Impact Factor
ISSN: 1837-9664
J Cancer 2022; 13(11):3221-3233. doi:10.7150/jca.73051 This issue Cite
Research Paper
Sex differences in hepatocellular carcinoma indicated BEX4 as a potential target to improve efficacy of lenvatinib plus immune checkpoint inhibitors
1. Department of Infectious Diseases, Shanghai Key Laboratory of Infectious Diseases and Biosafety Emergency Response, National Medical Center for Infectious Diseases, Huashan Hospital, Fudan University, Shanghai 200040, China.
2. Department of Infectious Diseases, The First Affiliated Hospital of Bengbu Medical College, Bengbu 233000, China.
3. Department of General Surgery, Huashan Hospital, Cancer Metastasis Institute, Fudan University, Shanghai 200040, China.
4. Department of Radiology, Huashan Hospital, Fudan University, Shanghai 200040, China.
*These authors contributed equally to this work.
Received 2022-3-20; Accepted 2022-8-20; Published 2022-9-6
Abstract
Background: Hepatocellular carcinoma (HCC) is the most common form of liver cancer, and significant sex disparities have been observed in HCC. We aim to explore the potential sex-biased mechanisms involved in hepatocarcinogenesis.
Methods: Based on TCGA data, we compared clinical features, genetic alterations, and immune cell infiltrations between male and female HCC patients. In addition, we performed sex-based differential expression analysis and functional enrichment analysis. Finally, GSE64041 dataset and another HCC cohort were engaged to validate our findings.
Results: Significant differences of genetic alterations and TME were observed between male and female HCC patients. Enhanced metabolism of lipids was associated with hepatocarcinogenesis in men, while ECM-organization-related pathways were correlated to HCC development in women. BEX4 was upregulated in female but downregulated in male HCC patients, and was positively correlated with immune checkpoint molecules and infiltrated immune cell. These findings were further validated in dataset GSE64041 and our HCC cohort. More importantly, a negative correlation was found between BEX4 expression and lenvatinib sensitivity.
Conclusion: Distinct biological processes were involved in sex-biased tumorigenesis of HCC. BEX4 can be targeted to improve the efficacy of lenvatinib plus immune checkpoint inhibitors.
Keywords: hepatocellular carcinoma, sex difference, immunity, metabolism, therapy
Introduction
Liver cancer is one of the most common cancers and the third leading cause of cancer-related deaths worldwide, with hepatocellular carcinoma (HCC) as the most common type [1, 2]. According to the latest global cancer statistics, liver cancer accounts for 4.7% of the 19.3 million new cases and 8.3% of the 9.9 million new deaths globally [1]. Specifically, men account for 70.1% of new cases and 70.3% of new deaths [1], significantly higher incidence and mortality in males reflect that HCC has sex disparities, which was considered as an effect dependent on sex hormones [3, 4]. Consistent with this, it has been reported that estrogen could inhibit secretion of interleukin-6, a multifunctional cytokine that causes inflammation, thus risk of inflammation-induced liver cancer in women was reduced [5]. In addition, Foxa1/2 and their targets have been demonstrated as central players in sexual dimorphism of HCC by interacting with estrogen receptor [3]. However, the comprehensive mechanisms of sexual dimorphism in hepatocarcinogenesis remains not fully understood.
Arranged in tandem on chromosome X, Brain-expressed X-linked 4 (BEX4) is a member of BEX family which was involved in different signaling pathways and played important roles in physiological and pathophysiological conditions [6, 7]. BEX4-mediated YAP/TAZ activation was reported to promote the tumor growth and radioresistance in Glioblastoma [8]. In addition, BEX4 was implicated in oncogenic microtubule hyperacetylation by interacting with sirtuin 2 [9]. On the contrary, BEX4 may act as a tumor suppressor in oral squamous cell carcinoma and ovarian cancer [10, 11]. Still, the function of BEX4 in HCC remains obscure to date.
In this study, based on TCGA data, we compared clinical features of HCC between male and female patients, and evaluated the differences of genetic alterations and immune cell infiltrations between male and female patients. In addition, we performed differential expression analysis and enrichment analysis to reveal potential biological processes and pathways that are implicated in sex-biased hepatocarcinogenesis. Finally, we focused on BEX4, the unique gene upregulated in female but downregulated in male HCC patients. We found BEX4 expression positively correlated with immune cell infiltration as well as immune checkpoint molecule expression. These results were further validated in dataset GSE64041. Our findings provided new insight into sex differences in tumorigenesis of HCC and were helpful to the development of sex-based therapeutic strategies.
Materials and methods
Data download and analysis
RNA-seq, genetic alteration, and clinicopathological data of HCC patients were downloaded from cBioPortal (https://www.cbioportal.org) [12]. The enrollment criteria for this study were as follows: First, all patients were diagnosed as having HCC by pathology. Second, BEX4 mRNA expression data and clinical features including gender, age, race, TNM stage, tumor grade, clinical stage were available. Third, informed consent was acquired from all patients by the TCGA Research Network. A total of 371 patients who met these criteria were enrolled. Differential expression analysis was performed using R package LIMMA with online tool BART [13], genes with |fold change|≥1.5 and adjusted p<0.05 were considered to be differentially expressed genes. Immune cell infiltration levels inferred by CIBERSORT algorithm [14] were downloaded from GDC database (https://gdc.cancer.gov/). GSE64041 dataset was downloaded from GEO database (https://www.ncbi.nlm.nih.gov/geo/).
CVCDAP database analysis
Cancer virtual cohort discovery analysis platform (CVCDAP, https://omics.bjcancer.org/cvcdap/home.do) is an integrated web tool for molecular and clinical analysis of cancer cohorts [15]. CVCDAP was engaged to analyze and visualize the top 5 most frequently mutated genes in HCC.
GEPIA database analysis
Gene expression profiling interactive analysis (GEPIA2.0, http://gepia2.cancer-pku.cn/#index) is a web-based platform which allow investigators to analyze RNA sequencing expression data deposited in TCGA and GTEx [16]. GEPIA was used to perform correlation analysis between BEX4 and immune checkpoint-related molecules.
Enrichment analysis
Gene set enrichment analysis (GSEA) [17] was applied to identify functions and pathways associated with hepatocarcinogenesis. The differentially expressed genes were subjected to Metascape (https://metascape.org/gp/index.html) to perform network enrichment analysis [18]. Protein-protein interaction (PPI) network was analyzed by STRING database (https://string-db.org/) [19] and visualized by Cytoscape (version 3.7.2) [20].
TISIDB database analysis
TISIDB (http://cis.hku.hk/TISIDB/index.php) is a web portal for tumor and immune system interactions [21]. We employed TISIDB to explore the distributions of BEX4 expression across immune/molecular subtypes of HCC. In addition, correlations between BEX4 expression and clinical stage/tumor grade of HCC were also evaluated.
TIMER database analysis
TIMER (https://cistrome.shinyapps.io/timer/) is a web resource for systematical assessment of the clinical impact of tumor-infiltrating immune cells in various human cancers [22, 23]. TIMER was engaged here to estimate the correlation between BEX4 expression and infiltration levels of immune cells. Besides, correlations between expression of BEX4 and immune checkpoint molecules as well as immune cell markers were also assessed.
GSCA database analysis
Gene set cancer analysis (GSCA, http://bioinfo.life.hust.edu.cn/GSCA/#/) is an integrated online resource for genomic and immunogenomic gene set cancer analysis [24]. GSCA also integrated small molecule drugs from the Cancer Therapeutics Response Portal. GSCA was engaged to evaluate correlation between BEX4 expression and small molecule drug sensitivity.
RT-qPCR
Validation study utilized 40 paired human HCC samples collected in previous studies [25]. RT-qPCR reactions were performed using SYBR premix Ex Taq™ II kit (TAKARA BIO INC.) according to the manufacturer's instructions. Specific primers for each gene were listed as followed:
GAPDH forward: TGCGAGTACTCAACACCAACA; GAPDH reverse: GCATATATTCGGCCCACA; BEX4 forward: AAAGAGGAACTAGCGGCAAAC; BEX4 reverse: CCAAATGGCGGGATTCTTCTTC.
Immunohistochemistry (IHC)
For validation cohort, BEX4 expression was detected by IHC. IHC staining was carried out using an anti-BEX4 antibody (rabbit polyclonal, Sinobiological) at 1:50 dilution as previously described [25]. BEX4 intensity score was qualified as no staining (score 0), weak (score 1), medium (score 2), and strong (score 3). The percentage of positive cells was divided into 4 scores: 1 = <25%, 2 = 25% to 50%, 3 = 50% to 75%, 4= >75%. The final IHC scores = intensity score × percentage score.
Flow Cytometry
32 HCC tissue were collected for flow cytometry analysis in previous studies. The tissue digestion, grinding and flow cytometry staining were performed as previously described [26]. CD4+ T cells, CD8+ T cells, B cells and macrophages were determined as CD45+CD3+CD4+CD8-, CD45+CD3+CD4-CD8+, CD45+CD3-CD19+ and CD45+CD14+CD68+, respectively.
Statistical analysis
Statistical difference was analyzed using t-test between two groups. Chi-square and Fisher tests were performed for categorical variables. Spearman correlation was used for correlation analysis. Statistical analysis for figures generated from online databases was calculated automatically. p value <0.05 was considered statistically significant.
Results
Clinicopathological characteristics
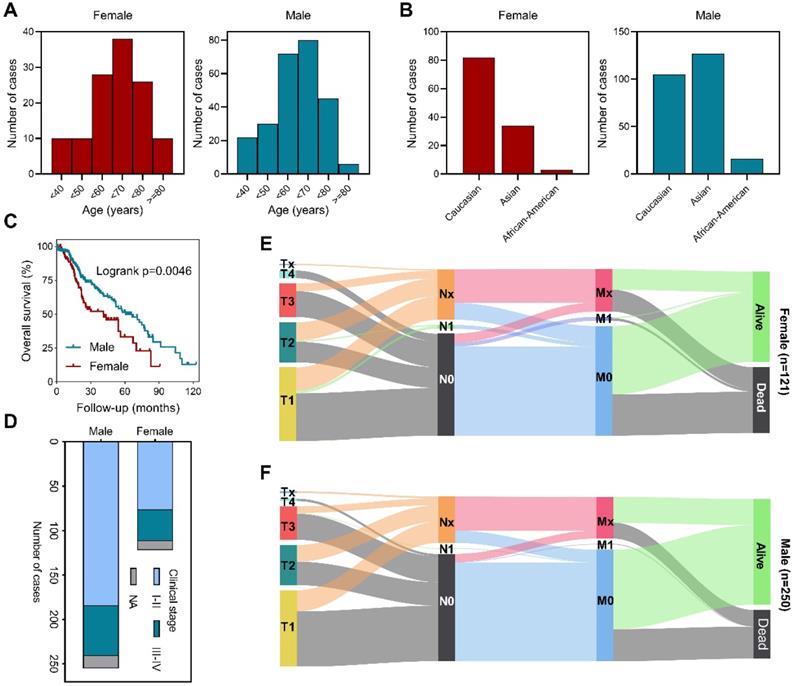
We first analyzed the age distribution of male and female patients and found that most patients were aged between fifty to eighty in both groups (Figure 1A). Exploration based on race showed that female patients are predominantly Caucasian, while male patients are mostly Asian and Caucasian (Figure 1B). Survival analysis revealed that male patients had longer overall survival (Figure 1C). Further investigation verified that stage I-II patients were more common in male (Figure 1D). Detailed primary tumor, lymph node, metastasis (TNM stage) distribution analysis demonstrated that male patients had lower T stage, less lymph node involvement and distal metastasis than female patients (Figures 1E,F). These data showed that in TCGA cohort, there was no significant difference in the age distribution of HCC patients by gender, but more male patients were in early stages of the disease. Clinicopathologic characteristics of enrolled cohort and the correlation between clinicopathologic characteristics and BEX4 were further summarized (Table S1 and Table S2).
Sex-differential genetic alterations
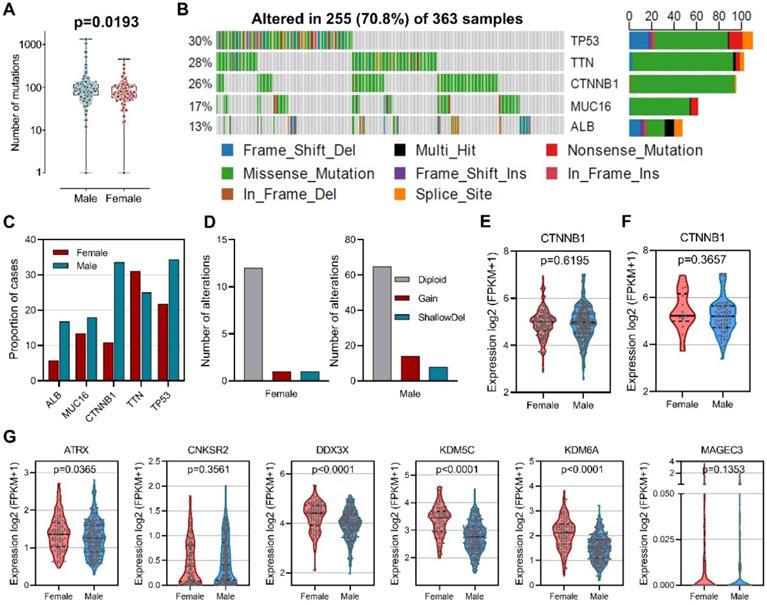
To explore the difference of genetic alterations between males and females in HCC, we determined mutated genes using cBioPortal database. Our results showed that male HCC patients had higher numbers of mutations than female HCC patients (Figure 2A). Among the 363 samples, the top five most frequently mutated genes in HCC were TP53 (30%), TTN (28%), CTNNB1 (26%), MUC16 (17%), and ALB (13%) (Figure 2B). Mutation frequencies were higher in male patients than in female patients for TP53 (34.4% versus 21.8%), CTNNB1 (33.6% versus 10.9%), MUC16 (18.0% versus 13.4%), and ALB (16.8% versus 5.8%), while mutation frequency of TTN (25.0% versus 31.1%) was lower (Figure 2C). CTNNB1 mutation has been identified as a sex-biased driver mutation in HCC [27-29], we then analyzed copy number alteration of CTNNB1 and found increased copy number gain in male-derived samples (Figure 2D). However, no differences in CTNNB1 transcript levels were observed between male and female in both total patients (Figure 2E) and CTNNB1 mutated patients (Figure 2F). In addition, a group of tumor suppressors that escape from X-inactivation have been implicated in cancer sex bias [30], our analysis did reveal higher expression of ATRX, DDX3X, KDM5C and KDM6A in female than male patients (Figure 2G). These results indicated that gene alterations are associated with sex-biased carcinogenesis of HCC.
Sex-differential immune alterations
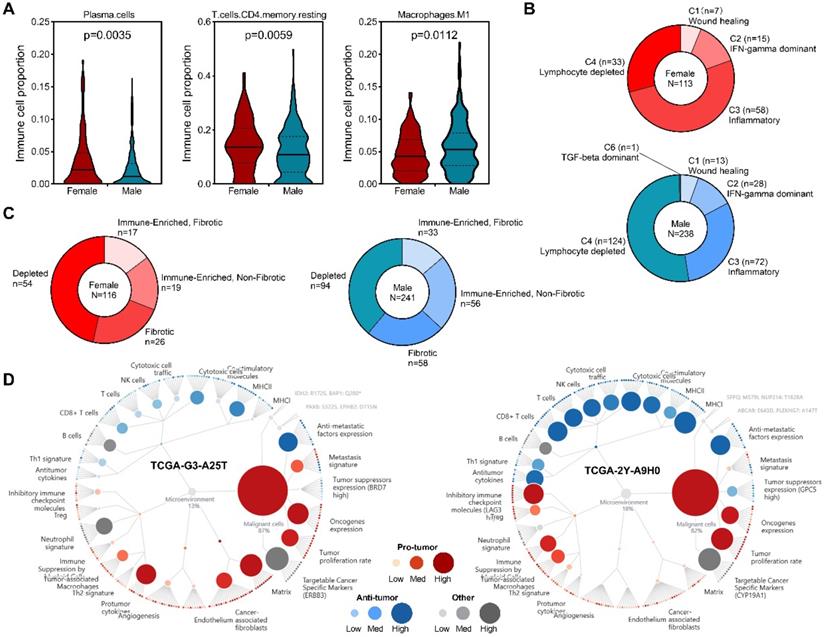
To get insight into sex differences of immune cell infiltration in HCC, RNA-seq data from TCGA project was subjected to CIBERSORT to infer immune cell populations. Overview of tumor-infiltrating immune cell populations arrange by sex was presented (Figure S1). Our results demonstrated that compared with male patients, female patients had higher infiltration levels of plasma cells and CD4 memory resting T cells, but lower levels of M1 macrophages (Figure 3A). A previous study identified six immune clusters in human malignancies that are associated with tumor microenvironment (TME) and patient's prognosis [31]. We found that in female patients, inflammatory subtype was the dominant cluster, followed by lymphocyte depleted subtype (Figure 3B). In male patients, however, most patients belonged to lymphocyte depleted subtype, while inflammatory subtype was the second largest cluster (Figure 3B). Recently, a study identified four TME subtypes (Immune-enriched, fibrotic, IE/F; Immune-enriched, non-fibrotic, IE; Fibrotic, F; Depleted, D) that correlated with patient response to immunotherapy [32], our analysis revealed increased immune-enriched, non-fibrotic subtype and decreased depleted subtype in male patients (Figure 3C), and representative molecular function portraits of HCC with TME subtype IE and D were displayed. These findings suggested sex-biased TME in HCC.
Sex-based clinicopathological analysis of HCC patients. (A) Age distribution among male and female patients. (B) Race distribution among male and female patients. (C) Sex-based survival analysis. (D) Clinical stage distribution among male and female patients. (E) Distribution of TNM stages in female patients. (F) Distribution of TNM stages in male patients.
Sex-based gene set enrichment analysis
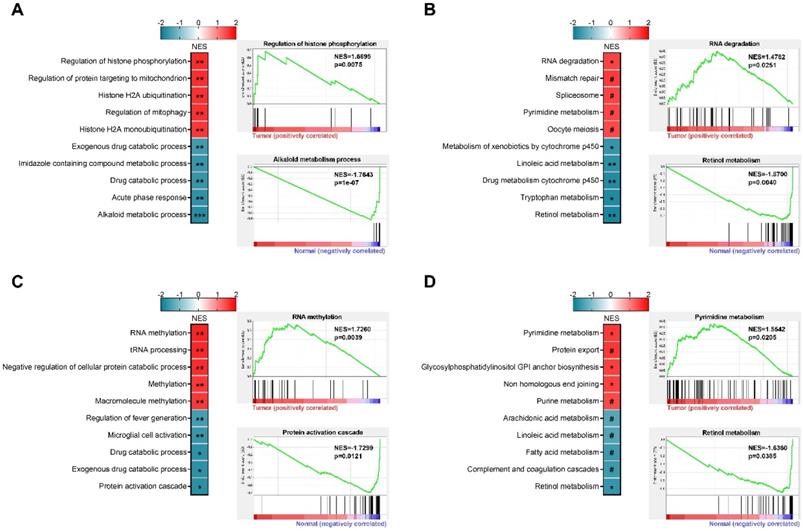
Based on paired RNA-seq data of male and female patients from TCGA project, we performed GSEA analysis and the top 5 positively and negatively enriched gene sets were presented (Figures 4A-D). In female patients, Alkaloid metabolic process, Acute phase response, Drug catabolic process, Imidazole containing compound metabolic process, and Exogenous drug catabolic process were the top 5 negatively enriched GO biological processes (Figure 4A), while Regulation of histone phosphorylation, Regulation of protein targeting to mitochondrion, Histone H2A ubiquitination, Regulation of mitophagy, and Histone H2A monoubiquitination were the top 5 positively enriched GO biological processes (Figure 4A). The top 5 negatively enriched KEGG pathways were Retinol metabolism, Tryptophan metabolism, Drug metabolism cytochrome P450, Linoleic acid metabolism, and Metabolism of xenobiotics by cytochrome P450 (Figure 4B). RNA degradation, Mismatch repair, Spliceosome, Pyrimidine metabolism, and Oocyte meiosis were the top 5 positively enriched KEGG pathways (Figure 4B). In male patients, the top 5 negatively enriched GO biological processes were Regulation of fever generation, Microglial cell activation, Drug catabolic process, Exogenous drug catabolic process, and Protein activation cascade (Figure 4C), while RNA methylation, tRNA processing, Negative regulation of cellular protein catabolic process, Methylation, and Macromolecule methylation were the top 5 positively enriched GO biological processes (Figure 4C). The top 5 negatively enriched KEGG pathways were Retinol metabolism, Complement and coagulation cascades, Fatty acid metabolism, Arachidonic acid metabolism, and Linoleic acid metabolism (Figure 4D), while the top 5 positively enriched KEGG pathways were Pyrimidine metabolism, Protein export, Glycosylphosphatidylinositol anchor biosynthesis, Non-homologous end joining, and Purine metabolism (Figure 4D. These findings highlighted sex-biased metabolism reprogramming in hepatocarcinogenesis, which may serve as regions of interest for further exploration.
Sex-based genetic alteration analysis. (A) Comparison of total mutation numbers between male and female patients. (B) Mutation plots of the top 5 most frequently mutated genes in HCC. (C) Mutation frequencies of the top 5 mutated genes in male and female patients. (D) Copy number alteration of CTNNB1 in male and female patients. (E) Comparison of CTNNB1 expression in male and female patients. (F) Comparison of CTNNB1 expression in CTNNB1 mutated male and female patients. (G) Expression comparison of tumor suppressor genes that escape from X-inactivation.
Sex-based immune alteration analysis. (A) Immune cells with different infiltration levels between male and female patients. (B) Immune cluster distribution among male and female patients. (C) TME subtype distribution among male and female patients. (D) Representative TME subtypes, Depleted (left) and Immune-Enriched, Non-Fibrotic [37].
Functional enrichment analysis of DEGs between male and female patients
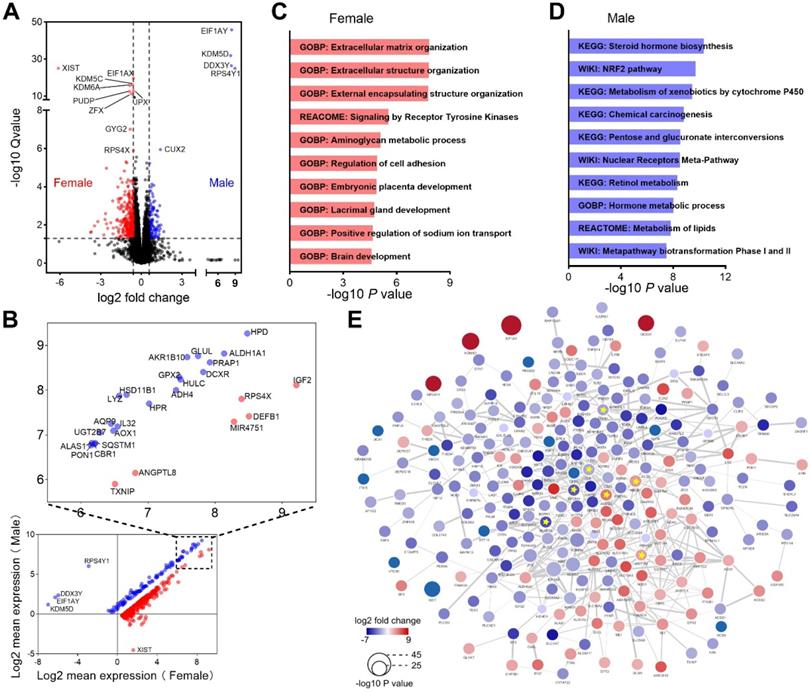
We next evaluated genes differentially expressed between male and female HCC patients. The comparison revealed that 110 genes were upregulated, while 279 genes were downregulated in male patients (Figure 5A). Most of these dysregulated genes possessed higher expression levels both in male and female patients, and the top expressed genes were labeled (Figure 5B). Enrichment analysis demonstrated that genes upregulated in female patients were mainly enriched in extracellular matrix organization and receptor tyrosine kinase signaling (Figure 5C). In contrast, genes upregulated in male patients were mainly involved in metabolism of lipids and carbohydrates (Figure 5D). We also performed PPI network analysis and identified 7 highly connected hub genes, including 4 genes (CDH1, EPCAM, SNAP25 and TGFB1) upregulated in female and 3 genes (ABCB1, ABCG2 and UGT1A9) upregulated in male (Figure 5E). CDH1, EPCAM and TGFB1 play important roles in epithelial-mesenchymal transition (EMT), a process associated with tumorigenesis [33]. ABCB1, ABCG2 and UGT1A9 were critical molecules that mediate chemotherapeutic drug resistance [34-36]. These results reflected that different genes and pathways were implicated in the development and progression of HCC in men and women.
Functional enrichment analysis of sex-specific dysregulated genes
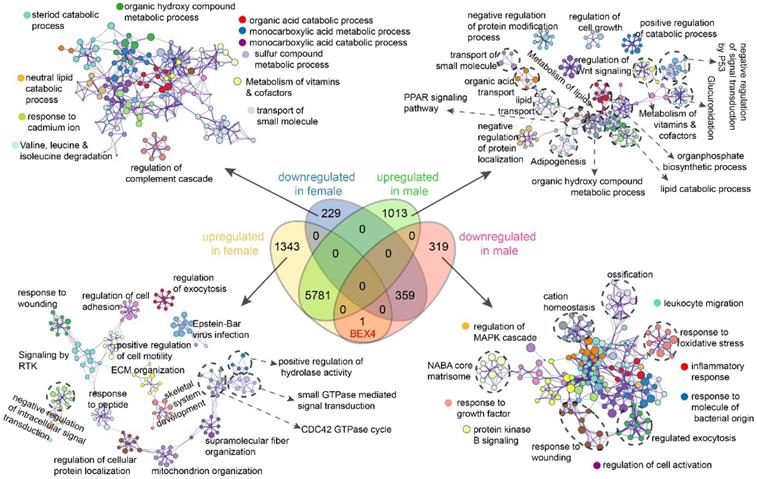
We then performed differential expression analysis based on these paired RNA-seq data and found that dysregulated genes in female patients were significantly fewer than that in male patients (Figure S2). Next, functional enrichment analyses of genes exclusively up-/down-regulated in male or female patients were implemented (Figure 6, Figure S3). Our results showed that genes exclusively downregulated in female patients were enriched in terms related to metabolic or catabolic processes of acids, lipid, steroid, and vitamins (Figure 6). Genes exclusively upregulated in female patients were mainly enriched in terms associated with signal transduction, ECM organization, mitochondrion organization, and protein localization (Figure 6). While in male patients, upregulated genes were enriched in terms related to signal transduction, material transport, protein modification/localization, metabolic and catabolic and biosynthetic processes, especially lipid metabolism related processes and signal transduction by P53 (Figure 6). In contrast, downregulated genes were mainly enriched in immune related processes including leukocyte migration, inflammatory response, response to molecule of bacterial origin, and cell activation (Figure 6). These results further emphasized the differences between male and female in hepatocarcinogenesis.
Potential role of BEX4 in HCC development
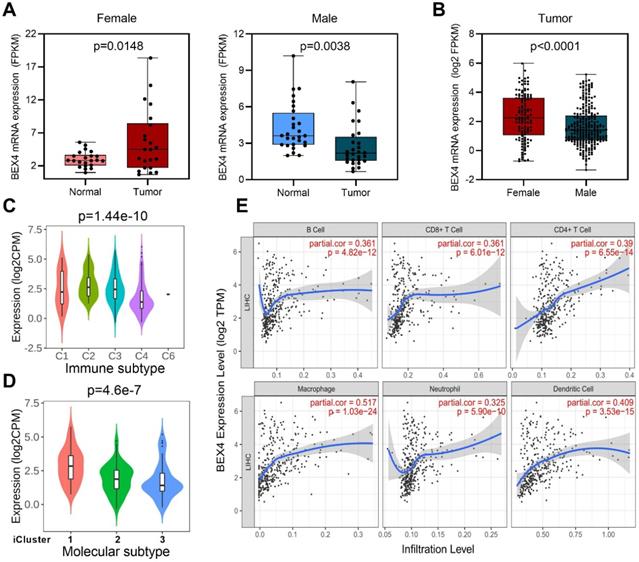
We then focused on Brain-expressed X-linked 4 (BEX4), the unique gene which upregulated in female but downregulated in male (Figure 6, Figure 7A). Expression analysis revealed that female HCC patients possessed higher BEX4 transcriptional levels than male patients (Figure 7B). Although no strong correlation was observed between BEX4 mRNA level and tumor grade (Figure S4A) or disease stage (Figure S4B), significantly differential expression of BEX4 was observed across immune and molecular subtypes of HCC (Figures 7C-D). Moreover, we evaluated the correlation between BEX4 expression and immune cell infiltration. The results revealed a robust positive association between mRNA levels of BEX4 and infiltrating immune cells, especially macrophage (Figure 7E). Similarly, strong positive correlations were found between immune checkpoint molecules and BEX4 expression (Figure S4C). Further validated in GSE64041 (Figure S5), these findings suggested that BEX4 can regulate anti-tumor immunity by modulating TME. Interestingly, data mined from the Cancer Therapeutics Response Portal (CTRP) [37] via GSCA indicate that BEX4 expression negatively correlates with sensitivity to multiple receptor tyrosine kinases (lenvatinib, axitinib, and AZD4547) (Figure S4D). Given that lenvatinib plus anti-PD-1 antibody can enhance antitumor activity in HCC by decreasing tumor-associated macrophage [38, 39], targeting BEX4 may further improve the efficacy of the combination therapy.
Sex-based gene set enrichment analysis. (A) Top 5 positively and negatively enriched GO terms in female patients, representative enrichment plots were presented. (B) Top 5 positively and negatively enriched KEGG pathways in female patients, representative enrichment plots were presented. (C) Top 5 positively and negatively enriched GO terms in male patients, representative enrichment plots were presented. (D) Top 5 positively and negatively enriched KEGG pathways in male patients, representative enrichment plots were presented. NES, normalized enrichment score; ***p<0.001, **p<0.01, *p<0.05, #p>0.05.
Functional enrichment analysis of DEGs between male and female patients. (A) Heatmap shows DEGs between male and female patients. (B) Average expression of the DEGs in male and female patients. (C) Top 10 pathways enriched in female patients. (D) Top 10 pathways enriched in male patients. (E) PPI interaction network of the DEGs. Hub genes were marked with yellow star, edge thickness indicates interaction score (range from 0.400 to 0.996).
Validation analyses using our HCC cohort
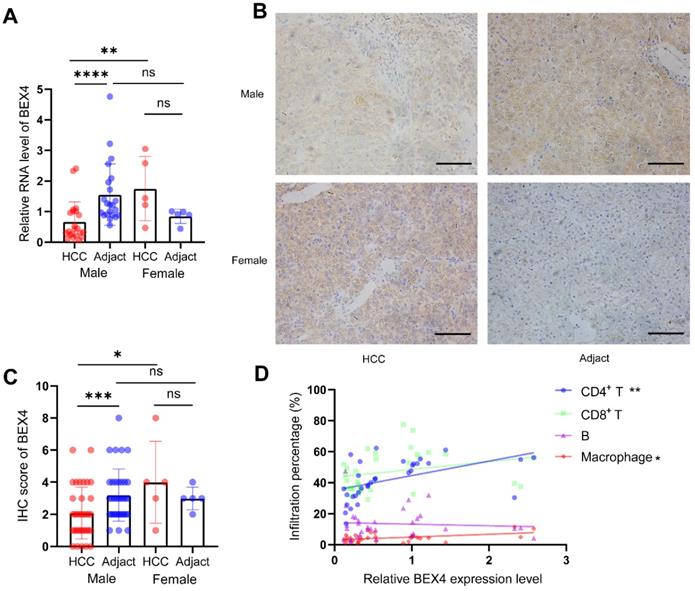
To validate our findings based on bioinformatic analyses, 40 HCC tissues and their adjacent tissues were collected. RT-qPCR suggested female HCC patients had higher BEX4 expression than male patients. The level of BEX4 in the cancerous tissue was significantly reduced compared with the paracarcinoma tissue in male patients. However, probably due to the small sample size, no statistical significance was reached in female patients (Figure 8A). BEX4 protein level was measured by Immunohistochemistry, similar results were found, which were consistent with our RT-qPCR results (Figure 8C). Representative images were shown in Figure 8B. Moreover, to validate the correlation between infiltrated immune cells and BEX4 level, flow cytometry was performed in 32 HCC samples. The results, as shown in Figure 8D, indicated that although no significant correlations were observed in CD8+ T cells and B cells group, BEX4 was positively correlated with CD4+ T cells (r=0.4914, P<0.01) and macrophages (r=0.4436, P<0.05).
Discussion
In cancer-related studies, sex as a critical biological variable is often understudied [40-42]. It is consensus that there are sex differences in incidence across human cancers, especially HCC [41, 42]. HCC is one of the tumors with the worst prognosis, and men show significantly higher incidence than women [3, 43]. Although sex hormones have been suggested as major contributors [3, 4], the exact mechanisms remain unclear. A better understanding of the potential impact of sex on hepatocarcinogenesis is of great importance for precision medicine in HCC.
Sex-based functional enrichment analysis. Enrichment pathway networks of genes exclusively downregulated in female, upregulated in female, downregulated in male, and upregulated in male.
Function exploration of BEX4 in HCC. (A) Comparison of BEX4 expression between paired normal and tumor samples in male and female patients. (B) Comparison of BEX4 expression between male and female patients. (C) Comparison of BEX4 expression across six immune subtypes. (D) Comparison of BEX4 expression across three molecular subtypes. (E) Correlation between BEX4 expression and immune cell infiltration.
Validation analyses using our HCC cohort. (A) RT-qPCR validation of BEX4 expression. (B,C) IHC validation of BEX4 expression. Scale bar=200 um. (D) Correlations between CD4+ T cells, CD8+ T cells, B cells, macrophages and BEX4 level. ****P<0.0001, ***P<0.001, **P<0.01, *P<0.05.
In the current study, we found that most HCC patients were aged between 50 to 80, predominantly Caucasian and Asian. Survival analysis revealed that male patients had better outcome than female patients. Further exploration showed that, in TCGA dataset, male HCC patients are mostly in the early stage of the disease.
Cancer has long been regarded as a disease of the genome and gene mutation has been implicated in carcinogenesis [44, 45], knowledge about sex-specific genetic and genome-wide influences in cancer has been reviewed elsewhere [45]. We here compared mutation numbers between male patients and female patients and found that male patients had significantly higher mutation numbers than female patients. Consistent with this, the top 5 most mutated genes in HCC showed different mutation frequency in male and female patients. Higher mutation frequencies of TP53, CTNNB1, MUC16, and ALB were observed in male patients, while higher mutation frequency of TTN was found in female patients. TTN is the largest protein in human, and genetic mutation of TTN was associated hereditary heart diseases [46]. In muscle cells, the product of TTN plays an important role in providing connections at individual microfilament level, while in non-muscle cells it contributes to condensation and segregation of chromosome during mitosis [47, 48]. Chromosome condensation leads to insulation of transcriptional factors and epigenetic regulators and reduces transcriptional activity, resulting in suppressed gene expression. Epigenetic regulation of gene expression was shown sex-biased pattern [40]. Lately, it is reported that solid stress generated by lesion growth can impair infiltration of cancer-specific T cells into lymph node metastases [49]. Further investigations are needed to explore whether TTN plays roles in sex-specific chromatin regulation and/or solid stress derived from tumor growth. CTNNB1 mutation is a sex-biased driver mutation in HCC [27, 29], although increased copy number gain was observed in male patients, no significant difference in the transcript levels of CTNNB1 was detected between male and female, the function of mutated CTNNB1 therefore warrants further investigation. In addition, higher expression of ATRX, DDX3X, KDM5C and KDM6A was observed in female patients, indicating that tumor-suppressor genes that escape from X-inactivation may contribute to HCC sex bias.
Tumors are not simply collections of cancer cells, coupled with stromal cells and extracellular matrix components they established an environment called tumor microenvironment (TME) [32, 50]. TME plays a critical role in tumor development and progression, as well as clinical outcomes and response to anti-cancer therapies [50, 51]. We found increased infiltration levels of plasma cells and CD4 memory resting T cells in female patients, and M1 macrophages in male patients. Among the six immune clusters associated with TME, female patients are predominantly inflammatory cluster, while male patients are predominantly lymphocyte depleted cluster. Moreover, TME subtype analysis revealed more immune-enriched, non-fibrotic subtype patients in male and more depleted subtype patients in female. These findings highlighted that the TME differs between male and female patients, and taking gender and TME into account will help to guide making precise treatment decision.
Understanding the potential molecular mechanisms of sex-biased cancer biology is vital to improve patient care [27, 52, 53]. Comparison between male and female HCC patients uncovered several hub genes and demonstrated distinct signaling pathway activity alteration, especially diminished EMT in men and enhanced anticancer drug resistance in women. In addition, sex-based differential expression analysis revealed that men had more dysregulated genes than women. In female patients, GSEA demonstrated that alkaloid metabolic process was the most negatively enriched biological process, while the most positively enriched biological process was regulation of histone phosphorylation. Besides, retinol metabolism was the most negatively enriched pathway and RNA degradation was the most positively enriched pathway. In male patients, our analysis showed that protein activation cascade and RNA methylation were the most negatively and positively enriched biological processes, respectively. In addition, retinol metabolism and pyrimidine metabolism were the most negatively and positively enriched pathways, respectively. Similarly, functional enrichment analyses demonstrated that genes exclusively up-/down-regulated in male or female patients were involved in almost totally different biological network. In female patients, downregulated genes were mainly involved in various metabolic and catabolic processes, while upregulated genes were chiefly implicated in intracellular signal transduction and extracellular matrix organization-related pathways. In male patients, upregulated genes were enriched in molecule transport as well as adipogenesis-related processes and for downregulated genes, immune response-related processes were the most enriched pathway networks. Sex differences in adipose distribution, adipose tissue and liver substrate metabolism have been well documented, and adipose tissue has been identified as contributor to sex differences in HCC incidence [43, 54]. These data improved our understanding of sex-biased molecular mechanisms of HCC.
Brain-expressed X-linked 4 (BEX4) has been associated with several cancers, but whether it exerts pro- or anti-cancer effects depending on the tumor type [9-11, 55]. Here we found BEX4 was upregulated in female patients and expression of BEX4 differed significantly among immune subtypes as well as molecular subtypes of HCC. Moreover, we observed strong positive correlations between transcript levels of BEX4 and infiltration of various immune cells as well as immune checkpoint molecules. Interestingly, BEX4 expression was positively correlated with resistance to multiple receptor tyrosine kinases, including lenvatinib. Thus targeting BEX4 may represent a potential approach to enhance efficacy of lenvatinib plus anti-PD1 antibody combination therapy in HCC.
In summary, our study revealed significant differences of genetic alterations and TME between male and female HCC patients. Enrichment analyses revealed distinct biological processes were involved in sex-biased tumorigenesis of HCC. Enhanced metabolism of lipids was associated hepatocarcinogenesis in men, while ECM-organization-related pathways were correlated to HCC development in women. In addition, BEX4 was found upregulated in female but downregulated in male HCC patients, and was positively correlated with immune checkpoint molecules and infiltrated immune cell. These results deepened our understanding of sex differences in HCC, and will help to elucidate sex-biased molecular mechanism of tumorigenesis. Further exploration will contribute to the development of novel therapeutic strategies and improve efficacy of lenvatinib plus immune checkpoint blockade combination therapy.
Abbreviations
HCC: hepatocellular carcinoma; Bex4: brain-expressed x-linked 4; CVCDAP: cancer virtual cohort discovery analysis platform; GEPIA: gene expression profiling interactive analysis; GSEA: gene set enrichment analysis; GSCA: gene set cancer analysis; RT-qPCR: real-time quantitative polymerase chain reaction; IHC: immunohistochemistry; DEGs: differentially expressed genes; TCGA: the cancer genome atlas; KEGG: kyoto encyclopedia of genes and genomes; TME: tumor microenvironment; ECM: extracellular matrix; EMT: epithelial-mesenchymal transition.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This study was supported by National Natural Science Foundation of China (82000576 and 81972703).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: a cancer journal for clinicians. 2021;71:209-49
2. Kanwal F, Singal AG. Surveillance for Hepatocellular Carcinoma: Current Best Practice and Future Direction. Gastroenterology. 2019;157:54-64
3. Li Z, Tuteja G, Schug J, Kaestner KH. Foxa1 and Foxa2 are essential for sexual dimorphism in liver cancer. Cell. 2012;148:72-83
4. Zheng B, Zhu YJ, Wang HY, Chen L. Gender disparity in hepatocellular carcinoma (HCC): multiple underlying mechanisms. Science China Life sciences. 2017;60:575-84
5. Naugler WE, Sakurai T, Kim S, Maeda S, Kim K, Elsharkawy AM. et al. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 2007;317:121-4
6. Fernandez EM, Diaz-Ceso MD, Vilar M. Brain expressed and X-linked (Bex) proteins are intrinsically disordered proteins (IDPs) and form new signaling hubs. PLoS One. 2015;10:e0117206
7. Kazi JU, Kabir NN, Ronnstrand L. Brain-Expressed X-linked (BEX) proteins in human cancers. Biochim Biophys Acta. 2015;1856:226-33
8. Lee S, Kang H, Shin E, Jeon J, Youn H, Youn B. BEX1 and BEX4 Induce GBM Progression through Regulation of Actin Polymerization and Activation of YAP/TAZ Signaling. Int J Mol Sci. 2021 22
9. Lee JK, Lee J, Go H, Lee CG, Kim S, Kim HS. et al. Oncogenic microtubule hyperacetylation through BEX4-mediated sirtuin 2 inhibition. Cell Death Dis. 2016;7:e2336
10. Chien J, Staub J, Avula R, Zhang H, Liu W, Hartmann LC. et al. Epigenetic silencing of TCEAL7 (Bex4) in ovarian cancer. Oncogene. 2005;24:5089-100
11. Gao W, Li JZ, Chen SQ, Chu CY, Chan JY, Wong TS. Decreased brain-expressed X-linked 4 (BEX4) expression promotes growth of oral squamous cell carcinoma. J Exp Clin Cancer Res. 2016;35:92
12. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6:pl1
13. Amaral ML, Erikson GA, Shokhirev MN. BART: bioinformatics array research tool. BMC Bioinformatics. 2018;19:296
14. Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453-7
15. Guan X, Cai M, Du Y, Yang E, Ji J, Wu J. CVCDAP: an integrated platform for molecular and clinical analysis of cancer virtual cohorts. Nucleic Acids Res. 2020;48:W463-W71
16. Li C, Tang Z, Zhang W, Ye Z, Liu F. GEPIA2021: integrating multiple deconvolution-based analysis into GEPIA. Nucleic Acids Res. 2021;49:W242-W6
17. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545-50
18. Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nature communications. 2019;10:1523
19. Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S. et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605-D12
20. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498-504
21. Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC. et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35:4200-2
22. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS. et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017;77:e108-e10
23. Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q. et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509-W14
24. Liu CJ, Hu FF, Xia MX, Han L, Zhang Q, Guo AY. GSCALite: a web server for gene set cancer analysis. Bioinformatics. 2018;34:3771-2
25. Zheng Y, Huang C, Lu L, Yu K, Zhao J, Chen M. et al. STOML2 potentiates metastasis of hepatocellular carcinoma by promoting PINK1-mediated mitophagy and regulates sensitivity to lenvatinib. J Hematol Oncol. 2021;14:16
26. Chen J, Lin Z, Liu L, Zhang R, Geng Y, Fan M. et al. GOLM1 exacerbates CD8(+) T cell suppression in hepatocellular carcinoma by promoting exosomal PD-L1 transport into tumor-associated macrophages. Signal Transduct Target Ther. 2021;6:397
27. Li CH, Prokopec SD, Sun RX, Yousif F, Schmitz N, Subtypes PT. et al. Sex differences in oncogenic mutational processes. Nature communications. 2020;11:4330
28. Li CH, Haider S, Shiah YJ, Thai K, Boutros PC. Sex Differences in Cancer Driver Genes and Biomarkers. Cancer Res. 2018;78:5527-37
29. Yuan Y, Liu L, Chen H, Wang Y, Xu Y, Mao H. et al. Comprehensive Characterization of Molecular Differences in Cancer between Male and Female Patients. Cancer Cell. 2016;29:711-22
30. Dunford A, Weinstock DM, Savova V, Schumacher SE, Cleary JP, Yoda A. et al. Tumor-suppressor genes that escape from X-inactivation contribute to cancer sex bias. Nat Genet. 2017;49:10-6
31. Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH. et al. The Immune Landscape of Cancer. Immunity. 2018;48:812-30 e14
32. Bagaev A, Kotlov N, Nomie K, Svekolkin V, Gafurov A, Isaeva O. et al. Conserved pan-cancer microenvironment subtypes predict response to immunotherapy. Cancer Cell. 2021;39:845-65 e7
33. Dituri F, Mancarella S, Cigliano A, Chieti A, Giannelli G. TGF-beta as Multifaceted Orchestrator in HCC Progression: Signaling, EMT, Immune Microenvironment, and Novel Therapeutic Perspectives. Seminars in liver disease. 2019;39:53-69
34. Lepper ER, Nooter K, Verweij J, Acharya MR, Figg WD, Sparreboom A. Mechanisms of resistance to anticancer drugs: the role of the polymorphic ABC transporters ABCB1 and ABCG2. Pharmacogenomics. 2005;6:115-38
35. Peer CJ, Sissung TM, Kim A, Jain L, Woo S, Gardner ER. et al. Sorafenib is an inhibitor of UGT1A1 but is metabolized by UGT1A9: implications of genetic variants on pharmacokinetics and hyperbilirubinemia. Clin Cancer Res. 2012;18:2099-107
36. Shi Z, Tiwari AK, Shukla S, Robey RW, Singh S, Kim IW. et al. Sildenafil reverses ABCB1- and ABCG2-mediated chemotherapeutic drug resistance. Cancer Res. 2011;71:3029-41
37. Basu A, Bodycombe NE, Cheah JH, Price EV, Liu K, Schaefer GI. et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013;154:1151-61
38. Kato Y, Tabata K, Kimura T, Yachie-Kinoshita A, Ozawa Y, Yamada K. et al. Lenvatinib plus anti-PD-1 antibody combination treatment activates CD8+ T cells through reduction of tumor-associated macrophage and activation of the interferon pathway. PloS one. 2019;14:e0212513
39. Kimura T, Kato Y, Ozawa Y, Kodama K, Ito J, Ichikawa K. et al. Immunomodulatory activity of lenvatinib contributes to antitumor activity in the Hepa1-6 hepatocellular carcinoma model. Cancer Sci. 2018;109:3993-4002
40. Tricarico R, Nicolas E, Hall MJ, Golemis EA. X- and Y-Linked Chromatin-Modifying Genes as Regulators of Sex-Specific Cancer Incidence and Prognosis. Clin Cancer Res. 2020;26:5567-78
41. Wilson MA, Buetow KH. Novel Mechanisms of Cancer Emerge When Accounting for Sex as a Biological Variable. Cancer Res. 2020;80:27-9
42. Haupt S, Caramia F, Klein SL, Rubin JB, Haupt Y. Sex disparities matter in cancer development and therapy. Nat Rev Cancer. 2021;21:393-407
43. Manieri E, Herrera-Melle L, Mora A, Tomas-Loba A, Leiva-Vega L, Fernandez DI. et al. Adiponectin accounts for gender differences in hepatocellular carcinoma incidence. J Exp Med. 2019;216:1108-19
44. Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17-37
45. Lopes-Ramos CM, Quackenbush J, DeMeo DL. Genome-Wide Sex and Gender Differences in Cancer. Frontiers in oncology. 2020;10:597788
46. Tijsen AJ, Cocera Ortega L, Reckman YJ, Zhang X, van der Made I, Aufiero S. et al. Titin Circular RNAs Create a Back-Splice Motif Essential for SRSF10 Splicing. Circulation. 2021;143:1502-12
47. Labeit S, Kolmerer B. Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science. 1995;270:293-6
48. Simon DN, Wilson KL. The nucleoskeleton as a genome-associated dynamic 'network of networks'. Nat Rev Mol Cell Biol. 2011;12:695-708
49. Jones D, Wang Z, Chen IX, Zhang S, Banerji R, Lei PJ. et al. Solid stress impairs lymphocyte infiltration into lymph-node metastases. Nature biomedical engineering. 2021;5:1426-36
50. Anderson NM, Simon MC. The tumor microenvironment. Curr Biol. 2020;30:R921-R5
51. Subramanian M, Marelli-Berg FM. CD36 pumps fat to defang killer T cells in tumors. Cell metabolism. 2021;33:1509-11
52. Clocchiatti A, Cora E, Zhang Y, Dotto GP. Sexual dimorphism in cancer. Nat Rev Cancer. 2016;16:330-9
53. Castro A, Pyke RM, Zhang X, Thompson WK, Day CP, Alexandrov LB. et al. Strength of immune selection in tumors varies with sex and age. Nature communications. 2020;11:4128
54. Goossens GH, Jocken JWE, Blaak EE. Sexual dimorphism in cardiometabolic health: the role of adipose tissue, muscle and liver. Nature reviews Endocrinology. 2021;17:47-66
55. Lee JK, Ha GH, Kim HS, Lee CW. Oncogenic potential of BEX4 is conferred by Polo-like kinase 1-mediated phosphorylation. Experimental & molecular medicine. 2018;50:1-12
Author contact
![]() Corresponding authors: Jubo Zhang, Department of Infectious Diseases, Huashan Hospital, Fudan University, Shanghai 200040, China. E-mail: drzhangjuboedu.cn; Huaping Sun, Department of Radiology, Huashan Hospital, Fudan University, Shanghai 200040, China. E-mail: 452812668com; Lu Lu, Department of General Surgery, Huashan Hospital, Cancer Metastasis Institute, Fudan University, Shanghai 200040, China. E-mail: luluorg.cn; Chuanmiao Liu, Department of Infectious Diseases, The First Affiliated Hospital of Bengbu Medical College, Bengbu 233000, China. E-mail: liuchuanmiao119com.
Corresponding authors: Jubo Zhang, Department of Infectious Diseases, Huashan Hospital, Fudan University, Shanghai 200040, China. E-mail: drzhangjuboedu.cn; Huaping Sun, Department of Radiology, Huashan Hospital, Fudan University, Shanghai 200040, China. E-mail: 452812668com; Lu Lu, Department of General Surgery, Huashan Hospital, Cancer Metastasis Institute, Fudan University, Shanghai 200040, China. E-mail: luluorg.cn; Chuanmiao Liu, Department of Infectious Diseases, The First Affiliated Hospital of Bengbu Medical College, Bengbu 233000, China. E-mail: liuchuanmiao119com.

 Global reach, higher impact
Global reach, higher impact