3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(24):7477-7487. doi:10.7150/jca.63244 This issue Cite
Research Paper
High-performance Collective Biomarker from Liquid Biopsy for Diagnosis of Pancreatic Cancer Based on Mass Spectrometry and Machine Learning
1. Department of Anatomy and Cell Biology, Faculty of Medicine, University of Yamanashi, Chuo, Yamanashi, Japan
2. Hepato-Biliary-Pancreatic Surgery Division, Department of Surgery, Graduate School of Medicine, The University of Tokyo, Tokyo, Japan
3. First Department of Surgery, Faculty of Medicine, University of Yamanashi, Chuo, Yamanashi, Japan
4. Department of Emergency and Critical Care Medicine, Faculty of Medicine, University of Yamanashi, Chuo, Yamanashi, Japan
5. Shimadzu Corporation, Nakagyo, Kyoto, Japan
Abstract
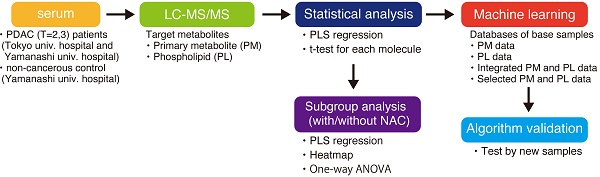
Background: Most pancreatic cancers are found at progressive stages when they cannot be surgically removed. Therefore, a highly accurate early detection method is urgently needed.
Methods: This study analyzed serum from Japanese patients who suffered from pancreatic ductal adenocarcinoma (PDAC) and aimed to establish a PDAC-diagnostic system with metabolites in serum. Two groups of metabolites, primary metabolites (PM) and phospholipids (PL), were analyzed using liquid chromatography/electrospray ionization mass spectrometry. A support vector machine was employed to establish a machine learning-based diagnostic algorithm.
Results: Integrating PM and PL databases improved cancer diagnostic accuracy and the area under the receiver operating characteristic curve. It was more effective than the algorithm based on either PM or PL database, or single metabolites as a biomarker. Subsequently, 36 statistically significant metabolites were fed into the algorithm as a collective biomarker, which improved results by accomplishing 97.4% and was further validated by additional serum. Interestingly, specific clusters of metabolites from patients with preoperative neoadjuvant chemotherapy (NAC) showed different patterns from those without NAC and were somewhat comparable to those of the control.
Conclusion: We propose an efficient screening system for PDAC with high accuracy by liquid biopsy and potential biomarkers useful for assessing NAC performance.
Keywords: Pancreatic ductal adenocarcinoma, Liquid biopsy, Metabolome, Machine leargning, Neoadjuvant chemotherapy

 Global reach, higher impact
Global reach, higher impact