Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(24):7326-7333. doi:10.7150/jca.64613 This issue Cite
Research Paper
Helicobacter pylori associated aberrant methylation genes in blood leukocyte and gastric mucosa
1. Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Cancer Epidemiology, Peking University Cancer Hospital and Institute, Beijing, 100142, People's Republic of China.
2. Linqu Public Health Bureau, Linqu, Shandong, 262600, People's Republic of China.
#These authors contributed equally to this work.
Received 2021-7-6; Accepted 2021-10-19; Published 2021-10-28
Abstract
Background and Aim: Methylation alterations may be involved in Helicobacter pylori-associated gastric carcinogenesis. This study aims to explore the potential H.pylori-associated methylation biomarkers in blood leukocyte and gastric mucosa.
Methods: Five candidate H.pylori-associated aberrant methylation genes were selected from the previous genome-wide profiling panels and validated in blood leukocyte and gastric mucosa in multi-stages (case-control validation between H.pylori positive and negative subjects and self-control validation before and after anti-H.pylori treatment).
Results: GNAS methylation level was decreased in blood leukocyte (62.07% v.s. 46.33%, p<0.001) and gastric mucosa (56.30% v.s. 32.42%, p<0.001) of H.pylori positive subjects compared to negative controls. While, MTERF1 methylation level was increased significantly in blood leukocyte (29.57% v.s. 56.02%, p<0.001) and gastric mucosa (31.10% v.s. 47.50%, p<0.001) of positive subjects compared to controls. After successful H.pylori eradication, the methylation levels were increased from 44.87% to 60.88% (p<0.001) for GNAS and decreased from 46.19% to 34.56% (p<0.001) for MTERF1 in blood leukocyte. Similar increasing and decreasing methylation alterations were also found for the two genes after successful eradication in paired gastric mucosa. In TCGA database, an inverse relationship was found between GNAS methylation and mRNA expression (r=-0.12, p=0.027). The GC cases with higher GNAS expression levels showed significantly worse survival (HR, 2.09, 95%CI, 1.22-3.57, p=0.007) compared to lower expression subjects.
Conclusions: GNAS and MTERF1 methylation levels may be affected by H.pylori infection in gastric mucosa and blood leukocyte. GNAS may be involved in advanced stage of GC development, although the possible mechanism still needs further study in precancerous lesions.
Keywords: Gastric cancer, Helicobacter pylori, Methylation biomarker, Blood leukocyte, Gastric mucosa
Introduction
According to the cancer statistics from GLOBOCAN 2018, 2.2 million new cancer cases were attributable to carcinogenic infections. Helicobacter pylori (H.pylori) is the most important infectious agents and responsible for 810 000 new cancer cases. High burden of infection-attributable cancer driven by H.pylori was observed in eastern Asia (480 000 cases), especially in China (340 000 cases) [1].
Evidences suggest that H.pylori-induced chronic inflammation and epigenetic alterations may serve as one of the most important mechanisms in the process of gastric carcinogenesis [2]. The degree of exposure to H.pylori infection may influence the accumulation of aberrant methylation in various genes, which produces an “epigenetic field of cancerization” in normal-appearing tissues. The severity of this field was found to be correlated with the predisposition to gastric cancer (GC) and recently was used to predict the risk of metachronous GC [3, 4].
Aberrant methylations in blood leukocyte DNA are considered as surrogate biomarkers for their easy accessibility and correlations with the risk of various cancers or endogenous and exogenous risk factors [5, 6]. For example, global hypomethylation (ALU, LINE1 and Sat2) [7, 8] and hypermethylation of specific genes (IGFII and TUSC3) [9, 10] were reported to be associated with GC or H.pylori infection. Our previous genome-wide profiling study has found a high correlation of methylation levels between blood leukocyte and matched gastric mucosa, which supports the methylation of blood leukocyte as a potential surrogate biomarker for the epigenetic alteration in gastric carcinogenesis. Furthermore, the panel of H.pylori-associated differentially methylated genes was preliminarily identified in blood leukocyte and in gastric tissue [11].
From the previous candidate H.pylori-associated aberrant methylation gene panel in blood leukocyte, our pilot validation found decreasing trends of GNAS and LTBR methylation levels after H.pylori eradication by denaturing high performance liquid chromatography (DHPLC) in a small sample size comparison [11]. Besides GNAS and LTBR, DKK3 was selected from the candidate panel according to literatures [11-13] for further validation in the present study. MTERF1 and HAUS5 were also selected from the candidate aberrant methylation panel overlapped between blood leukocyte and gastric tissue, in which the differentially methylated CpG sites were located both near CpG island and promoter regions [11].
Depending on the above selection strategy, the present multi-stage study validated the five candidate H.pylori-associated aberrant methylation genes by methylation specific real-time PCR in blood leukocyte as well as in gastric mucosa. We further evaluated the relationship between the potential non-invasive H.pylori-associated epigenetic biomarkers and precancerous gastric lesions. Possible clinical significance was also preliminarily investigated in GC using TCGA database.
Methods
Study design and population
Linqu county in Shandong province, China, is a high-risk area of GC [14]. From 2011 to 2013, a large community-based randomized intervention trial to prevent GC was launched in Linqu county using 13C-urea breath test (13C-UBT) for H.pylori infection screening and a 10-day quadruple anti-H.pylori treatment for positive subjects [15]. In 2012 and 2013, the National Upper Gastrointestinal Cancer Early Detection Project conducted endoscopic examinations and collected blood and fresh gastric biopsy samples in about 1500 Linqu county residents annually according to the inclusion criteria (aged 40-69, male or non-pregnant, non-lactating female, informed consent form signed). The exclusion criteria are as follows: blood clotting disorders, high blood pressure, liver disease, chronic obstructive pulmonary disease and inability to provide an informed consent.
For the current study, a total of 140 H.pylori positive subjects who were both treated in the intervention trial and received endoscopic examinations prior to and six months after treatment were enrolled in a two-step self-control validation. We identified the efficacy of intervention in the 140 subjects by a second 13C-UBT after treatment as successful and unsuccessful eradication regardless of what kind of therapies they were assigned. The two-step self-control validation includes a preliminary self-control comparison (18 successfully and eight unsuccessfully eradicated cases) and a secondary self-control comparison (66 successfully and 48 unsuccessfully eradicated cases). In addition, 47 H.pylori negative and 47 positive subjects were randomly selected from the endoscopic examination participants with baseline H.pylori infection screening result in 2013 for case-control validation.
A written informed consent was obtained from each subject. The study was approved by Institutional Review Board of Peking University Cancer Hospital (2015KT67).
Upper endoscopic examination and histopathology
The detailed procedure of endoscopy has been described elsewhere [16]. Briefly, gastric mucosa was examined by experienced gastroenterologist using video endoscopes (Olympus). At least two biopsies were obtained from less curve of antrum, one for pathological diagnosis, one for DNA extraction and methylation detection. The gastric biopsy specimens were reviewed blindly by two pathologists according to the criteria proposed by the Chinese Association of Gastric Cancer [17] and Updated Sydney System [18]. Each biopsy was diagnosed as normal, superficial gastritis (SG), chronic atrophic gastritis (CAG), intestinal metaplasia (IM) or dysplasia (DYS) based on the most severe histology.
DNA preparation and methylation measurement
Blood leukocyte and gastric biopsy samples were frozen at -80 °C until DNA extraction. Genomic DNA was extracted from gastric biopsies using the QIAamp DNA Mini Kit (Qiagen, California, USA) and from blood leukocyte by standard proteinase K digestion and phenol-chloroform extraction. The extracted DNA was bisulfite-converted using EZ DNA methylation kit (Zymo Research) according to the manufacturer's instructions.
The methylation levels of the five candidate genes were detected by quantitative methylation-specific PCR using a StepOne Real-Time PCR System (Applied Biosystems) with the primers and probes as described in Table S1. The 20μl PCR reaction mixture contained 1×TaqMan Fast Universal PCR Master Mix (Applied Biosystems), 0.9 mmol/L each of forward and reverse primers, 0.25 mmol/L each of probes, and 20 ng of DNA at the following conditions: 95 °C for 20 seconds, followed by 40 cycles at 95 °C for one seconds, and 60 °C for 30 seconds. The efficiencies of all the genes PCR amplification were confirmed as nearly 100%. To normalize the input DNA, ACTB was used as the reference. The methylation levels of the candidate genes were calculated by dividing target gene/ACTB ratio of a sample by the target genes/ACTB ratio of positive control DNA treated by CpG methyltransferase (M.Sssl).
GNAS and MTERF1 methylation and mRNA expression in TCGA data
Because of the lack of GC cases and mRNA expression in our multi-stage validation, we accessed TCGA stomach cancer study data (http://www.xenabrowser.net) for the methylation and mRNA expression levels of GNAS and MTERF1 in GC cases. A total of 11 CpG sites in the promoter region of GNAS (from cg19640589 to cg21625881) and 15 CpG sites in MTERF1 (from cg10872641 to cg22709100) were selected from the Illumina HumanMethylation 450 DNA methylation data. The mRNA expression data were retrieved from Illumina HiSeq RNA-SeqV2 platform and log2 (RPKM+1) transformed. Other relevant clinical information was obtained including age, gender, pathological stage, H.pylori infection, targeted molecular therapy and radiation therapy.
Statistical analysis
Wilcoxon Singed Ranks Test was used to compare the methylation levels before and after anti-H.pylori treatment. Mann-Whitney test was used to compare age and methylation levels between H.pylori positive and negative groups. We compared methylation levels among SG, CAG and IM/DYS groups by Kruskal-Wallis test. Odds ratios (ORs) with corresponding 95% confidence intervals (CI) were calculated by unconditional logistic regression to evaluate the relationships between methylation status and H.pylori infection or gastric lesion groups adjusting for age, sex, smoking, drinking and baseline pathological diagnosis. Spearman correlation analysis was performed to calculate the correlation coefficients (r values) between methylation levels in blood leukocyte and gastric mucosa, or between methylation and mRNA expression levels. The associations between methylation or expression with overall survival were assessed using hazard ratio (HR) and corresponding 95%CI by COX regression model adjusting for age, gender, pathological stage, H.pylori infection, targeted molecular therapy and radiation therapy.
Results
Characteristics of the subjects for multi-stage validation
For the two-step self-control validation, the distributions of age, gender, cigarette smoking, alcohol drinking and baseline pathology for the participants are shown in Table S2. No statistical differences were found between successful and unsuccessful treatment groups both in preliminary and secondary self-control validations (all p>0.05). For case-control validation, there are higher frequencies of advanced gastric lesions (CAG and IM/DYS) in H.pylori positive group compared with negative group (17.0%, 40.4% v.s. 8.5%, 12.8%, p=0.001), although no differences were found in age, gender, cigarette smoking and alcohol drinking (Table S3).
Preliminary self-control validation for candidate aberrant methylation genes
For preliminary self-control validation, we detected GNAS, LTBR and DKK3 methylation levels in blood leukocyte and MTERF1, HAUS5 in both blood leukocyte and gastric tissue (Table 1). GNAS and DKK3 methylation medians were increased in blood leukocyte after successful H.pylori eradication (GNAS: 46.42% v.s. 56.54%, p=0.035, DKK3: 25.68% v.s. 34.51%, p=0.035). No significant methylation alteration was found for LTBR in blood leukocyte before and after eradication (36.40% v.s. 43.92%, p=0.306). MTERF1 methylation was markedly decreased by successful eradication both in blood leukocyte (56.79% v.s. 30.96%, p=0.001) and gastric mucosa (45.38% v.s. 36.84%, p=0.031). HAUS5 methylation level was increased significantly in gastric mucosa (49.57% v.s. 60.84%, p=0.048). However, the increasing trend showed no statistical significance in blood leukocyte (35.74% v.s. 40.12%, p=0.528). All of the candidate genes showed no significant methylation changes before and after unsuccessful treatment (all p>0.05).
Preliminary self-control validation for candidate aberrant methylation genes
| Gene name | Methylation level medians (interquartile)% in successful eradication group (n=18) | Methylation level medians (interquartile)% in unsuccessful eradication group (n=8) | ||||
|---|---|---|---|---|---|---|
| Before treatment | After treatment | p† value | Before treatment | After treatment | p† value | |
| In blood leukocyte | ||||||
| GNAS | 46.42 (34.74-53.49) | 56.54 (39.91-69.82) | 0.035 | 49.80 (38.54-57.87) | 57.88 (49.66-61.39) | 0.161 |
| LTBR | 36.40 (23.83-50.43) | 43.92 (24.01-67.00) | 0.306 | 41.61 (30.10-55.65) | 48.04 (35.39-59.25) | 0.889 |
| DKK3 | 25.68 (12.68-37.20) | 34.51 (30.64-45.89) | 0.035 | 31.60 (18.63-38.55) | 19.97 (16.55-33.62) | 0.161 |
| In gastric mucosa and blood leukocyte | ||||||
| MTERF1 | ||||||
| Blood leukocyte | 56.79 (38.65-78.88) | 30.96 (16.12-44.13) | 0.001 | 62.90 (43.85-79.23) | 42.91 (13.94-88.23) | 0.401 |
| Gastric mucosa | 45.38 (36.28-61.15) | 36.84 (30.32-55.53) | 0.031 | 26.44 (23.00-52.78) | 35.69 (30.15-40.23) | 0.575 |
| HAUS5 | ||||||
| Blood leukocyte | 35.74 (24.06-62.05) | 40.12 (33.51-57.28) | 0.528 | 34.41 (22.65-58.73) | 37.89 (21.43-45.49) | 0.208 |
| Gastric mucosa | 49.57 (42.55-63.10) | 60.84 (46.30-68.78) | 0.048 | 31.64 (13.25-52.46) | 41.78 (15.69-58.53) | 0.484 |
† Wilcoxon Singed Ranks Test, comparing the methylation level before and after anti-H.pylori treatment.
Case-control validation for candidate aberrant methylation genes
| Genes | Sample source | Methylation level median (quartile) % | p value† | |
|---|---|---|---|---|
| H.pylori negative group (n=47) | H.pylori positive group (n=47) | |||
| GNAS | Blood leukocyte | 62.07 (51.69-75.86) | 46.33 (36.25-54.24) | <0.001 |
| Gastric mucosa | 56.30 (42.96-66.55) | 32.42 (25.87-42.81) | <0.001 | |
| DKK3 | Blood leukocyte | 32.22 (21.77-42.12) | 24.36 (18.59-36.17) | 0.076 |
| Gastric mucosa | 34.81 (23.16-45.77) | 29.31 (23.13-39.12) | 0.354 | |
| MTERF1 | Blood leukocyte | 29.57 (18.64-37.74) | 56.02 (39.91-80.62) | <0.001 |
| Gastric mucosa | 31.10 (20.95-42.17) | 47.50 (35.12-62.89) | <0.001 | |
| HAUS5 | Blood leukocyte | 57.91 (43.11-72.82) | 56.43 (26.10-69.74) | 0.255 |
| Gastric mucosa | 49.06 (30.28-64.64) | 46.17 (32.09-60.69) | 0.443 | |
† Mann-Whitney test.
Case-control validation for candidate aberrant methylation genes
The four significant genes in preliminary self-control validation were compared between H.pylori positive and negative cases (Table 2). GNAS showed marked methylation decreasing both in blood leukocyte (62.07% v.s. 46.33%, p<0.001) and gastric mucosa (56.30% v.s. 32.42%, p<0.001) in H.pylori positive subjects compared to negative ones. In addition, MTERF1 methylation levels were increased significantly both in blood leukocyte (29.57% v.s. 56.02%, p<0.001) and gastric mucosa (31.10% v.s. 47.50%, p<0.001) of positive subjects compared to controls. No statistical differences of methylation levels were found for DKK3 and HAUS5 between H.pylori positive and negative groups (all p>0.05).
The correlations of the methylation levels between blood leukocyte and gastric mucosa were further analyzed in the 94 subjects. MTERF1 methylation in blood leukocyte is correlated with that in gastric mucosa (r=0.50, p<0.001). Similar correlation can also be found for GNAS methylation levels in blood leukocyte and gastric mucosa (r=0.28, p=0.007). However, no significant correlations were found for DKK3 and HAUS5 methylation between the two kinds of samples (both p>0.05).
According to the methylation medians of GNAS (53.23%) and MTERF1 (39.51%) in blood leukocyte of the total subjects, we used 50% and 40% as cut-off values and divided methylation levels into hyper- and hypo-methylation categories. Methylation status of GNAS was decreased (in blood leukocyte: OR, 0.16; 95%CI, 0.06-0.45, in gastric mucosa: OR, 0.09; 95%CI, 0.03-0.28) and MTERF1 was increased (in blood leukocyte: OR, 25.96; 95%CI, 6.56-102.73, in gastric mucosa: OR, 8.70; 95%CI, 2.84-26.66) in H.pylori positive subjects compared to controls adjusting for age, sex, smoking, drinking and gastric pathology (Table 3).
Secondary self-control validation for candidate aberrant methylation genes
We validated GNAS and MTERF1 methylation in a secondary self-control comparison with larger sample size (Table 4). In 66 pairs of blood leukocyte samples, GNAS methylation median was increased from 44.87% to 60.88% (p<0.001) and MTERF1 was decreased from 46.19% to 34.56% (p<0.001) by successful eradication. In the 37 pairs of gastric mucosa obtained from successful eradication group, similar increasing and decreasing methylation alterations were also found for the two genes (GNAS: 55.56% v.s. 69.47%, p=0.002; MTERF1: 35.95% v.s. 19.84%, p<0.001). No significant methylation alterations of the two genes were found in 48 pairs of blood leukocyte or 13 pairs of gastric mucosa samples collected from unsuccessful eradication group (all p>0.05).
The associations between aberrant methylation genes and gastric lesions
We further investigated the relationship of the aberrant methylation genes with gastric lesions by combining the 94 case-control subjects and the 114 baseline subjects of the secondary self-control validation (Table 5). The total subjects consist of 103 SG, 74 CAG and 31 IM/DYS. In the blood leukocyte samples, GNAS methylation median showed decreasing trend with the severity of gastric lesions from 52.81% in SG to 46.20% in IM/DYS, p=0.053. In the 144 subjects possessing gastric mucosa samples, GNAS methylation median was also marginally decreased from 55.68% in SG to 37.87% in IM/DYS, p=0.053. While, MTERF1 methylation levels showed no difference in various lesion groups (p=0.407 in blood leukocyte and 0.547 in gastric mucosa). Multivariate unconditional logistic regression found no significant relationship between advanced gastric lesions (CAG/IM/DYS) and GNAS or MTERF1 methylation in blood leukocyte or gastric mucosa, all p>0.05 (Table S4).
The correlation of methylation with mRNA expression and clinical relevance investigation in GC by TCGA database
We used TCGA-STAD database to investigate the correlation of methylation and mRNA expression in GC tissue. The average promoter methylation and mRNA expression levels of GNAS and MTERF1 were retrieved from 372 gastric adenocarcinoma cases. Spearman correlation analysis revealed inverse relationships between promoter methylation and mRNA expression for GNAS (r=-0.12, p=0.027) and MTERF1 (r=-0.55, p<0.001) in GC tissues.
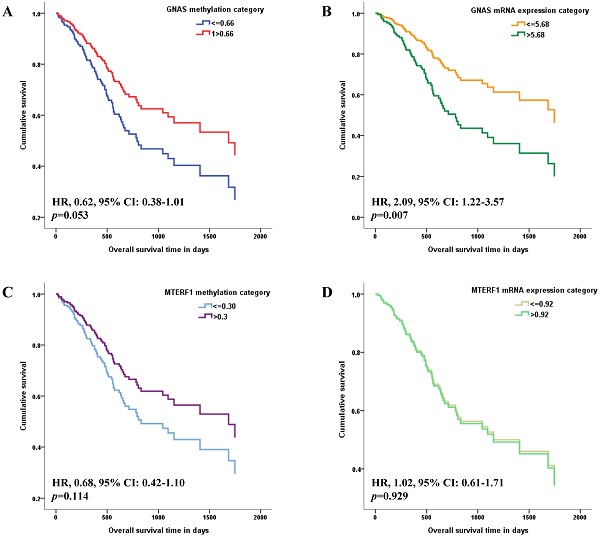
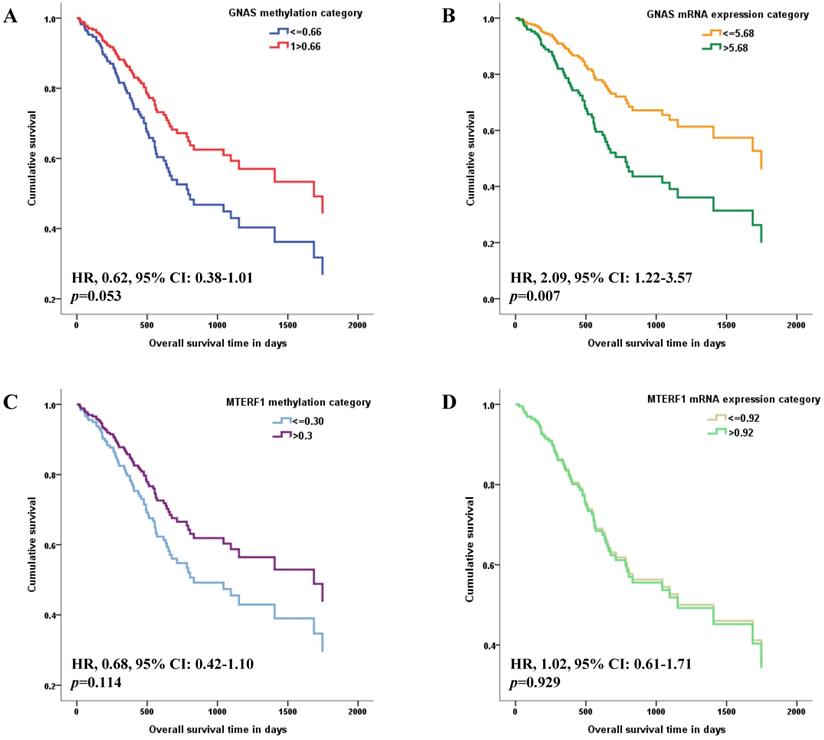
The methylation and mRNA expression levels of GNAS and MTERF1 were further evaluated with GC survival in TCGA-STAD. Methylation or mRNA expression medians for GNAS and MTERF1 in total subjects were used as cutoff values to divide GC cases into hyper- and hypo-methylation groups or higher and lower mRNA expression groups. Cox regression model found that the HR was 0.62 (95% CI: 0.38-1.01) for GC subjects with hypermethylated GNAS than hypomethylated subjects, although the p value showed no statistical significance (p=0.053, Figure 1A). In addition, the GC cases with higher GNAS mRNA expression levels showed significantly worse survival (HR, 2.09, 95% CI, 1.22-3.57, p=0.007) compared to lower expression subjects after adjusting for age, gender, pathological stage, H.pylori infection, targeted molecular therapy and radiation therapy (Figure 1B). No significant associations were found between GC survival and MTERF1 methylation (HR, 0.68, 95% CI: 0.42-1.10, p=0.114, Figure 1C) or mRNA expression (HR, 1.02, 95% CI: 0.61-1.71, p=0.929, Figure 1D).
Discussion
Depending on our previous genome-wide methylation profiles prior to and after anti-H.pylori treatment, the present study further validated that GNAS and MTERF1 methylation levels in blood leukocyte and gastric mucosa are correlated, and may be affected by H.pylori infection. Especially, GNAS may also be involved in advanced stage of GC development.
Case-control validation for candidate gene methylation status
| Genes | Sample source | Methylation status | H.pylori negative (n=47), N (%) | H.pylori positive (n=47), N (%) | OR (95% CI)† | p value † |
|---|---|---|---|---|---|---|
| GNAS | Blood leukocyte | Hypomethylation (≤50.00%) | 11 (23.4) | 29 (61.7) | 1.00 | |
| Hypermethylation (>50.00%) | 36 (76.6) | 18 (38.3) | 0.16 (0.06-0.45) | 0.001 | ||
| Gastric mucosa | Hypomethylation (≤50.00%) | 14 (29.8) | 39 (83.0) | 1.00 | ||
| Hypermethylation (>50.00%) | 33 (70.2) | 8 (17.0) | 0.09 (0.03-0.28) | <0.001 | ||
| MTERF1 | Blood leukocyte | Hypomethylation (≤40.00%) | 38 (80.9) | 12 (25.5) | 1.00 | |
| Hypermethylation (>40.00%) | 9 (19.1) | 35 (74.5) | 25.96 (6.56-102.73) | <0.001 | ||
| Gastric mucosa | Hypomethylation (≤40.00%) | 35 (74.5) | 14 (29.8) | 1.00 | ||
| Hypermethylation (>40.00%) | 12 (25.5) | 33 (70.2) | 8.70 (2.84-26.66) | <0.001 |
† Unconditional logistic regression analysis adjusted for age, sex, smoking, drinking and gastric pathology.
Secondary self-control validation for candidate gene methylation
| Methylation level medians (quartile)% | ||||||
|---|---|---|---|---|---|---|
| Successful eradication group | Unsuccessful eradication group | |||||
| Before treatment (n=66) | After treatment (n=66) | p† value | Before treatment (n=48) | After treatment (n=48) | p† value | |
| Blood leukocyte | ||||||
| GNAS | 44.87 (34.72-52.93) | 60.88 (48.17-70.67) | <0.001 | 47.50 (35.28-64.81) | 49.01 (39.80-65.93) | 0.356 |
| MTERF1 | 46.19 (34.62-69.71) | 34.56 (19.79-50.84) | <0.001 | 34.62 (27.97-46.07) | 40.68 (25.09-54.28) | 0.351 |
| Gastric mucosa | ||||||
| GNAS | 55.56 (39.74-69.45) | 69.47 (59.76-77.07) | 0.002 | 60.63 (58.40-77.88) | 60.25 (47.32-66.26) | 0.117 |
| MTERF1 | 35.95 (25.27-52.74) | 19.84 (15.61-31.81) | <0.001 | 41.73 (22.68-60.47) | 31.84 (22.32-55.40) | 0.753 |
† Wilcoxon Singed Ranks Test, comparing the methylation level before and after anti-H.pylori treatment.
Associations of candidate gene methylation and mRNA expression with overall survival of GC patients in TCGA database. The medians of methylation or mRNA expression levels for GNAS and MTERF1 in total subjects were used as cutoff values to divide GC cases into hyper- and hypo-methylation groups or higher and lower mRNA expression groups. Multivariate COX regression model found that overall survival is marginally better for GC subjects with hypermethylated GNAS promoter (A), and worse for GC subjects with higher GNAS mRNA expression levels (B) adjusting for age, gender, pathological stage, H.pylori infection, targeted molecular therapy and radiation therapy. However, no significant association was found between GC survival and MTERF1 methylation (C) or mRNA expression status (D).
Associations between candidate gene methylation and gastric lesions
| Genes | Methylation level medians (quartile)% | p value† | ||
|---|---|---|---|---|
| SG (n=103) | CAG (n=74) | IM/DYS (n=31) | ||
| Blood leukocyte | ||||
| GNAS | 52.81 (42.54-64.87) | 46.12 (36.28-53.78) | 46.20 (38.33-60.35) | 0.053 |
| MTERF1 | 37.40 (27.83-53.15) | 42.34 (30.37-62.27) | 42.09 (30.96-62.78) | 0.407 |
| Gastric mucosa | ||||
| GNAS | 55.68 (38.62-65.36) | 47.03 (32.42-62.03) | 37.87 (26.07-56.37) | 0.053 |
| MTERF1 | 35.85 (25.43-55.39) | 37.47 (23.93-49.88) | 40.31 (30.62-57.15) | 0.547 |
Abbreviations: CAG, chronic atrophic gastritis; DYS, dysplasia; IM, intestinal metaplasia; SG, superficial gastritis.
† Kruskal Wallis test.
In the preliminary self-control validation, the five candidate aberrant methylation genes were detected by quantitative methylation-specific PCR in 18 successful and eight unsuccessful H.pylori eradiation cases. Although the change of LTBR methylation shows no statistical significance, the methylation alterations of DKK3, GNAS, MTERF1 and HAUS5 are consistent with the previous methylation chip analysis [11], which suggests the high accuracy of our validation method.
H.pylori infection was previously reported to be associated with aberrant methylation in gastric mucosa or blood leukocyte DNA [19-21], however only few study evaluated the two kinds of samples at the same time. Our previous array analysis has compared the methylation levels between blood leukocyte and gastric mucosa in large-scale and found a high correlation [11], which suggests blood leukocyte methylation as a surrogate biomarker for gastric mucosa methylation. The present study further identified the decreasing GNAS and increasing MTERF1 methylation levels in both blood leukocyte and gastric mucosa of H.pylori positive subjects compared to negative controls. The correlated methylation levels in the two tissues support GNAS and MTERF1 methylation levels in blood leukocyte as potential surrogate biomarkers for H.pylori-associated methylation alterations in gastric mucosa. Chronic inflammation induced by H.pylori infection was reported to promote aberrant methylation in gastric mucosa [22], which may also be responsible for the correlated methylation alterations in blood leukocyte.
The relationship between H.pylori infection and methylation alterations can be further confirmed by anti-H.pylori intervention in addition to the case-control study. Some specific methylation changes (such as CDH1, COX-2, and LOX) were already reported in gastric mucosa after H.pylori eradication [23-25], which may be involved in gastric carcinogenesis. While no methylation alterations before and after eradication have been systemically evaluated both in gastric mucosa and in blood leukocyte. Our study identified that the hypomethylated GNAS and the hypermethylated MTERF1 in H.pylori positive subjects may be reversed by successful eradication in the two kinds of samples. Furthermore, no significant methylation changes in unsuccessful treatment group may also help to suggest the methylation alterations of GNAS and MTERF1 as the consequences of H.pylori infection.
The levels of aberrant methylation burden induced by H.pylori may increase with the period and density of the infection and finally accumulate in precancerous tissues producing an epigenetic field defect for cancerization [26]. Our validation identified that aberrant GNAS and MTERF1 methylation alterations can be induced by H.pylori infection. However, no independent relationships were found between GNAS or MTERF1 methylation levels and advanced gastric lesions (such as CAG or IM/DYS) in the present study. Further confirmation in a larger number of precancerous lesion subjects is still needed.
Although no GC patients were included in our case-control and self-control validations, we accessed TCGA data and found inverse relationships between promoter methylation and mRNA expression for GNAS and MTERF1 in GC tissues. Our result suggests that promoter methylation of GNAS and MTERF1 may serve as one of the mechanisms for expression regulation, although the detailed functions still need further investigation. Worse overall GC survival was found to be associated with higher expression of GNAS in GC patients, which may imply an important role of GNAS expression activation in the advanced stage of GC development. The hypomethylation induced by H.pylori may serve as the epigenetic prerequisite and provide a potential alternation to promote GNAS expression in addition to the well-studied activating mutations via the Wnt/β-catenin pathway or the ERK1/2 MAPK pathway [27, 28]. The correlated methylation alterations of GNAS in blood leukocyte and gastric mucosa may also suggest a potential non-invasive indicator for gastric epigenetic regulation, while the clinical significance still needs further validation.
The major strengths of our study lie in complementary design of combining case-control and self-control validations for H.pylori-associated differential methylation genes selected from the previous methylation chip screening. The significant H.pylori-associated methylation alterations were verified in blood leukocyte as well as in gastric mucosa, which may provide potential non-invasive indicator for gastric epigenetic regulation. A limitation in our study is the lack of GC subjects when we investigated the aberrant methylation status in various gastric lesions. However, TCGA database can provide us helpful information about gene methylation and expression in GC tissue to support our validation results.
For the five candidate methylation genes selected from our previous methylation chip screening, our case-control and self-control validations confirmed that GNAS and MTERF1 methylation levels may be affected by H.pylori infection both in gastric mucosa and in blood leukocyte. Methylation of GNAS may serve as one of the mechanisms for expression regulation. GNAS may be involved in advanced stage of GC development, although the possible mechanism still needs further study in precancerous lesions.
Abbreviations
13C-UBT: 13C-urea breath test; CAG: chronic atrophic gastritis; CI: confidence interval; DYS: dysplasia; GC: gastric cancer; HR: hazard ratio; H.pylori: Helicobacter pylori; IM: intestinal metaplasia; OR: odds ratio; SG: superficial gastritis.
Supplementary Material
Supplementary tables.
Acknowledgements
Funding
This work was supported by National Key R&D Program of China (2018YFC1313100), The third batch of public welfare development and reform pilot projects of Beijing Municipal Medical Research Institutes (Beijing Medical Research Institute, 2019-1), Beijing Natural Science Foundation (7182032), National Natural Science Foundation of China (81572774), A3 Foresight Program from Natural Science Foundation of China (30921140311), International (regional) Cooperation and Exchange Project (NSFC-DFG, 81861138041).
Competing Interests
The authors have declared that no competing interest exists.
References
1. de Martel C, Georges D, Bray F, Ferlay J, Clifford GM. Global burden of cancer attributable to infections in 2018: a worldwide incidence analysis. Lancet Glob Health. 2020;8:e180-e90
2. Chiba T, Marusawa H, Ushijima T. Inflammation-associated cancer development in digestive organs: mechanisms and roles for genetic and epigenetic modulation. Gastroenterology. 2012;143:550-63
3. Ushijima T. Epigenetic field for cancerization: its cause and clinical implications. BMC Proc. 2013;7(Suppl 2):K22
4. Hattori N, Ushijima T. Epigenetic impact of infection on carcinogenesis: mechanisms and applications. Genome Med. 2016;8:10
5. Ge A, Gao S, Liu Y, Zhang H, Wang X, Zhang L. et al. Methylation of WT1, CA10 in peripheral blood leukocyte is associated with breast cancer risk: a case-control study. BMC Cancer. 2020;20:713
6. Woo HD, Kim J. Global DNA hypomethylation in peripheral blood leukocytes as a biomarker for cancer risk: a meta-analysis. PLoS One. 2012;7:e34615
7. Chen D, Zhang XR, Zhang Y, Zhang L, Ma JL, You WC. et al. Hypomethylation of repetitive elements in blood leukocyte DNA and risk of gastric lesions in a Chinese population. Cancer Epidemiol. 2016;41:122-8
8. Hou L, Wang H, Sartori S, Gawron A, Lissowska J, Bollati V. et al. Blood leukocyte DNA hypomethylation and gastric cancer risk in a high-risk Polish population. Int J Cancer. 2010;127:1866-74
9. Zhang Y, Su HJ, Pan KF, Zhang L, Ma JL, Shen L. et al. Methylation status of blood leukocyte DNA and risk of gastric cancer in a high-risk Chinese population. Cancer Epidemiol Biomarkers Prev. 2014;23:2019-26
10. Su HJ, Zhang Y, Zhang L, Ma JL, Li JY, Pan KF. et al. Methylation status of COX-2 in blood leukocyte DNA and risk of gastric cancer in a high-risk Chinese population. BMC Cancer. 2015;15:979
11. Zhang Y, Zhang XR, Park JL, Kim JH, Zhang L, Ma JL. et al. Genome-wide DNA methylation profiles altered by Helicobacter pylori in gastric mucosa and blood leukocyte DNA. Oncotarget. 2016;7:37132-44
12. Pei Y, Tang Z, Cai M, Yao Q, Xie B, Zhang X. The E2F3/miR-125a/DKK3 regulatory axis promotes the development and progression of gastric cancer. Cancer Cell Int. 2019;19:212
13. Yang HJ, Kim SG, Lim JH, Choi JM, Kim WH, Jung HC. Helicobacter pylori-induced modulation of the promoter methylation of Wnt antagonist genes in gastric carcinogenesis. Gastric Cancer. 2018;21:237-48
14. You WC, Blot WJ, Chang YS, Ershow AG, Yang ZT, An Q. et al. Diet and high risk of stomach cancer in Shandong, China. Cancer Res. 1988;48:3518-23
15. Pan KF, Zhang L, Gerhard M, Ma JL, Liu WD, Ulm K. et al. A large randomised controlled intervention trial to prevent gastric cancer by eradication of Helicobacter pylori in Linqu County, China: baseline results and factors affecting the eradication. Gut. 2016;65:9-18
16. Guo Y, Zhang Y, Gerhard M, Gao JJ, Mejias-Luque R, Zhang L. et al. Effect of Helicobacter pylori on gastrointestinal microbiota: a population-based study in Linqu, a high-risk area of gastric cancer. Gut. 2020;69:1598-607
17. You WC, Blot WJ, Li JY, Chang YS, Jin ML, Kneller R. et al. Precancerous gastric lesions in a population at high risk of stomach cancer. Cancer Res. 1993;53:1317-21
18. Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am J Surg Pathol. 1996;20:1161-81
19. Michigami Y, Watari J, Ito C, Nakai K, Yamasaki T, Kondo T. et al. Long-term effects of H. pylori eradication on epigenetic alterations related to gastric carcinogenesis. Sci Rep. 2018;8:14369
20. Choi JM, Kim SG, Yang HJ, Lim JH, Cho NY, Kim WH. et al. Helicobacter pylori Eradication Can Reverse the Methylation-Associated Regulation of miR-200a/b in Gastric Carcinogenesis. Gut Liver. 2020;14:571-80
21. Zhou H, Sun H, Liu X, Chen J, Zhang L, Lin S. et al. Combined effect between WT1 methylation and Helicobacter pylori infection, smoking, and alcohol consumption on the risk of gastric cancer. Helicobacter. 2019;24:e12650
22. Schneider BG, Piazuelo MB, Sicinschi LA, Mera R, Peng DF, Roa JC. et al. Virulence of infecting Helicobacter pylori strains and intensity of mononuclear cell infiltration are associated with levels of DNA hypermethylation in gastric mucosae. Epigenetics. 2013;8:1153-61
23. Zhang Y, Zeng HM, Nie XR, Zhang L, Ma JL, Li JY. et al. Alterations of Cyclooxygenase-2 Methylation Levels Before and After Intervention Trial to Prevent Gastric Cancer in a Chinese Population. Cancer Prev Res (Phila). 2016;9:484-90
24. Shin CM, Kim N, Lee HS, Park JH, Ahn S, Kang GH. et al. Changes in aberrant DNA methylation after Helicobacter pylori eradication: a long-term follow-up study. Int J Cancer. 2013;133:2034-42
25. Kim HJ, Kim N, Kim HW, Park JH, Shin CM, Lee DH. Promising aberrant DNA methylation marker to predict gastric cancer development in individuals with family history and long-term effects of H. pylori eradication on DNA methylation. Gastric Cancer. 2021;24:302-13
26. Takeshima H, Niwa T, Toyoda T, Wakabayashi M, Yamashita S, Ushijima T. Degree of methylation burden is determined by the exposure period to carcinogenic factors. Cancer Sci. 2017;108:316-21
27. Nomura R, Saito T, Mitomi H, Hidaka Y, Lee SY, Watanabe S. et al. GNAS mutation as an alternative mechanism of activation of the Wnt/β-catenin signaling pathway in gastric adenocarcinoma of the fundic gland type. Hum Pathol. 2014;45:2488-96
28. Matsubara A, Sekine S, Kushima R, Ogawa R, Taniguchi H, Tsuda H. et al. Frequent GNAS and KRAS mutations in pyloric gland adenoma of the stomach and duodenum. J Pathol. 2013;229:579-87
Author contact
![]() Corresponding authors: Kai-Feng Pan, Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Cancer Epidemiology, Peking University Cancer Hospital & Institute, 52 Fucheng Rd, Haidian District, Beijing, 100142, P. R. China, Tel.: +86 10-88196701, Fax: +86 10-88122437, E-mail: pankaifeng2002com. Wei-Cheng You, Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Department of Cancer Epidemiology, Peking University Cancer Hospital & Institute, 52 Fucheng Rd, Haidian District, Beijing, 100142, P. R. China, Tel.: +86 10-88196761, Fax: +86 10-88122437, E-mail: weichengyoucom.
Corresponding authors: Kai-Feng Pan, Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Cancer Epidemiology, Peking University Cancer Hospital & Institute, 52 Fucheng Rd, Haidian District, Beijing, 100142, P. R. China, Tel.: +86 10-88196701, Fax: +86 10-88122437, E-mail: pankaifeng2002com. Wei-Cheng You, Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Department of Cancer Epidemiology, Peking University Cancer Hospital & Institute, 52 Fucheng Rd, Haidian District, Beijing, 100142, P. R. China, Tel.: +86 10-88196761, Fax: +86 10-88122437, E-mail: weichengyoucom.

 Global reach, higher impact
Global reach, higher impact