu X, Liu X, Song B. et al. Novel ESCC-related gene ZNF750 as potential Prognostic biomarker and inhibits Epithelial-Mesenchymal Transition through directly depressing SNAI1 promoter in ESCC. Theranostics. 2020;10:1798-813
Author contact
![]() Corresponding author: Hai-Ying Chen and Li Pan, Central laboratory of Liaocheng People's Hospital, Dong Chang West Road 67#, Liaocheng, 252000, P.R. China. E-mail: chenhycn; plly0805com.
Corresponding author: Hai-Ying Chen and Li Pan, Central laboratory of Liaocheng People's Hospital, Dong Chang West Road 67#, Liaocheng, 252000, P.R. China. E-mail: chenhycn; plly0805com.

 Global reach, higher impact
Global reach, higher impact