3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(24):7454-7464. doi:10.7150/jca.61618 This issue Cite
Review
The Regulatory Network of MicroRNA in the Metabolism of Colorectal Cancer
1. Department of Gastrointestinal Surgery, Shunde Hospital, Southern Medical University (The First People's Hospital of Shunde Foshan), Shunde, Foshan, Guangdong Province, 528300, China.
2. The Second School of Clinical Medicine, Southern Medical University, Guangzhou, Guangdong Province, 510080, China.
#These authors contributed equally to this work.
Received 2021-4-14; Accepted 2021-10-24; Published 2021-11-4
Abstract
Colorectal cancer (CRC) is the third most common malignant tumor in the world. During the progression of CRC, the entire metabolic network undergoes reprogramming, including marked changes in the regulation of glucose, lipid and amino acid metabolism. Although microRNAs (miRNAs) account for only 1% of the entire human genome, they play an important role in almost all physiological and pathological processes in the body. MiRNAs can react directly with key enzymes in the metabolic processes. MiRNAs also interact with other ncRNAs, as a member of non-coding RNA (ncRNA), to form their own regulatory network in various oncogenic pathways of CRC metabolism. The progression of colorectal cancer is closely related to the intestinal flora, where miRNAs act as important mediators. Understanding how miRNAs act in the regulatory network of CRC metabolism is helpful to elucidate the characteristics of tumor occurrence, proliferation, metastasis and drug resistance. This review summarizes the role of miRNAs in the metabolism of CRC and how miRNAs interact with key enzymes, ncRNA and intestinal flora to further discuss how miRNAs affect CRC and realize some new strategies for the early diagnosis and treatment of CRC.
Keywords: colorectal cancer, intestinal flora, metabolism, microRNA, molecular biomarkers, non-coding RNA, regulatory network, therapeutic targets
Introduction
Colorectal cancer is the second leading cause of cancer-related death worldwide and its incidence is increasing yearly [1,2], which is a multi-step, multi-stage and multi-gene involved cytogenetic disease [3]. In the progression of colorectal cancer, many factors contribute to dysregulation of the CRC signal pathway that promote tumor spread, metastasis and cause drug resistance [4]. The onset of CRC is insidious, and definitive diagnosis usually relies on colonoscopic biopsy and related tumor markers. Tumor markers play a role in heralding tumor recurrence or metastasis but lack specificity. Currently, the principal treatment options for CRC include surgical resection, radiotherapy, chemotherapy and immunotherapy. Improvements in these treatment methods have significantly impacted and enhanced the prognosis of CRC, especially in early-stage disease. Even with the notable advances in the management of CRC, further progress is both achievable and necessary. Therefore, it is important to search for molecular markers that can more effectively detect tumor progression, but perhaps even more importantly, understand their role in CRC and their specific actions in their regulatory networks. The metabolism of CRC mainly includes glycolysis, lipid metabolism and amino acid metabolism. The metabolic reprogramming of CRC represents changes made by tumor cells to adapt to the environment. And the key enzymes and signaling pathways involved in the metabolic pathways have become a hot topic in CRC study.
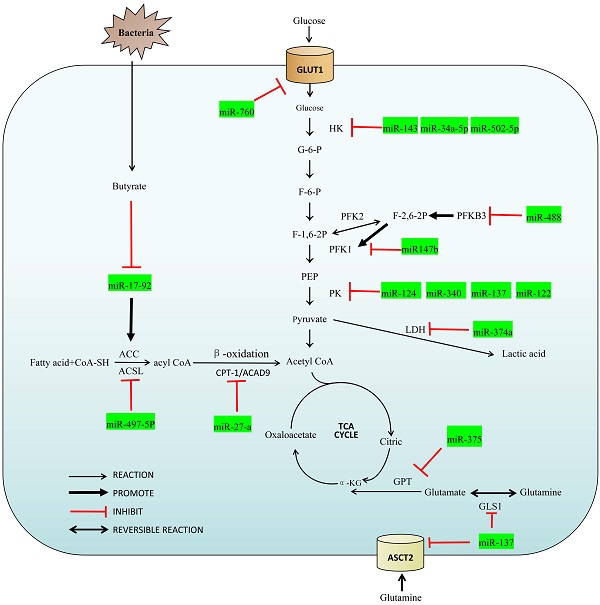
MicroRNAs, members of the non-coding RNA family, are single-stranded RNA molecules of about 22 nucleotides in length. Although miRNAs account for only 1% of the entire human genome, they play an important role in the regulatory network for colorectal cancer [5,6]. They can be coupled completely or incompletely by miRNA response elements (MRE) to the 3' untranslated region (3' UTR) of the target mRNA that drives the RNA induced silencing complex (RISC) to degrade the target mRNA, and inhibit expression of the target mRNA at the post-transcriptional level [5,7]. MiRNAs can also interact with other ncRNAs to form their own regulatory effects in various oncogenic CRC pathways [8]. In addition, we also summarized the relationship between miRNA and intestinal flora metabolism, which is a special part of colorectal tumors. With this overview of their influence on metabolic pathways, a more in-depth description of these and how miRNAs can be utilized in the molecular diagnosis, treatment and postoperative monitoring. A summary of the miRNAs and their target enzymes as well as other molecular interactions are shown in Figure 1 and Table 1 respectively.
Overview of the miRNAs in the regulatory network of colorectal cancer.
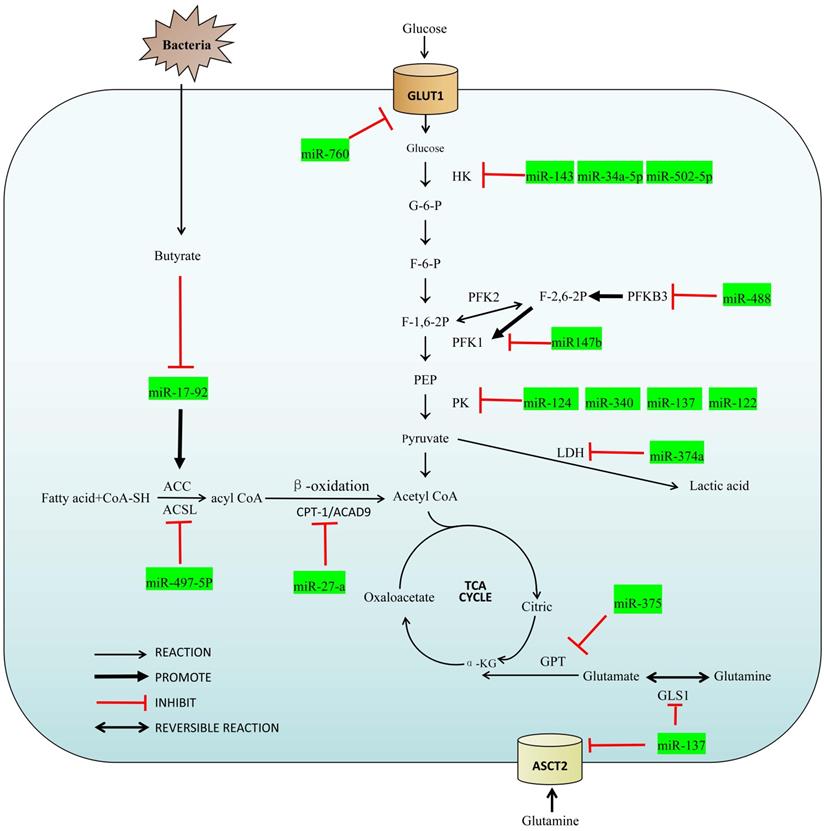
List of miRNAs involved in metabolism and its molecular interaction of CRC
| MicroRNA | Expression in CRC | Target/Pathway | Description | Effect on CRC | Sample | Refs |
|---|---|---|---|---|---|---|
| miR-143 | ↓ | HK2 | Inhibit aerobic glycolysis | Proliferation | Cell line | [21] |
| miR-513a-3p | ↓ | HK2 | Inhibit aerobic glycolysis | Proliferation | Cell line | [24] |
| miR-488 | ↓ | PFKFB3/PFK-1 | Inhibit aerobic glycolysis | Proliferation, migration and invasion | CRC tissues and cell line | [25] |
| miR-124 | ↓ | PKM2, PKM1 | Inhibit aerobic glycolysis, inhibited by lncRNA HNF1A-AS1 and circHIPK3 | Cancer growth | Cell line | [29,62] |
| ↓ | MYO6/HK2 | Migration and invasion | CRC tissues, paracancerous tissues and cell line | [54] | ||
| miR-137 | ↓ | PKM2, PKM1 | Inhibit aerobic glycolysis Suppress glutamine uptake and utilization | Cancer growth | Cell line | [29,40] |
| ↓ | ASCT2, GLS1 | Tumorigenesis | Cell line | |||
| miR-340 | ↓ | PKM2, PKM1 | Inhibit aerobic glycolysis | Cancer growth | Cell line | [29] |
| miR-16-5p/miR-15b-5p | ↑ | ALDH1A3/PKM2 | Inhibit aerobic glycolysis | Cancer growth and apoptosis | Cell line | [31] |
| miR-374a | ↓ | LDHA | Suppress lactate production | Cell growth and tumorigenesis | Cell line | [32] |
| miR-497-5p | ↓ | ACSL5 | Suppress fatty acyl-CoA production | Proliferation, metastasis and invasion | CRC tissues and cell line | [34] |
| miR-27a | ↑ | PGC-1α, PPARγ, CPT1A and ACAD9 | Inhibit fatty acids oxidation | Proliferation, migration, invasion and drug resistance | Cell line | [35] |
| miR-375 | ↓ | GPT2 via PI3K/Akt pathway | Suppress glutamine utilization | Cancer growth | Cell line | [42,43] |
| miR-34a-5p | ↓ | HK1 | Inhibit aerobic glycolysis, can be inhibited by lncARSR | Invasion and metastasis | Cell line | [23] |
| miR-147b | ↓ | NDUFA4, PFK-1, PKM2 | Inhibit aerobic glycolysis, can be inhibited by lncRNA MAFG-AS1 | Proliferation | Cell line | [55] |
| miR-222-3P | ↑ | PTEN/PI3K/Akt | Promote metabolic reprogramming, can be inhibited by IncRNA GAS5 | Migration and invasion | Cell line | [56] |
| miR-760 | ↓ | GLUT1 | Suppress glucose transport, can be inhibited by circDENND4C | Proliferation and migration | Cell line | [17] |
| miR-502-5p | ↓ | MYO6/HK2 | Inhibit aerobic glycolysis, can be inhibited by Hsa_circ_0000231 | Proliferation, migration and invasion | CRC tissues, paracancerous tissues and cell line | [22] |
| miR-455-3p | ↓ | MYO6 | Inhibit aerobic glycolysis, can be inhibited by circCSNK1G1 | Cell growth and metastasis | CRC tissues, paracancerous tissues and cell line | [59] |
| miR-7 | ↓ | IGF1R via PI3K/Akt pathway | Inhibit metabolic reprogramming, can be inhibited by circHIPK3 | Proliferation, migration and invasion | Cell line | [62] |
| miR-637 | ↓ | Beclin-1 via STAT3 pathway | Inhibit the autophagy of tumor cell, can be inhibited by circHIPK3 | Cell growth and apoptosis | CRC tissues, paracancerous tissues and cell line | [63] |
| miR-122 | ↓ | PKM2 | Inhibit aerobic glycolysis, can be inhibited by ciRS-122 | Apoptosis | Cell line | [64] |
| miR-133a | ↓ | GEF-H1/RhoA | Promote tumor invasion and metastasis, can be inhibited by circ-133 | Metastasis | Cell line | [66] |
| miR-17-92 | ↑ | ACC, HMGCR via LKB1/AMPK pathway | Promote the synthesis of fatty acids and cholesterol, can be inhibited by butyrate | Proliferation | Cell line | [74,75] |
| miR-106b | ↑ | p21 | Promote the proliferation of tumor cell, can be inhibited by butyrate | Proliferation | Cell line | [78] |
| miR-203 | ↓ | HEF1 | Inhibit tumor invasion and metastasis, can be promoted by butyrate | Proliferation, invasion, apoptosis | Cell line | [79] |
MicroRNA and glucose metabolism
Metabolic reprogramming is a hallmark of malignancy, and treatment resistance and tumor metastasis are facilitated as a consequence of these altered pathways [7]. In the metabolic reprogramming of colorectal cancer, the abnormal performance of glucose metabolism is particularly prominent. Cancers still conduct glycolysis, but at a rate 200 times that of normal cells. This is necessary because cancers rely principally on glycolysis for energy production, yielding only 2 ATP, a rather inefficient method of energy production [9,10]. In contrast, normal cells finish the process (Krebs cycle and the electron transport chain) that generates about 36 ATP. This dramatic reliance on aerobic glycolysis is named the Warburg effect [11,12], and is seen in almost all solid tumors cells. Current research regarding miRNA on the basis of this abnormal tumor-related aerobic glycolysis may provide new ideas for the early diagnosis and therapies of colorectal cancer [13]. CRC regulates the uptake and utilization of glucose by adjusting the activity of glucose metabolism-related receptors or enzymes [14]. Following, we summarize enzymes and transporter involved in the regulation of miRNA during glucose metabolism.
Glucose transporter 1
Glucose transporter 1 (GLUT1) is a transmembrane protein on the cell membrane and a key transporter for the metabolism of glucose in mammalian cells, which is usually up-regulated in tumor cells [15]. Extracellular glucose cannot pass freely through the cell membrane, it must be entered into the cell with the help of GLUT1 to participate in glucose metabolism. Santasusagna et al. [16] found that miR-328 can bind to the SLC2A1-3'UTR and reduce the level of GLUT1 through SLC2A1/ GLUT1 pathway in CRC. Zhang et al. [17] found that GLUT1 was the target of miR-760, and the level of GLUT1 increased when miR-760 was decreased. Both miR-328 and miR-760 are negative correlation with the level of GLUT1 so that the up-regulate of these miRNAs can consequently down-regulate GLUT1-mediated glycolytic activity in CRC.
Hexokinase
The function of Hexokinase (HK) is to convert glucose into G-6-P, which is a rate-limiting enzyme in glycolysis. There are four important subtypes of mammalian hexokinase, named HK1 to HK4. In particular, HK2 is dysregulated in a variety of cancers including CRC and is often found upregulated in CRC as to adapt to the high rate of glucose metabolism necessary [18-20]. Gregersen et al. [21] found miRNA-143 can target the HK2-3'UTR to reduce its expression in colorectal cancer. Liu et al. [22] found miR-502-5p inhibited the expression of HK2 and Li et al. [23] found miR-34a-5 inhibited the expression of HK1. These miRNAs are all impeded aerobic glycolysis in CRC by inhibit the expression of HK and act as a tumor suppression factor. Chen et al. [24] found low expression of miR-513a-3p in CRC cells increased HK2 expression and promoted proliferation and metabolism in colorectal cancer cells.
Phosphofructokinase-1
The function of Phosphofructokinase-1 (PFK-1) is to convert G-6-P into fructose-1,6-bisphosphate (F-1,6-2P) and it is also a rate-limiting enzyme in glycolysis. The function of Phosphofructokinase-2 (PFK-2) is to convert G-6-P into fructose-2,6-bisphosphate (F-2,6-2P) which is a strong activator of PFK-1. PFKFB3, a protein expressed by pfkfb3 gene and often highly expressed in CRC, can promote the synthesis of F-2,6-2P. Deng et al. [25] found PFKFB3 may be downstream of miR-488 and is negatively regulated by miR-488. The increasing expression of PFKFB3 can reverse the inhibition of miR-488 on glycolysis suggesting that the miR-488/ PFKFB3 axis may stimulate FPK1 by influencing F-2,6-2P, and ultimately affect the glycolysis process.
Pyruvate kinase
Pyruvate kinase (PK) is to covert phosphoenolpyruvate (PEP) to pyruvate and it is the third rate-limiting enzyme in glycolysis including PKM1 and PKM2. PKM1 is expressed in most well-differentiated tissues and mainly promotes oxidative phosphorylation. PKM2, however, is only expressed in rapidly proliferating cells such as embryonic cells and cancer cells, and can promote glycolysis even under aerobic conditions [26-28]. Sun et al. [29] and Clower et al. [30] found that miR-124, miR-137 and miR-340 could down-regulate the selective splice protein acting on the PKM gene (PTB1/hnRNAPA1/ hnRNAPA2), thereby promoting the conversion of PKM2 to PKM1. High ratios of PKM1/PKM2 can inhibit aerobic glycolysis so that miR-124, miR-137 and miR-340 can act as regulator of glucose metabolism in CRC. Huang et al. [31] found that CuET, as a metabolite of an alcoholism treatment drug DSF, is capable of regulating the miR-16-5p, miR-15b-5p/ALDH1A3/PKM2 axis. CuET inhibits ALDH1A3, an important isoform of the aldehyde metabolic enzyme system ALDH family, by selectively enhancing expressions of miR-16-5p and miR-15b-5p and then inhibits the ubiquitination of PKM2, so as to alleviate the glycolysis of CRC cells.
Lactate dehydrogenase
Lactate dehydrogenase (LDH) is to convert pyruvate to lactic acid, lactate levels in cells are also an important factor regulating glycolysis. Janney et al. [32] found that LDHA expression in CRC, one isoform of LDH, might have a negative correlation with miR-34a, miR-34c, miR-369-3p, miR-374-a and miR-4524-a/b expression. The 3'UTR region of LDHA relative gene can match with miR-374a. When miR-374a is overexpression in CRC, the decrease of LDHA cells will lead to the accumulation of pyruvic acid. Thus, miRNA regulate glycolysis by affecting the conversion of pyruvate to lactic acid.
MicroRNA and lipid metabolism
Lipid metabolism, an important and complex biochemical reaction in the body, is the process of digestion, absorption, synthesis and decomposition of lipid in cells. Lipid metabolism not only provides energy, but lipids are also an integrate component of the cell membrane. MiRNAs frequently act on enzymes in lipid metabolism, are related to the proliferation and progression of tumor cells, so the regulatory network for lipid metabolism is summarized below.
Acyl-CoA synthetase
Fatty acids must be activated before being oxidized to generate acyl-coenzyme A (acyl CoA) under the catalysis of Acyl-CoA synthetase (ACS). ACLS5, one of the subtypes of ACS, is highly expressed in colorectal cancer and has been proved to be regulated by miRNA[33]. Gharib et al. [34] found that miR-497-5p can target the ACSL5-3'UTR and reduce its expression in colorectal cancer so that fatty acids cannot synthesize acyl-CoA normally, resulting in abnormal β-oxidation. MiR-497-5p reduces the energy produced by colorectal cancer from lipid metabolism by inhibiting ACSL, suppressing the development of CRC. This has also stimulated a new treatment strategy: the starvation therapy.
Carnitine palmitoy transterase-1 and Acyl-CoA Dehydrogenase 9
Carnitine palmitoy transterase-1 (CPT-1) and Acyl-CoA Dehydrogenase 9 (ACAD9) are the two important enzymes in β-oxidation. Barisciano et al. [35] found that CPT-1 and ACAD9 may be potential targets for miR-27a. When colorectal cancer cells have miR-27a knockout, the levels of both proteins increased; in contrast, both proteins decreased with overexpression of miR-27a. Of note, the effect of miR-27a on glucose metabolism was reviewed previously, and it can also regulate lipid metabolism in CRC [36]. Thus, the regulation of miR-27a on metabolic reprogramming can affect tumor invasiveness and drug resistance in CRC.
MicroRNA and amino acid metabolism
Amino acid metabolism has been a hot spot in cancer research recently. When tumor cells proliferate and synthesize proteins rapidly, a large number of essential and non-essential amino acids are required [37]. In the metabolic regulation network of CRC, glutamine is an important molecule. It can be converted into α-ketoglutaric acid (α-KG) to enter the tricarboxylic acid cycle (TCA cycle, Krebs cycle) as a key replenishment component of the TCA. Similarly, α-KG is also a raw material for the synthesis of nucleotides, fatty acids and other non-essential amino acids [38]. Several enzymes and the glutamine transporter participate in this process are regulated by miRNA.
ASC family transporter 2
ASC family transporter (ASCT) is a glutamine transporter, which mainly transports extracellular glutamine into cells and participates in amino acid metabolism. Among them ASC family transporter 2 (ASCT2, also known as SLC1A5) is generally up-regulated in human cancers and involved in metabolic reprogramming [39]. Dong et al. [40] found that the level of ASCT2 in most tumor cells, including CRC cells, is regulated by miR-137. MiR-137 can directly target the ASCT2-3'UTR, resulting in down-regulation of ASCT2 expression, thereby reducing the uptake of glutamine in CRC.
Glutaminase 1
Glutaminase 1 (GLS1) is an enzyme that catalyzes the formation of glutamate from glutamine. It has been previously noted that miR-137 can inhibit ASCT and, in addition, it can also reduce the GLS1 levels. Li et al. [41] found that miR-137 may be a GLS1-targeting miRNA, regulated by HSF1. The miR-137 can be inhibited by HSF1 because of promoter methylation of the miR-137 upstream gene mir137hg that then reduces expression of miR-137. Inhibition of GLS1 by miR-137 therefore inhibits the decomposition and transformation of glutamine. Because glutamine is very important to energy supply and molecular synthesis, miR-137 reduced the growth of CRC cells through this regulatory network.
Glutamic pyruvic transaminase
Glutamic pyruvic transaminase (GPT) converts glutamate to α-ketoglutaric acid, which is a critical component of the TCA. CRC cells with mutations in pik3ca have a higher requirement for glutamine due to the catalytic subunit P110α. This change results in an increase in the expression of glutamic pyruvic transaminase 2 (GPT2) as an adaptation to the energy requirement for the rapid growth and proliferation of CRC [42]. The miR-375 can bind to the PIK3CA-3'UTR and inhibit the expression of PIK3CA in CRC, thereby inhibiting its growth through the PI3K/Akt pathway [43]. The lower expression of PI3K leads to a consequent decrease in the expression of GPT2, thus limiting the utilization of glutamine in CRC.
MicroRNA and other non-coding RNA
Non-coding RNAs can be linked to the regulation network of tumor cell metabolism, proliferation, metastasis and apoptosis through miRNA [8]. These regulatory non-coding RNAs include small non-coding RNA (sncRNA), long non-coding RNA (lncRNA), and circle RNA (circRNA). Some lncRNAs and circ RNAs act as competitive endogenous RNAs (ceRNA) to affect the post-transcriptional regulation of genes by binding to miRNA directly. They usually have miRNA binding sites and play a role of miRNA sponges to produce the corresponding biological effects [44-46]. A deep understanding of the regulatory network for non-coding RNA interactions may help to elucidate the role of miRNA in the regulatory network and provide a new direction for the prevention, early diagnosis and therapy of CRC. The following section focuses on the role of these ncRNAs and their interaction in the lncRNA-miRNA-mRNA regulatory network.
MicroRNA and lncRNA
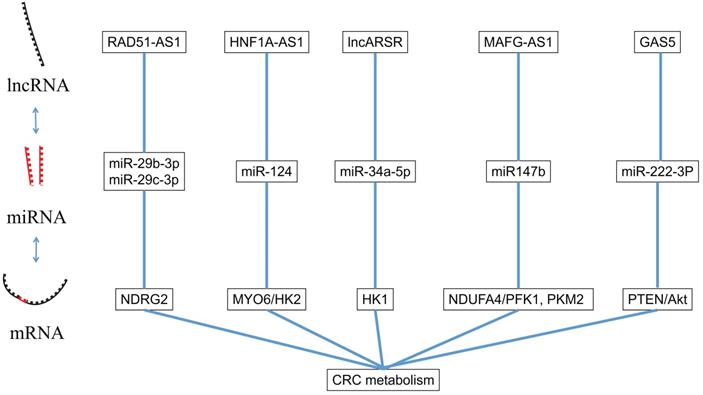
LncRNA is a kind of RNA molecule with a length of 200~100000nt and its regulation in colorectal cancer is varied [47]. LncRNA regulates the level of chromatin in the nucleus [48], and also regulates the transcription of its adjacent mRNA through the cis-acting element [49]. In the following sections, we will summarize several lncRNAs involved in the proliferation, apoptosis, invasion and migration of CRC (Figure 2).
LncRNA RAD51-AS1
The dysregulation of LncRNA RAD51-AS1 has been reported as closely related to a variety of cancers [50]. Li et al. [51] found that LncRNA RAD51-AS1 acts as a tumor suppressor in CRC, yet LncRNA RAD51-AS1 can also affect the glycolysis of CRC by affecting HK2 and GLUT. Furthermore, LncRNA RAD51-AS1 can bind to miR-29b-3p and miR-29c-3p through sponge effect and up-regulates NDRG2, which has been reportedly involved in the regulation of glycolytic metabolism [52]. LncRNA RAD51-AS1 exerts its tumor suppressor effect through miR-29b/c-3p/NDRG2, and inhibits the proliferation, invasion and aerobic glycolysis of CRC.
Hepatocyte nuclear clear factor receptor 1 homeobox A-antisense RNA 1
In colorectal cancer tissues and cell lines, the expression of lncRNA hepatocyte nuclear clear factor receptor 1 homeobox A-antisense RNA 1 (HNF1A-AS1) was significantly upregulated [53]. Guo et al. [54] found that miR-124 was a target of HNF1A-AS1, and MYO6 was a target mRNA of miR-124 in CRC cells. The reduction of HNF1A-AS1 or up-regulation of miR-124 resulted in a similar biological effect: inhibition of migration and invasion of CRC cells and glycolysis. Further experiments demonstrated that down-regulation of miR-124 promoted expression of MYO6, which was positively associated with HK2 in CRC cells. Therefore, HNF1A-AS1 can influence cell migration, invasion, and glycolysis by regulating the miR-124/ MYO6 axis of colorectal cancer cells.
LncARSR and MAFG-AS1
Li et al. [23] found that lncARSR could promote the expression of HK1 in colorectal cancer through target inhibition of miR-34a-5p, and Cui et al. [55] found that lncRNA MAFG-AS1 could promote the expression of PFK1 and PKM2 in colorectal cancer through the miR-147b/NDUFA4 axis. These lncRNAs can regulate the key enzymes of glycolysis in CRC through miRNA.
Growth arrest-specific 5
Liu et al. [56] found that lncRNA growth arrest-specific 5 (GAS5) can competitively bind to miR-222-3P, which can target the PTEN-3'UTR gene and inhibit its expression. PTEN is an anti-oncogene gene which was reported to be related to cancer cell proliferation and migration. The tumor suppressor protein expressed by PTEN can dephosphorylate Akt and reduce its activation. It is a negative regulator of the PI3K/Akt pathway which can prevent all downstream signaling events regulated by Akt [57]. Therefore, lncRNA GAS5 led to the inactivation of Akt through miR-222-3P/ PTEN axis and affected the proliferation and migration of CRC.
The interaction between miRNA and lncRNA.
The interaction between miRNA and circRNA.
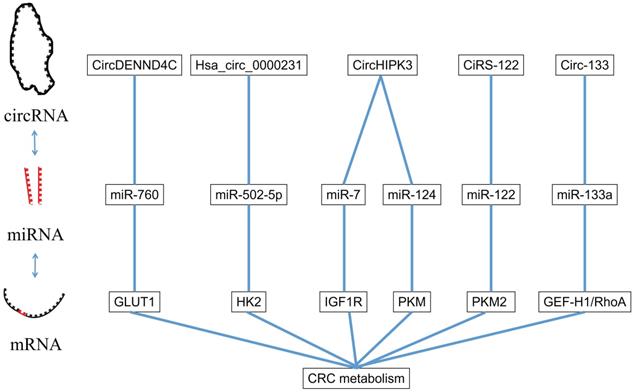
MicroRNA and circRNA
CircRNA is a kind of special RNA molecule with a closed ring structure, without either a 5'-end or 3'-end. It is not affected by RNA exonuclease and its expression is more stable and it is not easily degraded [58]. The biological function of circRNA in the human body was gradually revealed with the development of high-throughput sequencing technology. The circRNA acts as a ceRNA to target the downstream gene of miRNA. In the following sections, we will summarize several circRNAs involved in the proliferation, apoptosis, invasion and migration of CRC and the pathway they involve (Figure 3).
CircDENND4C
We have demonstrated that miR-760 can act as a regulator in the glycolysis by targeting the GLUT1, and circDENND4C can also take part in the miR-760/GLUT1 axis. Zhang et al. [17] proved that circDENND4C could target miRNA-760, reduce its inhibitory effect on GLUT1 by the action of a miRNA sponge, and finally promote glycolysis in CRC cells.
Hsa_circ_0000231
MYO6 is a key substance that links circRNA, miRNA and glucose metabolism in CRC. Huang et al. [59] found that Hsa_circ_0000231 could compete with miR-502-5p as a ceRNA. The miR-502-5p can promote the expression of MYO6 which is positively correlated with HK2 in CRC cells. So Hsa_Circ_0000231 acts on MYO6 through miR-502-5p to regulate glycolysis in colorectal cancer.
CircHIPK3
Transcription factor c-Myb regulates metabolism, proliferation, and metastasis in CRC through a variety of signaling pathways [60]. It has been shown that c-Myb can improve the transcription of circHIPK3 in colorectal cancer [61-63]. Zeng et al. [61] found that miR-7 could inhibit the expression of insulin-like growth factor receptor 1 (IGF1R) in CRC cells, whereas overexpression of circHIPK3 could effectively reverse the protective effect of miR-7 in CRC. IGF1R mediates metabolic changes, cell proliferation, and invasion and metastasis of colorectal cancer cells by activating PI3K/Akt pathway, which has been reported to be related to cancer cell metabolism, proliferation and migration. Zheng et al. [62] found that circHIPK3 could also competitively inhibited miR-124, thereby inhibiting the PKM2 to PKM1 transition, ultimately promoting glycolysis in CRC. Zhang et al. [63] found that circHIPK3 inhibits autophagy in CRC cells by competitively binding to miR-637, activating the STAT3 signaling pathway, increasing Bcl-2 expression, and blocking Beclin-1 expression, resulting in increased drug resistance to oxaliplatin based chemotherapy.
CiRS-122
CircRNA is widely present in exosomes due to its structural stability and achieves connection to the regulatory network between CRC cells through its miRNA sponge effect. Wang et al. [64] found that exosomes from colorectal cancer cells that have developed oxaliplatin resistance can promote glycolysis in chemotherapy-sensitive cells and reduce their drug sensitivity. These exosomes contain ciRS-122, which can be transmitted to sensitive cells to inhibit miR-122 through the miRNA sponge effect. The miR-122 can bind to the PKM2-3'UTR and reduce PKM2 expression, resulting in upregulation of glycolysis, thus producing resistance to oxaliplatin.
Circ-133
Oxygen supply in CRC is unbalanced, resulting in altered metabolic balance within tumor cells, and the development of hypoxic and normoxic cancers [65]. This difference results in an environment that is not conducive for hypoxic cancer cells to proliferate and grow. However, a strong regulatory network for colorectal cancer appears to alter this condition. Yang et al. [66] found that tumor cells derived from an environment of low partial pressure of oxygen, were able to secrete exosomes containing circ-133. These exosomes promoted the invasion and metastasis of tumor cells by acting on the miR-133a/GEF-H1/RhoA signaling pathway in tumor cells derived from normal partial pressure of oxygen. This intercellular regulation redistributes oxygen resources in the TME and promotes the invasion and metastasis of CRC.
MicroRNA and intestinal flora metabolism
The development of colorectal cancer is closely related to the intestinal flora [67]. High levels of dietary fibre have been reported to reduce the risk of colon cancer in humans and animal models [68], attributed to short-chain fatty acids (SCFA) [69,70]. Butyrate is one of the major components of SCFAs, involved in the metabolism of normal colorectal epithelial cells, and it is an important component of energy metabolism [71,72]. In CRC cells, most of the butyrate is not metabolized due to the Warburg effect. The accumulated butyrate can act as a histone deacetylase inhibitor (HDAC), which is involved in regulating the metabolism, proliferation and apoptosis of tumor cells [73].
miR-17-92
Hu et al. [74] found that butyrate can decrease the expression of c-Myc and hinder the combination of c-Myc and E-box elements (E3) in the core promoter region, resulting in a decreased expression of miR-17-92. Izreig et al. [75] found that miR-17-92 binds to the 3'-UTR region of tumor suppressor gene liver kinase B1 (LKB1). LKB1 can rapidly inactivate acetyl-CoA carboxylase (ACC) and HMG-coenzyme A reductase (HMGCR) by activating AMP-dependent protein kinase (AMPK), thus inhibiting the synthesis of fatty acids and cholesterol in tumor cells, but also slightly upregulating the activity of PFK-1 and promoting glycolysis [76]. Thus, butyrate from the intestinal flora can remodel the metabolic regulatory network of CRC through negative regulation of the c-Myc/miR-17-92 axis, reduce its energy source overall, resulting in inhibition of tumorigenesis.
miR-106b
In addition, butyrate can also play an anti-tumor role by regulating the proliferation, metastasis and apoptosis of colorectal cancer through miRNA. p21 is a cyclin dependent kinase (CDK) inhibitor located downstream of p53. It can form a G1 checkpoint in the cell cycle with p53, thus reducing the replication and accumulation of damaged DNA, thereby exerting an anti-cancer effect [77]. Hu et al. [78] found that butyrate increased p21 protein expression by inhibiting miR-106b.
miR-203
HEF1 is a focal adhesion protein that proved to be important in the regulation of the CRC cell. Han et al. [79] found that miR-203 inhibits butyrate by promoting miR-203 expression, thereby prolonging the colorectal cancer cell cycle by inhibiting HEF1 and tumor cell colony formation. Although butyrate has been shown to have excellent antitumor effects, its short half-life and low bioavailability still limit its clinical application.
Conclusions and perspectives
Colorectal cancer is the third most common tumor worldwide, the second leading cause of cancer-related death, and its incidence is increasing yearly. It is valuable and becoming increasingly necessary to have a comprehensive understanding of miRNAs in the CRC regulatory network. In the review, we summarize the role and regulation of CRC-related miRNAs in the metabolism and the molecular interaction between miRNAs and other ncRNAs. Additionally, the regulatory network of miRNAs in intestinal flora metabolism is also included.
The research of miRNAs has been carried out for many years and has made excellent progression in drug application and biomarkers for rapid diagnosis. However, the research on its molecular mechanism is still relatively insufficient. Regarding the research on the mechanism of the regulatory network in miRNA and CRC metabolism is mostly based on the complete or incomplete complementary pairing with the mRNA of related metabolic genes after the miRNA binds to the AGO protein to form a RISC complex, thereby mediating its degradation or inhibiting translation. However, this mechanism is applicable mainly to miRNAs distributed in the cytoplasm. Other mechanisms in CRC metabolisms, such as interactions with other RNA binding proteins, negative regulation of RNA precursor transcripts, combine with the promoter in regulation transcription levels and mediate the distribution of RNA inside and outside the nucleus. However, the current research on this area is still not deep enough. More molecular mechanisms of miRNA in CRC metabolism need to be further elucidated.
In clinical treatment, the diagnosis of CRC mainly relies on biopsy under colonoscopy and the treatment is mainly surgery, supplemented by radiotherapy, chemotherapy, and targeted therapy. Although the accuracy of pathological diagnosis is very high at present, the surgical treatment of colorectal cancer is becoming more and more mature in technology, and there are still some patients who cannot benefit from it. Lack of specific diagnostic methods due to its insidious symptoms, many CRC patients are already in the advanced stage at the time of diagnosis. Drug therapy is the primary treatment for unresectable stage and extensively metastasized patients, but tumor drug resistance makes the treatment effect in some patients poor. Most studies have mentioned that non-coding RNA has great potential in tumor diagnosis and drug treatment. However, according to the clinical trial database of NIH (ClinicalTrials.gov), there are only a few items about CRC-related ncRNA trials, indicating the inadequacy in clinical translational. Non-coding RNA is an ideal choice as a marker for early diagnosis of CRC due to its stability, specificity and non-invasiveness of detection methods. It is essential to promote non-coding RNA research transform into clinical trials or clinical applications.
At the same time, research on new uses for existing drugs is also eye-catching. It seems to be a short-term solution to the current clinical dilemma due to the long development cycle of new anti-tumor drugs. Metabolism reprogramming is common in various tumors. Since metformin and some traditional nonsteroidal anti-inflammatory drugs (NSAIDs) have also been found anti-tumor effects by affecting tumor metabolism. Recently, the anti-alcohol metabolism drug DSF has also been found can affects the glycolysis of tumors by targeting miRNAs, which provides new ideas for non-coding RNA research.
Therefore, from the perspective of the miRNA regulatory network, exploring the metabolic reprogramming mechanism of CRC may be able to solve the problem of poor specificity and resistance of antimetabolites in the treatment of CRC, but there is still a long way to go.
Abbreviations
CRC: Colorectal cancer; miRNA: microRNA; GLUT1: Glucose Transporter 1; CuET: Diethyldithiocarbamate-copper complex; DSF: Disulfiram; HK: Hexokinase; HSF1: heat shock factor 1; PFK-1: Phosphofructokinase-1; F-1,6-2P: fructose-1,6-bisphosphate; PFK-2: Phosphofructokinase-2; F-2,6-2P: fructose-2,6-bisphosphate; PK: Pyruvate kinase; PEP: phosphoenolpyruvate; LDH: Lactate dehydrogenase; ACS: Acyl-CoA synthetase; acyl CoA: acyl-coenzyme A; CPT-1: Carnitine palmitoy transterase-1; ACAD9: Acyl-CoA Dehydrogenase 9; α-KG: α-ketoglutaric acid; ASCT: ASC family transporter; GLS1: Glutaminase 1; GPT: Glutamic pyruvic transaminase; sncRNA: small non-coding RNA; lncRNA: long non-coding RNA; circRNA: circle RNA; ceRNA: competitive endogenous RNAs; HNF1A-AS1: hepatocyte nuclear clear factor receptor 1 homeobox A-antisense RNA 1; GAS5: growth arrest-specific 5; ZEB: zinc finger protein; IGF1R: insulin-like growth factor receptor 1; SCFA: short-chain fatty acids; LKB1: liver kinase B1; ACC: acetyl-CoA carboxylase; HMGCR: HMG-coenzyme A reductase; AMPK: AMP-dependent protein kinase; CDK: cyclin dependent kinase.
Acknowledgements
The authors would like to express their gratitude to EditSprings (https://www.editsprings.com/) for the expert linguistic services provided.
Funding
This study was jointly supported by the 2021 Characteristic Innovation Project of Department of Education of Guangdong Province (2021KTSCX015), the 2021 Foshan City Innovation Drive Assisted Engineering Project (2021043), the 2020 Talent Project Supported by Competition in Shunde District, the 2018 Foshan City Outstanding Young Medical Talent Training Project (600009), Southern Medical University Shunde Hospital Scientific Research Startup Plan (SRSP2018001), Guangdong Medical Science and Technology Research Fund Project (A2019302), Science and Technology Planning Project of Foshan Science and Technology Bureau (2018AB000683), National Natural Science Foundation of China, National Natural Science Foundation of China for Youth (81802879), Southern Medical University's Scientific Research Startup Plan (PY2018N110) and Foshan 13th Five-Year Key Specialty Project (FSGSP2D135051).
Author Contributions
All authors have read and agreed to the published version of the manu-script. Conceptualization, Y LU; Methodology, Y LU; Software, WJ LI; Validation, Y LU and WJ LI; Formal analysis, WJ LI; Investigation, WJ LI. and CD Ye; Resources, W LI and CD Ye; data curation, MZ Ouyang and WJ LI; writing—original draft preparation, WJ LI; writing—review and editing, MZ Ouyang; visualization, WJ LI; supervision, MZ Ouyang; project administration, MZ Ouyang.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Keum NN, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nature Reviews Gastroenterology and Hepatology. 2019;16:713-32
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians. 2018;68:394-424
3. Fakih MG. Metastatic colorectal cancer: current state and future directions. J Clin Oncol. 2015;33:1809-24
4. Shen Y, Tong M, Liang Q. et al. Epigenomics alternations and dynamic transcriptional changes in responses to 5-fluorouracil stimulation reveal mechanisms of acquired drug resistance of colorectal cancer cells. Pharmacogenomics Journal. 2018;18:23-8
5. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-33
6. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509-24
7. Dragomir MP, Knutsen E, Calin GA. SnapShot: Unconventional miRNA Functions. Cell. 2018;174:1038-1038.e1
8. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5-18
9. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029-33
10. Lu J, Tan M, Cai Q. The Warburg effect in tumor progression: mitochondrial oxidative metabolism as an anti-metastasis mechanism. Cancer Lett. 2015;356:156-64
11. Warburg O. On the origin of cancer cells. Science. 1956;123:309-14
12. Potter M, Newport E, Morten KJ. The Warburg effect: 80 years on. Biochemical Society Transactions. 2016;44:1499-505
13. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203-22
14. Phan LM, Yeung SCJ, Lee MH. Cancer metabolic reprogramming: importance, main features, and potentials for precise targeted anti-cancer therapies. Cancer Biology and Medicine. 2014;11:1-19
15. Ancey PB, Contat C, Meylan E. Glucose transporters in cancer - from tumor cells to the tumor microenvironment. FEBS Journal. 2018;285:2926-43
16. Santasusagna S, Moreno I, Navarro A. et al. miR-328 mediates a metabolic shift in colon cancer cells by targeting SLC2A1/GLUT1. Clinical and Translational Oncology. 2018;20:1161-7
17. Zhang ZJ, Zhang YH, Qin XJ, Wang YX, Fu J. Circular RNA circDENND4C facilitates proliferation, migration and glycolysis of colorectal cancer cells through miR-760/GLUT1 axis. European Review for Medical and Pharmacological Sciences. 2020;24:2387-400
18. Kudryavtseva AV, Fedorova MS, Zhavoronkov A. et al. Effect of lentivirus-mediated shRNA inactivation of HK1, HK2, and HK3 genes in colorectal cancer and melanoma cells. BMC Genet. 2016;17:156
19. Ou B, Sun H, Zhao J. et al. Polo-like kinase 3 inhibits glucose metabolism in colorectal cancer by targeting HSP90/STAT3/HK2 signaling. J Exp Clin Cancer Res. 2019;38:426
20. Xu S, Herschman HR. A Tumor Agnostic Therapeutic Strategy for Hexokinase 1-Null/Hexokinase 2-Positive Cancers. Cancer Res. 2019;79:5907-14
21. Gregersen LH, Jacobsen A, Frankel LB, Wen J, Krogh A, Lund AH. MicroRNA-143 down-regulates Hexokinase 2 in colon cancer cells. BMC Cancer. 2012;12:232
22. Liu Y, Li H, Ye X. et al. Hsa_circ_0000231 knockdown inhibits the glycolysis and progression of colorectal cancer cells by regulating miR-502-5p/MYO6 axis. World Journal of Surgical Oncology. 2020;18:1-13
23. Li S, Zhu K, Liu L, Gu J, Niu H, Guo J. lncARSR sponges miR-34a-5p to promote colorectal cancer invasion and metastasis via hexokinase-1-mediated glycolysis. Cancer Science. 2020;111:3938-52
24. Li C, Yu Z, Ye J. MicroRNA-513a-3p regulates colorectal cancer cell metabolism via targeting hexokinase 2. Experimental and Therapeutic Medicine. 2020;20:572-80
25. Deng X, Li D, Ke X. et al. Mir-488 alleviates chemoresistance and glycolysis of colorectal cancer by targeting PFKFB3. Journal of Clinical Laboratory Analysis. 2020 p:1-11
26. Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181-6
27. Dayton TL, Jacks T, Vander Heiden MG. PKM 2, cancer metabolism, and the road ahead. EMBO reports. 2016;17:1721-30
28. Zhang Z, Deng X, Liu Y, Liu Y, Sun L, Chen F. PKM2, function and expression and regulation. Cell and Bioscience. 2019;9:1-25
29. Sun Y, Zhao X, Zhou Y, Hu Y. MiR-124, miR-137 and miR-340 regulate colorectal cancer growth via inhibition of the Warburg effect. Oncology Reports. 2012;28:1346-52
30. Clower C V, Chatterjee D, Wang Z, Cantley LC, Heidena MGV, Krainer AR. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1894-9
31. Huang X, Hou Y, Weng X. et al. Diethyldithiocarbamate-copper complex (CuET) inhibits colorectal cancer progression via miR-16-5p and 15b-5p/ALDH1A3/PKM2 axis-mediated aerobic glycolysis pathway. Oncogenesis. 2021;10:4
32. Wang J, Wang H, Liu A, Fang C, Hao J, Wang Z. Lactate dehydrogenase A negatively regulated by miRNAs promotes aerobic glycolysis and is increased in colorectal cancer. Oncotarget. 2015;6:19456-68
33. Chen WC, Wang CY, Hung YH, Weng TY, Yen MC, Lai MD. Systematic analysis of gene expression alterations and clinical outcomes for long-chain acyl-coenzyme A synthetase family in cancer. PLoS ONE. 2016;11:1-23
34. Gharib E, Nasri Nasrabadi P, Reza Zali M. miR-497-5p mediates starvation-induced death in colon cancer cells by targeting acyl-CoA synthetase-5 and modulation of lipid metabolism. Journal of Cellular Physiology. 2020;235:5570-89
35. Barisciano G, Colangelo T, Rosato V. et al. miR-27a is a master regulator of metabolic reprogramming and chemoresistance in colorectal cancer. British Journal of Cancer. 2020;122:1354-66
36. Francque S, Szabo G, Abdelmalek MF. et al. Nonalcoholic steatohepatitis: the role of peroxisome proliferator-activated receptors. Nat Rev Gastroenterol Hepatol. 2021;18:24-39
37. van der Meij BS, Teleni L, Engelen MPKJ, Deutz NEP. Amino acid kinetics and the response to nutrition in patients with cancer. International Journal of Radiation Biology. 2019;95:480-92
38. Altman BJ, Stine ZE, Dang C V. From Krebs to clinic: Glutamine metabolism to cancer therapy. Nature Reviews Cancer. 2016;16:619-34
39. Bhutia YD, Babu E, Ramachandran S, Ganapathy V. Amino acid transporters in cancer and their relevance to 'glutamine addiction': Novel Targets for the design of a new class of anticancer drugs. Cancer Research. 2015;75:1782-8
40. Dong J, Xiao D, Zhao Z. et al. Epigenetic silencing of microRNA-137 enhances ASCT2 expression and tumor glutamine metabolism. Oncogenesis. 2017;6:e356-e356
41. Li J, Song P, Jiang T. et al. Heat Shock Factor 1 Epigenetically Stimulates Glutaminase-1-Dependent mTOR Activation to Promote Colorectal Carcinogenesis. Molecular Therapy. 2018;26:1828-39
42. Hao Y, Samuels Y, Li Q. et al. Oncogenic PIK3CA mutations reprogram glutamine metabolism in colorectal cancer. Nat Commun. 2016;7:11971
43. Wang Y, Tang Q, Li M, Jiang S, Wang X. MicroRNA-375 inhibits colorectal cancer growth by targeting PIK3CA. Biochemical and Biophysical Research Communications. 2014;444:199-204
44. Hansen TB, Jensen TI, Clausen BH. et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-8
45. Thomson DW, Dinger ME. Endogenous microRNA sponges: Evidence and controversy. Nature Reviews Genetics. 2016;17:272-83
46. Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033-8
47. Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nature Cell Biology. 2019;21:542-51
48. Atianand MK, Hu W, Satpathy AT. et al. A Long Noncoding RNA lincRNA-EPS Acts as a Transcriptional Brake to Restrain Inflammation. Cell. 2016;165:1672-85
49. IIott NE, Heward JA, Roux B. et al. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat Commun. 2014;5:3979
50. Chen C-C, Chen C-Y, Wang S-H. et al. Melatonin Sensitizes Hepatocellular Carcinoma Cells to Chemotherapy Through Long Non-Coding RNA RAD51-AS1-Mediated Suppression of DNA Repair. Cancers (Basel). 2018;10:E320
51. Li C, Wang P, Du J, Chen J, Liu W, Ye K. LncRNA RAD51-AS1/miR-29b/c-3p/NDRG2 crosstalk repressed proliferation, invasion and glycolysis of colorectal cancer. IUBMB Life. 2021;73:286-98
52. Li X, Hou G, Zhu Z. et al. The tumor suppressor NDRG2 cooperates with an mTORC1 inhibitor to suppress the Warburg effect in renal cell carcinoma. Investigational New Drugs. 2020;38:956-66
53. Zhu W, Zhuang P, Song W. et al. Knockdown of lncRNA HNF1A-AS1 inhibits oncogenic phenotypes in colorectal carcinoma. Molecular Medicine Reports. 2017;16:4694-700
54. Guo X, Zhang Y, Liu L, Yang W, Zhang Q. Hnf1a-as1 regulates cell migration, invasion and glycolysis via modulating mir-124/myo6 in colorectal cancer cells. OncoTargets and Therapy. 2020;13:1507-18
55. Cui S, Yang X, Zhang L, Zhao Y, Yan W. LncRNA MAFG-AS1 promotes the progression of colorectal cancer by sponging miR-147b and activation of NDUFA4. Biochemical and Biophysical Research Communications. 2018;506:251-8
56. Liu L, Wang HJ, Meng T. et al. lncRNA GAS5 Inhibits Cell Migration and Invasion and Promotes Autophagy by Targeting miR-222-3p via the GAS5/PTEN-Signaling Pathway in CRC. Molecular Therapy - Nucleic Acids. 2019;17:644-56
57. Lee Y-R, Chen M, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor: new modes and prospects. Nat Rev Mol Cell Biol. 2018;19:547-62
58. Salzman J. Circular RNA Expression: Its Potential Regulation and Function. Trends in Genetics. 2016;32:309-16
59. Huang X, Shen X, Peng L, Mai W, Wan Y, Zhang H. Circcsnk1g1 contributes to the development of colorectal cancer by increasing the expression of myo6 via competitively targeting mir-455-3p. Cancer Management and Research. 2020;12:9563-75
60. Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat Rev Cancer. 2008;8:523-34
61. Zheng Q, Bao C, Guo W. et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215
62. Zeng K, Chen X, Xu M. et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7 article. Cell Death and Disease. 2018 p:9
63. Zhang Y, Li C, Liu X. et al. circHIPK3 promotes oxaliplatin-resistance in colorectal cancer through autophagy by sponging miR-637. EBioMedicine. 2019;48:277-88
64. Wang X, Zhang H, Yang H. et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Molecular Oncology. 2020;14:539-55
65. Hyochol Ahn, et al 2017. Reprogramming glucose metabolism in cancer: can it be exploited for cancer therapy? Physiology & behavior. 2017;176:139-48
66. Yang H, Zhang H, Yang Y. et al. Hypoxia induced exosomal circRNA promotes metastasis of colorectal cancer via targeting GEF-H1/RhoA axis. Theranostics. 2020;10:8211-26
67. Janney A, Powrie F, Mann EH. Host-microbiota maladaptation in colorectal cancer. Nature. 2020;585:509-17
68. Slavin JL. Position of the American Dietetic Association: health implications of dietary fiber. Journal of the American Dietetic Association. 2008;108:1716-31
69. Fukuda S, Toh H, Taylor TD, Ohno H, Hattori M. Acetate-producing bifidobacteria protect the host from enteropathogenic infection via carbohydrate transporters. Gut Microbes. 2012;3:449-54
70. Lopetuso LR, Scaldaferri F, Petito V, Gasbarrini A. Commensal Clostridia: leading players in the maintenance of gut homeostasis. Gut Pathog. 2013;5:23
71. Burns MB, Lynch J, Starr TK, Knights D, Blekhman R. Virulence genes are a signature of the microbiome in the colorectal tumor microenvironment. Genome Med. 2015;7:55
72. Zeng H, Hamlin SK, Safratowich BD, Cheng W-H, Johnson LK. Superior inhibitory efficacy of butyrate over propionate and acetate against human colon cancer cell proliferation via cell cycle arrest and apoptosis: linking dietary fiber to cancer prevention. Nutr Res. 2020;83:63-72
73. Wu S en, Hashimoto-Hill S, Woo V. et al. Microbiota-derived metabolite promotes HDAC3 activity in the gut. Nature. 2020;586:108-12
74. Hu S, Liu L, Chang EB, Wang JY, Raufman JP. Butyrate inhibits pro-proliferative miR-92a by diminishing c-Myc-induced miR-17-92a cluster transcription in human colon cancer cells. Molecular Cancer. 2015;14:1-15
75. Izreig S, Samborska B, Johnson RM. et al. The miR-17∼92 microRNA Cluster is a Global Regulator of Tumor Metabolism. Cell Reports. 2016;16:1915-28
76. Herzig S, Shaw RJ. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat Rev Mol Cell Biol. 2018;19:121-35
77. Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400-14
78. Hu S, Dong TS, Dalal SR. et al. The microbe-derived short chain fatty acid butyrate targets miRNA-dependent p21 gene expression in human colon cancer. PLoS One. 2011;6:e16221
79. Han R, Sun Q, Wu J, Zheng P, Zhao G. Sodium Butyrate Upregulates miR-203 Expression to Exert Anti-Proliferation Effect on Colorectal Cancer Cells. Cellular Physiology and Biochemistry. 2016;39:1919-29
Author contact
![]() Corresponding author: Manzhao Ouyang, E-mail: doctorkenedu.cn; Tel.:+86-139-2323-0723.
Corresponding author: Manzhao Ouyang, E-mail: doctorkenedu.cn; Tel.:+86-139-2323-0723.

 Global reach, higher impact
Global reach, higher impact