Impact Factor
ISSN: 1837-9664
J Cancer 2022; 13(11):3251-3257. doi:10.7150/jca.75754 This issue Cite
Research Paper
Analysis of MUC6 Genetic Variants on the Clinicopathologic Characteristics of Patients with Hepatocellular Carcinoma
1. School of Medicine, Chung Shan Medical University, Taichung 40201, Taiwan.
2. Institute of Medicine, Chung Shan Medical University, Taichung 40201, Taiwan.
3. Department of Surgery, Chung Shan Medical University Hospital, Taichung 40201, Taiwan.
4. Graduate Institute of Biomedical Sciences, China Medical University, Taichung 40402, Taiwan.
5. Ph.D. Program for Translational Medicine, China Medical University, Taichung 40402, Taiwan.
6. Institute of Translational Medicine and New Drug Development, Taichung 40402, Taiwan.
7. Drug Development Center, Research Center for Cancer Biology, China Medical University, Taichung 40402, Taiwan.
8. Center for Molecular Medicine, China Medical University Hospital, Taichung 40402, Taiwan.
9. Department of Beauty Science, National Taichung University of Science and Technology, Taichung 40404, Taiwan.
10. Department of Otorhinolaryngology Head and Neck Surgery, China Medical University Hospital, Taichung, Taiwan.
11. School of Medicine, College of Medicine, China Medical University, Taichung 40402, Taiwan.
12. Department of Surgery, China Medical University Hospital, Taichung 40402, Taiwan.
13. Department of Hematology and Oncology, China Medical University Hospital, Taichung 40402, Taiwan.
14. Institute of Medicine, Chung Shan Medical University, Taichung 40201, Taiwan.
15. Department of Medical Research, Chung Shan Medical University Hospital, Taichung 40201, Taiwan.
16. Department of Medical Laboratory Science and Biotechnology, Asia University, Taichung 41354, Taiwan.
#These authors contributed equally to this work.
Received 2022-6-2; Accepted 2022-7-27; Published 2022-9-6
Abstract
Hepatocellular carcinoma (HCC) is the leading malignancy associated with cancer-related deaths worldwide. Many studies have indicated that mucin (MUC) expression plays an important role in cancer metastasis and recurrence. MUC6 expression is observed in gastric and oncocytic phenotypes and may play an important role during cancer progression. We found the level of MUC6 is lower in HCC patients but did not affect the survival of HCC patients. Therefore, in this study, we investigated the combined effect of MUC6 polymorphisms and exposure to environmental carcinogens on the susceptibility to and clinicopathological characteristics of HCC. Three single-nucleotide polymorphisms (SNPs) of MUC6 (rs61869016, rs6597947, and rs7481521) from 1197 healthy controls and 423 HCC patients were analyzed using real-time PCR. After adjusting for other co-variants, we found that carrying a CC genotype at MUC6 rs61869016 had a lower risk of developing HCC than wildtype carriers. Moreover, patients with a smoking habit who carried the C allele of rs61869016 and T allele of rs7481521 had a higher (B or C) Child-Pugh score than other genotypes, suggesting significant functional compromise and decompensated disease. Therefore, our findings suggest that genetic variations in MUC6 may corelate to HCC and indicate progression in HCC patients.
Keywords: hepatocellular carcinoma, MUC6, Child-Pugh score, single-nucleotide polymorphisms
Introduction
The main risk factors for liver cancer are hepatitis virus infection and cirrhosis, as well as chronic hepatitis, which leads to cirrhosis and then to liver cancer. Most of the symptoms of cirrhosis are the result of the progression of viral hepatitis, drug-related hepatitis, and alcoholic hepatitis. Mucin (MUC) is the main component of any mucus secretion, providing the mucus with its biophysiochemical properties as a function of its characteristics and degree of glycosylation [1, 2]. Mucins play a role in both physiological and pathological conditions [3-7]. Aberrant expression of mucins can lead to loss of epithelial cell polarity and promote epithelial-mesenchymal transition (EMT), which leads to increased cell motility and invasion, a critically important step in tumorigenesis [3, 8, 9].
It is generally accepted that hepatocellular carcinoma (HCC) does not produce mucins, whereas cholangiocarcinoma (CC) or combined/mixed hepatocellular cholangiocarcinoma (cHCC-CC) may produce these glycoproteins [10, 11]. However, a growing number of reports have indicated that HCC cells that do not exhibit or that have not yet morphologically differentiated into the biliary phenotype can also produce mucins [12-15]. Mucin 6 (MUC6) is one of the main components of the mucus barrier in the stomach, and it is secreted by the pyloric gland cells of the gastric sinus and the mucus neck cells located in the lower layer of the gastric mucosa. MUC6 expression is observed in both gastric and cancer cell phenotypes. It has been reported that methylation of the MUC6 promoter may lead to significant downregulation of MUC6 in gastric cancer and promote the progression of gastric cancer [16]. Furthermore, high MUC6 expression is a characteristic in chronic viral hepatitis, which may induce hepatocellular carcinoma [17]. However, the detailed role of the tissue expression of mucins in HCC tumor cells is not well understood.
A number of studies have reported genetic susceptibility factors that may be involved in HCC. For example, single-nucleotide polymorphisms (SNPs) are the most common type of DNA sequence variation that have shown the potential to predict cancer risk [18, 19]. The expression of proteins or their functions may be altered by their SNPs, thus influencing the progression of cancer. The relationship between the expression of MUC6 SNPs and chronic atrophic gastritis was revealed [20]. However, the exact role of MUC6 SNPs in cancer progression and development in Taiwanese HCC patients remains poorly investigated. In the current study, we selected three MUC6 SNPs (rs61869016 (5'-UTR), rs6597947 (5'-UTR), and rs7481521 (exon)) with the aim of elucidating their correlations to Taiwanese HCC patients and cancer prognosis.
Materials and Methods
Study Participants and Specimen Collection
In this study, 423 HCC patients were recruited from Chung Shan Medical University Hospital in Taichung, Taiwan. All participants provided informed written consent during the registration process. HCC patients were clinically staged at the time of diagnosis according to the tumor/node/metastasis staging system of the American Joint Committee on Cancer (AJCC, 2002). The diagnosis of cirrhosis is based on liver biopsy or abdominal ultrasound. Clinical features, including liver cirrhosis, aspartate aminotransferase (AST), the levels of α-fetoprotein (AFP), alanine aminotransferase (ALT), tumor staging, tumor size, lymph-node metastasis, distant metastasis, presence of HBV surface antigen (HBsAg), and reactivity with antibody against HCV (anti-HCV), were collected from chart reviews. For the control group, 1197 individuals, between 20 and 70 years of age with no history of cancer, were selected from the Taiwan Biobank (https://www.twbiobank.org.tw).
The information on gender, age, cigarette smoking status, and alcohol drinking status was collected from each subject. An average of more than two drinks per day was considered alcohol consumption. Smoking of at least one cigarette per day in the latest 3 months was considered a persistent smoking habit. The research was approved by the Institutional Review Board of Chung Shan Medical University Hospital.
Comprehensive Analysis of MUC6 from The Cancer Genome Atlas (TCGA)
UALCAN is a comprehensive, user-friendly, and interactive web resource for analyzing cancer omics data (http://ualcan.path.uab.edu/index.html). UALCAN uses TCGA level 3 RNA-seq and clinical data from 31 cancer types [21]. Gene expression profile interactive analysis 2 (GEPIA2, http://gepia2.cancer-pku.cn/#index) is a updated version of GEPIA for analyzing the RNA sequencing expression data of 9,736 tumors and 8,587 normal samples from the TCGA and the GTEx projects, using a standard processing pipeline [22]. In this study, we used UALCAN and GEPIA2 for tumor/normal differential expression analysis and overall survial of MUC6 expression in HCC patients.
Selection of MUC6 Polymorphisms
A total of three SNPs in MUC6 (NM_005961.3) were selected from the International HapMap Project data for this study. We included the SNPs rs61869016 (5'-UTR), rs6597947 (5'-UTR), and rs7481521 (exon) of MUC6.
MUC6 Genotyping
Allelic discrimination of the MUC6 polymorphisms rs61869016, rs6597947, and rs7481521 was assessed using an ABI StepOne real-time polymerase chain reaction system (Applied Biosystems), SDS v3.0 software (Applied Biosystems), and the TaqMan assay [18].
Statistical Analyses
To evaluate the differences in age and demographic characteristics between the control groups and HCC patients, the Mann-Whitney U test was used. The odds ratios with 95% confidence intervals (CIs) were estimated using logistic regression models. A p-value <0.05 was considered significant. The data were analyzed using SAS statistical software.
Results
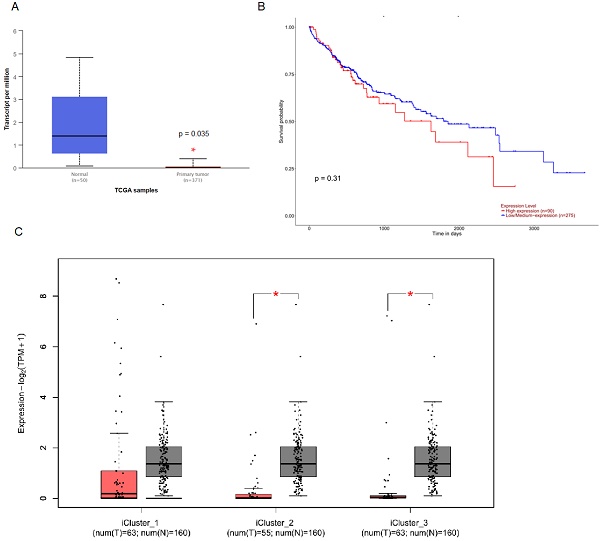
To investigate the clinical impact of MUC6 on HCC progression, we used UALCAN and GEPIA 2 to assess the relationship between cellular levels of MUC6 of normal people and HCC patients and the overall survival of HCC patients. The results indicated that the level of MUC6 in normal people was significantly much higher than in all and different subtypes HCC patients (Figure 1A and 1C). Interestingly, the expression of MUC6 did not affect the overall survival of HCC patients. This result implies that the regulation of MUC6 in HCC may have unknown mechanisms.
To identify possible factors causing HCC in clinical practice, a total of 1197 healthy controls and 423 HCC patients were recruited for this case cohort study. According to our analysis of HCC patients, we found significant differences in age (p < 0.001) and alcohol consumption (p < 0.001) between HCC patients and the healthy group (Table 1).
The level of MUC6 is correlated with HCC progression but not with the survival rate of HCC. (A) The level of MUC6 in normal control and hepatocellular carcinoma patients. (B) The overall survival of different levels of MUC6 in HCC patients as assessed with data from UALCAN. (C) The level of MUC6 in different subtypes of HCC patients. * p < 0.05.
Demographical characteristics of 1197 controls and 423 patients with HCC
| Variable | Controls (N = 1197) | Patients (N = 423) | p-value |
|---|---|---|---|
| Age (yrs) | Mean ± SD | Mean ± SD | |
| 59.4 ± 7.1 | 63.7 ± 11.2 | p < 0.001* | |
| Gender | |||
| Male | 838 (70.0%) | 298 (70.4%) | |
| Female | 359 (30.0%) | 125 (29.6%) | p = 0.865 |
| Cigarette smoking | |||
| No | 727 (60.7%) | 259 (61.2%) | |
| Yes | 470 (39.3%) | 164 (38.8%) | p = 0.858 |
| Alcohol drinking | |||
| No | 1028 (85.9%) | 279 (66.0%) | |
| Yes | 169 (14.1%) | 144 (34.0%) | p < 0.001* |
| HBsAg | |||
| Negative | 247 (58.4%) | ||
| Positive | 176 (41.6%) | ||
| Anti-HCV | |||
| Negative | 241 (57.0%) | ||
| Positive | 182 (43.0%) | ||
| Stage | |||
| I + II | 305 (72.1%) | ||
| III + IV | 118 (27.9%) | ||
| Tumor T status | |||
| T1 + T2 | 311 (73.5%) | ||
| T3 + T4 | 112 (26.5%) | ||
| Lymph node status | |||
| N0 | 412 (97.4%) | ||
| N1 + N2 + N3 | 11 (2.6%) | ||
| Metastasis | |||
| M0 | 400 (94.6%) | ||
| M1 | 23 (5.4%) | ||
| Vascular invasion | |||
| No | 359 (84.9%) | ||
| Yes | 64 (15.1%) | ||
| Child-Pugh score | |||
| A | 362 (85.6%) | ||
| B or C | 61 (14.4%) | ||
| Liver cirrhosis | |||
| Negative | 68 (16.1%) | ||
| Positive | 355 (83.9%) |
Mann-Whitney U test or Fisher's exact test was used between healthy controls and patients with HCC. * p < 0.05 was considered statistically significant.
To reduce possible confounding by several environmental factors, AORs and their corresponding 95% CIs were estimated by multivariate logistic regression models, after controlling for risks associated with age and alcohol consumption use. The genotype distributions and the associations between HCC and MUC6 SNPs are presented in Table 2. The alleles with the highest frequency of distribution in MUC6 rs61869016, rs6597947, and rs7481521 were homozygous T/T, homozygous C/C, and homozygous C/C, respectively, in HCC patients and controls. After adjusting for variables, individuals with rs61869016 C/C showed a 0.571-fold (95% CI: 0.380-0.858) lower risk of HCC. Individuals with the rs6597947 and rs7481521 polymorphisms showed no reduction in HCC risk compared to wildtype individuals.
Genotype and allele frequency of MUC6 single-nucleotide polymorphism (SNPs) in HCC patients and normal controls
| Variable | Controls (N = 1197) (%) | Patients (N = 423) (%) | OR (95% CI) | AOR (95% CI)a |
|---|---|---|---|---|
| rs61869016 | ||||
| TT | 497 (41.5%) | 191 (45.1%) | 1.000 (reference) | 1.000 (reference) |
| TC | 541 (45.2%) | 195 (46.1%) | 0.938 (0.742-1.185) | 0.912 (0.717-1.160) |
| CC | 159 (13.3%) | 37 (8.8%) | 0.606 (0.408-0.899)b | 0.571 (0.380-0.858)c |
| TC + CC | 700 (58.5%) | 232 (54.9%) | 0.862 (0.690-1.078) | 0.834 (0.662-1.050) |
| rs6597947 | ||||
| CC | 644 (53.8%) | 229 (54.1%) | 1.000 (reference) | 1.000 (reference) |
| CA | 460 (38.4%) | 165 (39.0%) | 1.009 (0.799-1.274) | 1.013 (0.797-1.288) |
| AA | 93 (7.8%) | 29 (6.9%) | 0.877 (0.563-1.366) | 0.931 (0.591-1.466) |
| CA + AA | 553 (46.2%) | 194 (45.9%) | 0.987 (0.790-1.232) | 0.999 (0.795-1.257) |
| rs7481521 | ||||
| CC | 605 (50.5%) | 204 (48.2%) | 1.000 (reference) | 1.000 (reference) |
| CT | 486 (40.6%) | 192 (45.4%) | 1.172 (0.931-1.475) | 1.137 (0.896-1.442) |
| TT | 106 (8.9%) | 27 (6.4%) | 0.756 (0.481-1.187) | 0.717 (0.451-1.141) |
| CT + TT | 592 (49.5%) | 219 (51.8%) | 1.097 (0.879-1.370) | 1.061 (0.844-1.333) |
a Adjusted for the effects of age and alcohol drinking; b p = 0.013; c p = 0.001.
In addition, the effect of the polymorphic genotypes of MUC6 rs61869016 and rs7481521 on the clinical status of HCC was investigated (Tables 3 and 4). The results showed that patients with the C/C genotype of the rs61869016 SNP (OR = 3.515, 95% CI: 1.040-11.878, p = 0.043) and the T/T genotype of the rs7481521 SNP (OR = 4.582, 95% CI: 1.061-19.778, p = 0.041) had a higher Child-Pugh score (B or C) compared to other genotypes, suggesting poor survival in patients with chronic liver disease.
Moreover, we analyzed the levels of AFP, AST, and ALT, common clinicopathological markers of HCC associated with MUC6 genotype frequency, to see how they related to the progression of clinical status in HCC patients. The homozygous genotype for the polymorphic allele of rs6597947 (C/A + A/A) had a significantly higher AST/ALT ratio compared with the C/C genotype in patients with HCC (Table 5).
Discussion
SNPs are single-nucleotide variants that occur at the DNA level in each human cell. Associated with environmental factors, SNPs can not only mimic the diversity of the human phenotype, but also indicate susceptibility to a variety of diseases, including cancer [23]. The association between SNPs and HCC has been tested in case-control and prospective cohort studies, which are based on hypothesis-driven, hypothetical genetic studies. For example, many of the changes affecting inflammatory pathways, oxidative stress, iron metabolism, or DNA repair mechanisms in hepatitis patients have been associated with the development of liver cancer [24].
Although the importance of MUC6 in cancer is well recognized, its exact role in tumorigenesis remains a controversial topic, as both oncogenic and inhibitory effects have been demonstrated [16, 25, 26]. For example, MUC6 is highly expressed at early noninvasive stages of pancreatic tumor progression and then suppressed or lost at invasive stages [27, 28]. The MUC6 SNP rs7481521 had a significant association with a decreased risk for homozygous carriers and a significant dose-response relation with the number of alleles in chronic atrophic gastritis patients [20]. However, the correlation between MUC6 polymorphisms and risk factors in HCC has not yet been clarified. The results of this study clarify the role of MUC6 SNPs in HCC susceptibility and other clinicopathological conditions.
Odds ratio (OR) and 95% confidence interval (CI) of clinical status and MUC6 rs61869016 genotypic frequencies in HCC patients among smokers
| Variable | OR (95% CI) | p-value | ||
|---|---|---|---|---|
| Clinical Stage | ||||
| rs61869016 | Stage I + II (n = 118) (%) | Stage III + IV (n = 46) (%) | ||
| TT | 44 (37.3%) | 23 (50.0%) | 1.00 | |
| TC | 60 (50.8%) | 20 (43.5%) | 0.638 (0.312-1.303) | p = 0.217 |
| CC | 14 (11.9%) | 3 (6.5%) | 0.410 (0.107-1.574) | p = 0.194 |
| Tumor size | ||||
| rs61869016 | ≤T2 (n = 118) (%) | >T2 (n = 46) (%) | ||
| TT | 43 (36.4%) | 24 (52.2%) | 1.00 | |
| TC | 61 (51.7%) | 19 (41.3%) | 0.588 (0.272-1.143) | p = 0.111 |
| CC | 14 (11.9%) | 3 (6.5%) | 0.384 (0.100-1.471) | p = 0.163 |
| Lymph node metastasis | ||||
| rs61869016 | No (n = 160) (%) | Yes (n = 4) (%) | ||
| TT | 66 (41.3%) | 1 (25.0%) | 1.00 | |
| TC | 77 (48.1%) | 3 (75.0%) | 2.571 (0.261-25.315) | p = 0.418 |
| CC | 17 (10.6%) | 0 (0.0%) | - | - |
| Distant metastasis | ||||
| rs61869016 | M0 (n = 156) (%) | M1 (n = 8) (%) | ||
| TT | 62 (39.7%) | 5 (62.5%) | 1.00 | |
| TC | 77 (49.4%) | 3 (37.5%) | 0.483 (0.111-2.101) | p = 0.332 |
| CC | 17 (10.9%) | 0 (0.0%) | - | - |
| Vascular invasion | ||||
| rs61869016 | No (n = 138) (%) | Yes (n = 26) (%) | ||
| TT | 59 (42.8%) | 8 (30.8%) | 1.00 | |
| TC | 66 (47.8%) | 14 (53.8%) | 1.564 (0.613-3.993) | p = 0.349 |
| CC | 13 (9.4%) | 4 (15.4%) | 2.269 (0.593-8.684) | p = 0.231 |
| Child-Pugh score | ||||
| rs61869016 | A (n = 141) (%) | B or C (n = 23) (%) | ||
| TT | 58 (41.1%) | 9 (39.1%) | 1.00 | |
| TC | 72 (51.1%) | 8 (34.8%) | 0.716 (0.260-1.972) | p = 0.518 |
| CC | 11 (7.8%) | 6 (26.1%) | 3.515 (1.040-11.878) | p = 0.043* |
| HBsAg | ||||
| rs61869016 | Negative (n = 93) (%) | Positive (n = 71) (%) | ||
| TT | 39 (41.9%) | 28 (39.4%) | 1.00 | |
| TC | 43 (46.3%) | 37 (52.1%) | 1.199 (0.623-2.307) | p = 0.588 |
| CC | 11 (11.8%) | 6 (8.5%) | 0.760 (0.251-2.298) | p = 0.627 |
| Anti-HCV | ||||
| rs61869016 | Negative (n = 92) (%) | Positive (n = 72) (%) | ||
| TT | 36 (39.1%) | 31 (43.1%) | 1.00 | |
| TC | 45 (48.9%) | 35 (48.6%) | 0.903 (0.470-1.734) | p = 0.760 |
| CC | 11 (12.0%) | 6 (8.3%) | 0.633 (0.210-1.912) | p = 0.418 |
| Liver cirrhosis | ||||
| rs61869016 | Negative (n = 26) (%) | Positive (n = 138) (%) | ||
| TT | 13 (50.0%) | 54 (39.1%) | 1.00 | |
| TC | 11 (42.3%) | 69 (50.0%) | 1.510 (0.627-3.635) | p = 0.358 |
| CC | 2 (7.7%) | 15 (10.9%) | 1.806 (0.366-8.897) | p = 0.468 |
The ORs analyzed by their 95% CIs were estimated by logistic regression models; >T2: multiple tumors more than 5 cm or tumor involving a major branch of the portal or hepatic vein(s); * p < 0.05 was considered statistically significant.
Odds ratio (OR) and 95% confidence interval (CI) of clinical status and MUC6 rs7481521 genotypic frequencies in HCC patients among smokers
| Variable | OR (95% CI) | p-value | ||
|---|---|---|---|---|
| Clinical Stage | ||||
| rs7481521 | Stage I + II (n = 118) (%) | Stage III + IV (n = 46) (%) | ||
| CC | 51 (43.2%) | 23 (50.0%) | 1.00 | |
| CT | 61 (51.7%) | 20 (43.5%) | 0.727 (0.359-1.472) | p = 0.376 |
| TT | 6 (5.1%) | 3 (6.5%) | 1.109 (0.255-4.826) | p = 0.891 |
| Tumor size | ||||
| rs7481521 | ≤T2 (n = 118) (%) | >T2 (n = 46) (%) | ||
| CC | 52 (44.1%) | 22 (47.8%) | 1.00 | |
| CT | 60 (50.8%) | 21 (45.6%) | 0.827 (0.409-1.672) | p = 0.598 |
| TT | 6 (5.1%) | 3 (6.6%) | 1.182 (0.271-5.155) | p = 0.824 |
| Lymph node metastasis | ||||
| rs7481521 | No (n = 160) (%) | Yes (n = 4) (%) | ||
| CC | 73 (45.6%) | 1 (25.0%) | 1.00 | |
| CT | 78 (48.8%) | 3 (75.0%) | 2.808 (0.286-27.603) | p = 0.376 |
| TT | 9 (5.6%) | 0 (0.0%) | - | - |
| Distant metastasis | ||||
| rs7481521 | M0 (n = 156) (%) | M1 (n = 8) (%) | ||
| CC | 70 (44.9%) | 4 (50.0%) | 1.00 | |
| CT | 77 (49.3%) | 4 (50.0%) | 0.909 (0.219-3.773) | p = 0.896 |
| TT | 9 (5.8%) | 0 (0.0%) | - | - |
| Vascular invasion | ||||
| rs7481521 | No (n = 138) (%) | Yes (n = 26) (%) | ||
| CC | 66 (47.8%) | 8 (30.8%) | 1.00 | |
| CT | 65 (47.1%) | 16 (61.5%) | 2.031 (0.813-5.071) | p = 0.129 |
| TT | 7 (5.1%) | 2 (7.7%) | 2.357 (0.416-13.353) | p = 0.333 |
| Child-Pugh score | ||||
| rs7481521 | A (n = 141) (%) | B or C (n = 23) (%) | ||
| CC | 63 (44.7%) | 11 (47.8%) | 1.00 | |
| CT | 73 (51.8%) | 8 (34.8%) | 0.628 (0.238-1.657) | p = 0.347 |
| TT | 5 (3.5%) | 4 (17.4%) | 4.582 (1.061-19.778) | p = 0.041* |
| HBsAg | ||||
| rs7481521 | Negative (n = 93) (%) | Positive (n = 71) (%) | ||
| CC | 41 (44.1%) | 33 (46.5%) | 1.00 | |
| CT | 47 (50.5%) | 34 (47.9%) | 0.899 (0.476-1.698) | p = 0.742 |
| TT | 5 (5.4%) | 4 (5.6%) | 0.994 (0.247-4.000) | p = 0.993 |
| Anti-HCV | ||||
| rs7481521 | Negative (n = 92) (%) | Positive (n = 72) (%) | ||
| CC | 45 (48.9%) | 29 (40.3%) | 1.00 | |
| CT | 40 (43.5) | 41 (56.9%) | 1.591 (0.840-3.012) | p = 0.154 |
| TT | 7 (7.6%) | 2 (2.8%) | 0.443 (0.086-2.284) | p = 0.331 |
| Liver cirrhosis | ||||
| rs7481521 | Negative (n = 26) (%) | Positive (n = 138) (%) | ||
| CC | 13 (50.0%) | 61 (44.2%) | 1.00 | |
| CT | 12 (46.2%) | 69 (50.0%) | 1.225 (0.520-2.887) | p = 0.642 |
| TT | 1 (3.8%) | 8 (5.8%) | 1.705 (0.196-14.833) | p = 0.629 |
The ORs analyzed by their 95% CIs were estimated by logistic regression models; >T2: multiple tumors more than 5 cm or tumor involving a major branch of the portal or hepatic vein(s); * p < 0.05 was considered statistically significant.
Association of MUC6 genotypic frequencies with the HCC laboratory findings
| Characteristic | α-Fetoproteina (ng/mL) | ASTa (IU/L) | ALTa (IU/L) | AST/ALTa ratio |
|---|---|---|---|---|
| rs61869016 | ||||
| TT | 996.4 ± 364.4 | 44.8 ± 5.3 | 42.0 ± 4.5 | 1.23 ± 0.04 |
| TC + CC | 990.8 ± 325.9 | 42.8 ± 3.7 | 79.7 ± 39.4 | 1.18 ± 0.02 |
| p-value | 0.991 | 0.761 | 0.342 | 0.248 |
| p-valueb | 0.935 | 0.779 | 0.398 | 0.214 |
| rs6597947 | ||||
| CC | 1084.0 ± 362.2 | 41.4 ± 2.7 | 85.5 ± 42.1 | 1.15 ± 0.02 |
| CA + AA | 886.9 ± 314.3 | 46.3 ± 5.9 | 38.2 ± 3.7 | 1.26 ± 0.04 |
| p-value | 0.681 | 0.445 | 0.264 | 0.004 |
| p-valueb | 0.705 | 0.424 | 0.297 | 0.002 |
| rs7481521 | ||||
| CC | 784.8 ± 306.2 | 44.6 ± 5.0 | 43.7 ± 4.7 | 1.22 ± 0.03 |
| CT + TT | 1201.0 ± 377.4 | 42.7 ± 3.6 | 83.6 ± 45.2 | 1.18 ± 0.02 |
| p-value | 0.392 | 0.751 | 0.380 | 0.281 |
| p-valueb | 0.471 | 0.784 | 0.354 | 0.275 |
The Mann-Whitney U test was used between two groups; a mean ± SE; b adjusted for age and alcohol drinking.
Taiwan is a region where viral hepatitis is very common; thus, if the AST/ALT ratio is high in Taiwan, the most likely cause is chronic hepatitis B, hepatitis C, or fatty liver disease [29-32]. In our results, we observed that MUC6 SNP rs6597947 was correlated with a significantly higher AST/ALT ratio in HCC patients. Moreover, the ALT value exceeded the normal value of 40 IU/L in patients with MUC6 SNP rs61869016 and rs7481521, indicating liver damage (Table 5). As shown in Table 4, patients with the C/C genotype of SNP rs61869016 and the T/T genotype of SNP rs7481521 had higher Child-Pugh scores (B or C), suggesting poor survival in patients with chronic liver disease. In chronic viral liver disease, including chronic viral hepatitis, chronic alcoholism, and nonalcoholic fatty liver disease, an elevated AST/ALT ratio can be interpreted as a predictor for assessing long-term complications such as fibrosis and cirrhosis. In recent years, chronic liver inflammation has been the subject of intense research and is thought to have the potential to progress to liver cancer [29]. Interestingly, after adjusting for variables, individuals with rs61869016 C/C showed a lower risk of HCC (Table 2). Unfortunately, the sample size of MUC6 polymorphism for C/C at rs61869016 is 37 patients and we cannot provide the prognosis data for MUC6 polymorphism for CC at rs61869016 in this current study. However, the detailed mechanisms of MUC6 SNPs in HCC require future elucidation.
Conclusions
In conclusion, our findings suggest that genetic variations in MUC6 may help to predict cancer susceptibility and hepatitis in HCC. This study provides new information about the relationship between MUC6 polymorphisms and the clinical pathology of HCC in the Taiwanese population.
Acknowledgements
Funding Statement
This research was funded by grants from the Ministry of Science and Technology, Taiwan (MOST 109-2320-B-039-013-MY3), the National Health Research Institute, Taiwan (NHRI-110A1-CACO-13222202), the China Medical University, Taiwan (CMU108-MF-01; CMU109-MF-03), the China Medical University Hospital, Taiwan (DMR-111-053; DMR-111-209; DMR-111-202), and the “Drug Development Center, China Medical University” from the Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE), Taiwan.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Author Contributions
Conceptualization and data curation, H.L.L., Y.C.C., and S.F.Y.; formal analysis, H.L.W., L.Y.B., C.H.H., and S.F.Y.; resources and software, C.H.H., L.C.L., and G.W.W.; writing—original draft and methodology, Y.C.C., C.H.H., and H.L.L.; supervision and writing—review and editing, L.Y.B., S.F.Y., and Y.L.Y. All authors read and agreed to the published version of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Corfield A. Eukaryotic protein glycosylation: a primer for histochemists and cell biologists. Histochem Cell Biol. 2017;147:119-47
2. Hanisch FG. O-glycosylation of the mucin type. Biol Chem. 2001;382:143-9
3. Dhanisha SS, Guruvayoorappan C, Drishya S, Abeesh P. Mucins: Structural diversity, biosynthesis, its role in pathogenesis and as possible therapeutic targets. Crit Rev Oncol Hematol. 2018;122:98-122
4. Moniaux N, Escande F, Porchet N, Aubert JP, Batra SK. Structural organization and classification of the human mucin genes. Front Biosci. 2001;6:D1192-206
5. Nath S, Mukherjee P. MUC1: a multifaceted oncoprotein with a key role in cancer progression. Trends Mol Med. 2014;20:332-42
6. Rachagani S, Torres MP, Moniaux N, Batra SK. Current status of mucins in the diagnosis and therapy of cancer. Biofactors. 2009;35:509-27
7. Yonezawa S, Higashi M, Yamada N, Yokoyama S, Kitamoto S, Kitajima S. et al. Mucins in human neoplasms: clinical pathology, gene expression and diagnostic application. Pathol Int. 2011;61:697-716
8. Hollingsworth MA, Swanson BJ. Mucins in cancer: protection and control of the cell surface. Nat Rev Cancer. 2004;4:45-60
9. Rajabi H, Kufe D. MUC1-C Oncoprotein Integrates a Program of EMT, Epigenetic Reprogramming and Immune Evasion in Human Carcinomas. Biochim Biophys Acta Rev Cancer. 2017;1868:117-22
10. Matull WR, Andreola F, Loh A, Adiguzel Z, Deheragoda M, Qureshi U. et al. MUC4 and MUC5AC are highly specific tumour-associated mucins in biliary tract cancer. Br J Cancer. 2008;98:1675-81
11. Uchida T, Yamamoto Y, Ito T, Okamura Y, Sugiura T, Uesaka K. et al. Cystic micropapillary neoplasm of peribiliary glands with concomitant perihilar cholangiocarcinoma. World J Gastroenterol. 2016;22:2391-7
12. Dai Y, Liu L, Zeng T, Liang JZ, Song Y, Chen K. et al. Overexpression of MUC13, a Poor Prognostic Predictor, Promotes Cell Growth by Activating Wnt Signaling in Hepatocellular Carcinoma. Am J Pathol. 2018;188:378-91
13. Ling Y, Zhu J, Gao L, Liu Y, Zhu C, Li R. et al. The silence of MUC2 mRNA induced by promoter hypermethylation associated with HBV in Hepatocellular Carcinoma. BMC Med Genet. 2013;14:14
14. Wang RY, Chen L, Chen HY, Hu L, Li L, Sun HY. et al. MUC15 inhibits dimerization of EGFR and PI3K-AKT signaling and is associated with aggressive hepatocellular carcinomas in patients. Gastroenterology. 2013;145:1436-48 e1-12
15. Bozkaya G, Korhan P, Cokakli M, Erdal E, Sagol O, Karademir S. et al. Cooperative interaction of MUC1 with the HGF/c-Met pathway during hepatocarcinogenesis. Mol Cancer. 2012;11:64
16. Shi D, Xi XX. Regulation of MUC6 Methylation Correlates with Progression of Gastric Cancer. Yonsei Med J. 2021;62:1005-15
17. Sasaki M, Nakanuma Y, Ho SB, Kim YS. Increased MUC6 apomucin expression is a characteristic of reactive biliary epithelium in chronic viral hepatitis. J Pathol. 1998;185:191-8
18. Shastry BS. SNP alleles in human disease and evolution. J Hum Genet. 2002;47:561-6
19. Yu YL, Su KJ, Hsieh YH, Lee HL, Chen TY, Hsiao PC. et al. Effects of EZH2 polymorphisms on susceptibility to and pathological development of hepatocellular carcinoma. PLoS One. 2013;8:e74870
20. Frank B, Weck MN, Muller H, Klopp N, Illig T, Raum E. et al. Polymorphisms in MUC1, MUC2, MUC5B and MUC6 genes are not associated with the risk of chronic atrophic gastritis. Eur J Cancer. 2012;48:114-20
21. Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B. et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649-58
22. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556-W60
23. Dong LM, Potter JD, White E, Ulrich CM, Cardon LR, Peters U. Genetic susceptibility to cancer: the role of polymorphisms in candidate genes. JAMA. 2008;299:2423-36
24. Nahon P, Zucman-Rossi J. Single nucleotide polymorphisms and risk of hepatocellular carcinoma in cirrhosis. J Hepatol. 2012;57:663-74
25. Yuki A, Fujii C, Yamanoi K, Matoba H, Harumiya S, Kawakubo M. et al. Glycosylation of MUC6 by alpha1,4-linked N-acetylglucosamine enhances suppression of pancreatic cancer malignancy. Cancer Sci. 2022;113:576-86
26. Reis CA, David L, Carvalho F, Mandel U, de Bolos C, Mirgorodskaya E. et al. Immunohistochemical study of the expression of MUC6 mucin and co-expression of other secreted mucins (MUC5AC and MUC2) in human gastric carcinomas. J Histochem Cytochem. 2000;48:377-88
27. Ohya A, Yamanoi K, Shimojo H, Fujii C, Nakayama J. Gastric gland mucin-specific O-glycan expression decreases with tumor progression from precursor lesions to pancreatic cancer. Cancer Sci. 2017;108:1897-902
28. Sierzega M, Mlynarski D, Tomaszewska R, Kulig J. Semiquantitative immunohistochemistry for mucin (MUC1, MUC2, MUC3, MUC4, MUC5AC, and MUC6) profiling of pancreatic ductal cell adenocarcinoma improves diagnostic and prognostic performance. Histopathology. 2016;69:582-91
29. Botros M, Sikaris KA. The de ritis ratio: the test of time. Clin Biochem Rev. 2013;34:117-30
30. De Ritis F, Coltorti M, Giusti G. An enzymic test for the diagnosis of viral hepatitis; the transaminase serum activities. Clin Chim Acta. 1957;2:70-4
31. Huang TY, Peng SF, Huang YP, Tsai CH, Tsai FJ, Huang CY. et al. Combinational treatment of all-trans retinoic acid (ATRA) and bisdemethoxycurcumin (BDMC)-induced apoptosis in liver cancer Hep3B cells. J Food Biochem. 2020;44:e13122
32. Bayram AA, Al-Dahmoshi HOM, Al-Khafaji NSK, Al Mammori RTO, Al-Shimmery AHS, Saki M. Study of the D-dimer, C-reactive protein, and autoantibodies markers among HBV infected patients in Babylon province, Iraq. Biomedicine (Taipei). 2021;11:67-72
Author contact
![]() Corresponding authors: lybai6com (L.-Y. B.); ysfedu.tw (S.-F.Y.); ylyucmu.edu.tw (Y.-L.Y.).
Corresponding authors: lybai6com (L.-Y. B.); ysfedu.tw (S.-F.Y.); ylyucmu.edu.tw (Y.-L.Y.).

 Global reach, higher impact
Global reach, higher impact