3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2022; 13(15):3701-3709. doi:10.7150/jca.78498 This issue Cite
Research Paper
Exonuclease 1 genetic variant is associated with clinical outcomes of pemetrexed chemotherapy in lung adenocarcinoma
1. Department of Biochemistry, School of Medicine, Kyungpook National University, Daegu, Republic of Korea.
2. Cell and Matrix Research Institute, School of Medicine, Kyungpook National University, Daegu, Korea.
3. Department of Internal Medicine, School of Medicine, Kyungpook National University, Daegu, Republic of Korea.
4. BK21 Plus KNU Biomedical Convergence Program, Department of Biomedical Science, Kyungpook National University, Daegu, Korea.
5. Department of Pathology, School of Medicine, Kyungpook National University, Daegu, Republic of Korea.
6. Department of Radiology School of Medicine, Kyungpook National University, Daegu, Republic of Korea.
7. Medical Research Collaboration Center in Kyungpook National University Hospital and School of Medicine, Kyungpook National University, Daegu, Republic of Korea.
8. Department of Thoracic Surgery, Soonchunhyang University Gumi Hospital, Gumi, Korea.
*These authors contributed equally to this work.
Received 2022-8-30; Accepted 2022-11-16; Published 2022-12-7
Abstract
Pemetrexed is an anti-folate agent which is one of the most frequently used chemotherapy agents for non-squamous non-small cell lung cancer (NSCLC) patients. However, clinical response to pemetrexed chemotherapy and survival outcome of patients varies significantly. We evaluated whether the genetic variants in miRNA target sites may affect the treatment outcome of pemetrexed chemotherapy in lung adenocarcinoma patients. One hundred SNPs in miRNA binding regions in cancer-related genes were obtained from the crosslinking, ligation, and sequencing of hybrids (CLASH) and CancerGenes database, and the associations with the response to pemetrexed chemotherapy and survival outcomes were investigated in 314 lung adenocarcinoma patients. Two polymorphisms, EXO1 rs1047840G>A and CAMKK2 rs1653586G>T, were significantly associated with worse chemotherapy response (adjusted odds ratio [aOR] = 0.41, 95% CI = 0.24-0.68, P = 0.001, under dominant model; and aOR = 0.33, 95% CI = 0.16-0.67, P = 0.002, under dominant model, respectively) and worse OS (adjusted hazard ratio [aHR] = 1.34, 95% CI = 1.01-1.77, P = 0.04, under dominant model; and aHR = 1.50, 95% CI = 1.06-2.13, P = 0.02, under dominant model, respectively) in multivariate analyses. Significantly increased luciferase activity was noted in EXO1 rs1047840 A allele compared to G allele. In conclusion, two SNPs in miRNA binding sites, especially EXO1 rs1047840G>A, were associated with the chemotherapy response and survival outcome in lung adenocarcinoma patients treated with pemetrexed.
Keywords: lung adenocarcinoma, miRNA target sites, genetic variants, chemotherapy, response, survival
Introduction
A huge effort has been made to improve the prognosis of lung cancer patients over the past decades, and the treatment of lung cancer has progressed remarkably by recent innovations of targeted therapy and immunotherapy [1-3]. However, cytotoxic chemotherapy alone or in combination with other cancer therapies remains as an important treatment modality for non-small cell lung cancer (NSCLC) patients. Pemetrexed is one of the most frequently used chemotherapeutic agents for non-squamous NSCLC patients. Recently, immune checkpoint blockade combined with pemetrexed and platinum doublet chemotherapy has been approved as a standard first-line treatment in patients without driver mutations, which significantly increased overall survival (OS) and progression-free survival (PFS) compared to previous standard of care chemotherapy in non-squamous NSCLC patients [4].
Pemetrexed is an anti-folate agent that inhibits cell replication and growth through interrupting synthesis of DNA and RNA by targeting three enzymes involved in purine and pyrimidine synthesis: thymidylate synthase (TS), dihydrofolate reductase (DHFR), and glycinamide ribonucleotide formyltransferase (GARFT) [5]. It is widely accepted that the efficacy of pemetrexed is worse in squamous NSCLC and small-cell lung cancer than non-squamous NSCLC [6], but therapeutic outcomes of pemetrexed varies significantly even in patients with adenocarcinoma. Despite much effort to discover and validate molecular biomarkers to predict efficacy of pemetrexed, no viable biomarkers have yet been clearly established in clinical practice [7-9].
MicroRNA (miRNA) is a small (containing about 22 nucleotides) single-stranded non-coding RNA molecule that functions as endogenous negative regulators by binding to the 3'-untranslated region (UTR) of target mRNA causing degradation or translational inhibition of the target [10, 11]. Comprehensive miRNA profiling demonstrated that miRNAs are involved in vital biological processes such as cell division and death, cellular metabolism, intracellular signaling, and immunity [12-15]. In addition, miRNAs are critically involved in the development and progression of diverse human cancers [16, 17]. Computational approach for miRNA target prediction have focused on canonical (seed-based) target sites, demonstrating that single nucleotide polymorphisms (SNPs) in miRNA binding sites could be used as cancer biomarkers for predicting cancer risk, therapeutic response and prognosis of patients [18]. However, recent advances in transcriptome-wide mapping of miRNA target sites by crosslinking, ligation, and sequencing of hybrid (CLASH), elucidated that considerable portion of miRNA-mRNA interactions are mediated not only through canonical seed sites, but also through non-canonical sites [19]. Based on the association between miRNA dysregulation and human malignancies, we hypothesized that SNPs in miRNA target sites may alter miRNA-mRNA binding and consequently gene expression, thus affecting the treatment response and survival outcomes of patients treated with pemetrexed. To test this hypothesis, we selected SNPs in miRNA target sites using CLASH data and analyzed the association with the clinical outcome of pemetrexed treatment in lung adenocarcinoma patients.
Materials and Methods
Study populations
A total of 314 lung adenocarcinoma patients with available genomic DNA samples who underwent pemetrexed based chemotherapy at Kyungpook National University Hospital (KNUH) in Daegu, Korea, between March 2007 and July 2015, were enrolled. We excluded patients who had any active malignancies ≤ 3 years before the diagnosis of lung cancer, except resected non-melanoma skin cancer and in situ cancers including carcinoma in situ of the cervix or breast. Patients who were diagnosed with stage III/IV or had recurred disease after curative surgery were treated with either pemetrexed plus cisplatin for up to 4 to 6 cycles as first line chemotherapy regimen with/without pemetrexed maintenance therapy, or pemetrexed alone as second or further line chemotherapy. Because pemetrexed maintenance therapy was approved in 2009 in Korea, patients who were treated with pemetrexed plus cisplatin before 2009 did not underwent pemetrexed maintenance therapy even if their disease had not progressed after four cycles of first-line pemetrexed/cisplatin chemotherapy [20]. Chemotherapy was discontinued upon disease progression, major toxicities, or according to patient's or physician's decision. Response to chemotherapy was assessed according to the Response Evaluation Criteria in Solid Tumors [21] and the best overall response for each patient was reported. Patients with complete response or partial response were defined as responders, and patients with stable disease or progressive disease were defined as nonresponders. Survival outcome was assessed with overall survival (OS), the time between the first date of chemotherapy and date of death or last follow-up. Genomic DNA samples were provided by the National Biobank of Korea, KNUH, which is supported by the Ministry of Health, Welfare and Family Affairs. Written informed consent was obtained from all patients and this study was approved by the Institutional Review Board of the KNUH.
SNP selection and genotyping
PolymiRTS database 3.0 (http://compbio.uthsc.edu/miRSNP) [22] was used to find potentially functional variants in miRNA binding region, and 24,027 SNPs were extracted from the CLASH data which has been integrated in PolymiRTS database. Out of these, 1,574 SNPs in genes involved in human cancer were selected using the CancerGenes database (http://cbio.mskcc.org/cancergenes) [23]. Lastly, 100 SNPs were collected after excluding those with minor allele frequency < 0.05 in the HapMap-JPT and those in strong linkage disequilibrium (LD, r2 ≥ 0.8). Genotyping was performed using the iPLEX® Assay and MassARRAY® System (Agena Bioscience, San Diego, CA, USA).
Cloning of the luciferase reporter gene and dual luciferase assay
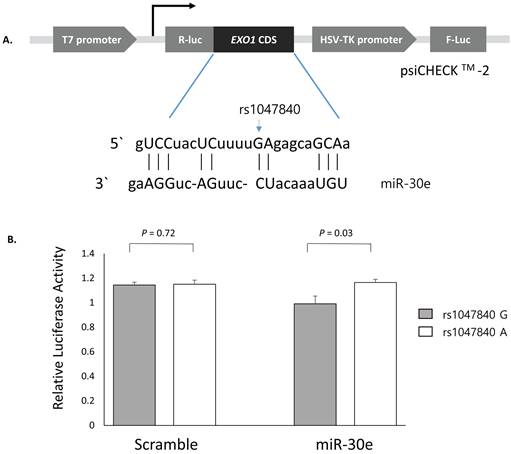
Luciferase reporter assay was conducted to determine whether EXO1 rs1047840G>A and CAMKK2 rs1653586G>T modulate the binding of miRNA and thus affects the expression level of target genes. Luciferase reporter plasmids was constructed using the psiCHECKTM-2 vector (Promega, Madison, WI, USA). EXO1 coding sequence containing rs1047840G or rs1047840A and CAMKK2 3'UTR sequence containing rs1653586G or rs1653586T were synthesized using PCR from human genomic DNA and cloned into the psiCHECKTM-2 vector. In the PolymiRTS database, we found that miR-30e and miR-185 were experimentally demonstrated to interact with the target sites on mRNAs of EXO1 and CAMKK2, respectively. Each construct was then co-transfected with miRNA (psiCHECKTM-2-EXO1 with miR-30e, and psiCHECKTM-2-CAMKK2 with miR-185) into H1299 cells following the manufacturer's instructions. After 24 hours of incubation, Renilla luciferase activities were measured using the firefly luciferase activities as a normalization control.
Statistical analysis
Deviations from Hardy-Weinberg equilibrium was measured by a goodness-of-fit χ2 test. Genotypes for SNPs were analyzed as three-group categorical variable, and analyzed under dominant, recessive, and codominant models. The association between clinical variables or genotypes and response to chemotherapy was tested by odds ratio (OR) and 95% confidence interval (CI) using unconditional logistic regression. Kaplan-Meier method was used to estimate survival analysis, and log-rank test was used to compare the difference in OS according to different clinical variables or genotypes. Hazard ratio (HR) and 95% CI were estimated using multivariate Cox proportional hazards model. Variables with P value < 0.05 were considered statistically significant. The statistical data were obtained using Statistical Analysis System for Windows, version 9.4 (SAS Institute, Cary, NC, USA).
Results
Associations between clinical parameters and the response to chemotherapy and survival outcomes are presented in Table 1. Overall response rate was 41.7% and median survival time was 24.1 months (95% CI = 21.7-25.9 months). Univariate analyses showed that age > 65, no benefit from tyrosine kinase inhibitor (TKI) treatment which include wild-type EGFR or ALK or no response to EGFR- or ALK-TKIs, pemetrexed maintenance therapy, and pemetrexed/cisplatin regimen were associated with better response. Regarding the OS, female sex, never-smoking status, postsurgical recurrence versus stage III/IV, ECOG performance status 0, benefit from TKI, pemetrexed maintenance therapy, and pemetrexed alone were significantly associated with the better OS. The conflicting results may be partly explained by first-line pemetrexed/cisplatin use in patients with wild-type EGFR or ALK and second-line pemetrexed monotherapy in patients with upfront EGFR- or ALK-TKI treatment, because the pemetrexed/cisplatin provides higher response rate than pemetrexed monotherapy but EGFR- or ALK- TKI is associated with better survival than chemotherapy alone.
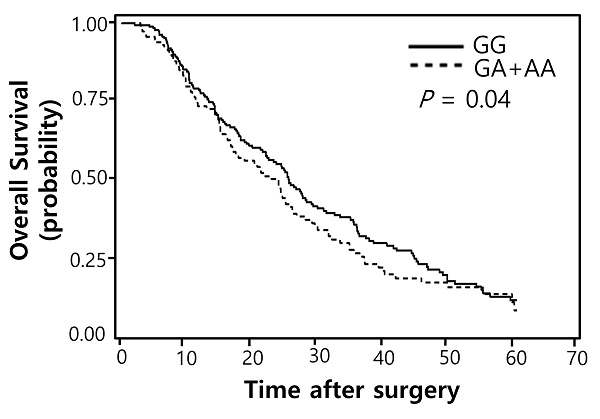
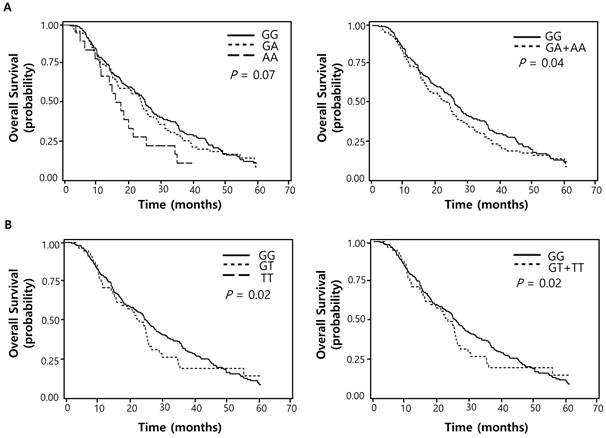
Of the 100 SNPs genotyped, 74 SNPs were further analyzed (Supplementary Table 1) after excluding 3 SNPs with genotyping failure and 23 SNPs with deviations from Hardy-Weinberg equilibrium (P < 0.05) or low call rates (< 95%). Among these, 7 SNPs were significantly associated with the response to pemetrexed (Table 2) while 16 SNPs with OS (Table 3). Two polymorphisms (EXO1 rs1047840G>A and CAMKK2 rs1653586G>T) were significantly associated with both chemotherapy response and survival outcome. The EXO1 rs1047840G>A and CAMKK2 rs1653586G>T were significantly associated with worse chemotherapy response (adjusted odds ratio [aOR] = 0.41, 95% CI = 0.24-0.68, P = 0.001, under a dominant model; and aOR = 0.33, 95% CI = 0.16-0.67, P = 0.002, under a dominant model, respectively) and worse OS (adjusted hazard ratio [aHR] = 1.34, 95% CI = 1.01-1.77, P = 0.04, under a dominant model; and aHR = 1.50, 95% CI = 1.06-2.13, P = 0.02, under a dominant model, respectively) in multivariate analyses (Table 4 and Figure 1).
Next, to investigate whether the EXO1 rs1047840G>A and CAMKK2 rs1653586G>T modulates the binding of miRNAs and consequently influence the expression level of EXO1 and CAMKK2, psiCHECKTM-2-EXO1 containing rs1047840G>A and psiCHECKTM-2-CAMKK2 containing rs1653586G>T were generated and co-transfected into H1299 cells with miR-30e and miR-185, respectively. The CLASH data showed that the binding between EXO1 and miR-30e was non-canonical (Figure 2A) and luciferase assay showed that the Renilla luciferase activity was significantly increased in EXO1 rs1047840A allele compared with rs1047840G allele (P = 0.03, Figure 2B). These results suggest that rs1047840G>A altered the binding of miR-30e and consequently increased the expression of EXO1. However, there was no difference in the luciferase activity between CAMKK2 rs1653586 G and T alleles (data not shown).
Overall survival curves according to (A) EXO1 rs1047840G>A (B) CAMKK2 rs1653586G>T genotypes. P values by multivariate Cox proportional hazard model.
Univariate analysis for response to chemotherapy and overall survival by clinical variables
| No. of | Response to chemotherapy | Overall survival | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| respondersa | nonrespondersa | OR (95% CI) | P | MST | 95% CI | Log-Rank | HR (95% CI) | P | ||
| Variables | cases | (CR+PR) | (SD+PD) | (months) | P | |||||
| Overall | 314 | 131 (41.7)a | 183 (58.3) | 24.1 | 21.7-25.9 | |||||
| Age (years) | ||||||||||
| ≤ 65 | 177 | 65 (36.7) | 112 (63.3) | 1.00 | 25.8 | 23.6-31.1 | 1.00 | |||
| >65 | 137 | 66 (48.2) | 71 (51.8) | 1.60 (1.02-2.52) | 0.04 | 21.1 | 17.2-25.1 | 0.08 | 1.26(0.97-1.63) | 0.08 |
| Sex | ||||||||||
| Male | 151 | 64 (42.4) | 87 (57.6) | 1.00 | 16.6 | 14.6-20.9 | 1.00 | |||
| Female | 163 | 67 (41.1) | 96 (58.9) | 0.95 (0.61-1.49) | 0.82 | 28.5 | 25.1-34.3 | <0.0001 | 0.60(0.46-0.78) | 0.0001 |
| Smoking status | ||||||||||
| Never | 165 | 70 (42.4) | 95 (57.6) | 1.00 | 28.6 | 24.7-54.4 | 1.00 | |||
| Ever | 149 | 61 (40.9) | 88 (59.1) | 0.94 (0.60-1.47) | 0.79 | 16.4 | 14.5-22.5 | 0.0002 | 1.64(1.27-2.12) | 0.0002 |
| Stage | ||||||||||
| III/IV | 244 | 102 (41.8) | 142 (58.2) | 1.00 | 22.9 | 17.9-24.6 | 1.00 | |||
| Postsurgical recurrence | 70 | 29 (41.4) | 41 (58.6) | 0.99 (0.57-1.69) | 0.96 | 34.9 | 25.9-44.6 | 0.001 | 0.59(0.43-0.82) | 0.001 |
| PS ECOG | ||||||||||
| 0 | 102 | 45 (44.1) | 57 (55.9) | 1.00 | 29.5 | 24.6-35.4 | 1.00 | |||
| 1-2 | 212 | 86 (40.6) | 126 (59.4) | 0.87 (0.54-1.39) | 0.55 | 21.7 | 17.4-24.6 | 0.01 | 1.46(1.1-1.94) | 0.01 |
| TKI benefit | ||||||||||
| No | 199 | 95 (47.7) | 104 (52.3) | 1.00 | 15.2 | 14.0-17.2 | 1.00 | |||
| Yes | 115 | 36 (31.3) | 79 (68.7) | 0.50 (0.31-0.81) | 0.01 | 39.4 | 34.4-44.3 | <0.0001 | 0.31(0.24-0.42) | <0.0001 |
| Maintenanceb | ||||||||||
| No | 113 | 52 (46.0) | 61 (54.0) | 1.00 | 14.3 | 10.8-17.1 | 1.00 | |||
| Yes | 67 | 45 (67.2) | 22 (32.8) | 2.40 (1.28-4.50) | 0.01 | 27.2 | 16.6- | <0.0001 | 0.41(0.27-0.62) | <0.0001 |
| Regimen | ||||||||||
| Pem/Cis | 180 | 97 (53.9) | 83 (46.1) | 1.00 | 16.6 | 14.6-20.9 | 1.00 | |||
| Pem alone | 134 | 34 (25.4) | 100 (74.6) | 0.29 (0.18-0.47) | <0.0001 | 28.8 | 25.4-35.4 | 0.0001 | 0.60(0.46-0.78) | 0.0001 |
Abbreviation: CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease; OR, odds ratio; MST, median survival time; CI, confidence interval; HR, hazard ratio; PS, performance status; ECOG, Eastern Cooperative Oncology Group; TKI, tyrosine kinase inhibitor; Pem, pemetrexed; Cis, cisplatin.
a Row percentage.
bAmong patients with Pem/Cis.
Summary of 7 SNPs and response to chemotherapy
| ID No.a | Target Gene | miRNA | Alleles | CR (%) | MAF | HWE-p | Pb for response | ||
|---|---|---|---|---|---|---|---|---|---|
| dominant | recessive | codominant | |||||||
| rs1047840 | EXO1 | hsa-miR-30e | GA | 97.77 | 0.22 | 0.17 | 0.001 | 0.38 | 0.002 |
| rs1653586 | CAMKK2 | hsa-miR-185 | GT | 99.68 | 0.09 | 0.08 | 0.002 | - | 0.002 |
| rs2295865 | SUPT16H | hsa-miR-186 | CA | 98.09 | 0.11 | 0.26 | 0.05 | 0.99 | 0.03 |
| rs11541557 | ARF1 | hsa-miR-92a | GT | 98.09 | 0.10 | 0.06 | 0.03 | - | 0.03 |
| rs6698826 | RAB3B | hsa-miR-27b | CA | 95.54 | 0.14 | 0.72 | 0.01 | 0.85 | 0.01 |
| rs3762158 | SUPT16H | has-miR-484 | GC | 93.95 | 0.12 | 0.27 | 0.05 | 0.99 | 0.04 |
| rs4705 | PDGFRL | hsa-miR-25 | CT | 98.73 | 0.47 | 0.41 | 0.87 | 0.04 | 0.27 |
Abbreviation: CR, call rate; MAF, minor allele frequency; and HWE, Hardy-Weinberg equilibrium.
aInformation about SNPs and SNP ID were obtained from NCBI database (https://www.ncbi.nlm.nih.gov/). The transcription start site was counted as +1 in reference sequences.
bP values were calculated by multivariate regression analysis, adjusted for age, sex, smoking status, stage, ECOG performance status, and chemotherapy regimen.
Summary of 16 SNPs and overall survival
| ID No.a | Target Gene | miRNA | Alleles | CR (%) | MAF | HWE-p | Pb for overall survival | ||
|---|---|---|---|---|---|---|---|---|---|
| dominant | recessive | codominant | |||||||
| rs1047840 | EXO1 | hsa-miR-30e | GA | 97.77 | 0.22 | 0.17 | 0.04 | 0.62 | 0.07 |
| rs1653586 | CAMKK2 | hsa-miR-185 | GT | 99.68 | 0.09 | 0.08 | 0.02 | 0.02 | |
| rs17445840 | CDH2 | hsa-miR-615-3p | CT | 98.73 | 0.04 | 0.52 | 0.04 | 0.19 | 0.03 |
| rs6934058 | CDC5L | hsa-miR-505 | TC | 97.77 | 0.41 | 0.46 | 0.04 | 0.26 | 0.05 |
| rs1056471 | HADHB | hsa-miR-99a | GC | 98.73 | 0.16 | 0.22 | 0.002 | 0.95 | 0.01 |
| rs296888 | HNRNPK | hsa-miR-615-3p | CT | 99.36 | 0.28 | 0.71 | 0.01 | 0.30 | 0.01 |
| rs3212986 | CD3EAP | hsa-miR-92a | GT | 98.09 | 0.27 | 0.24 | 0.02 | 0.45 | 0.03 |
| rs12449580 | AIPL1 | has-miR-3615 | CG | 98.41 | 0.39 | 0.29 | 0.04 | 0.05 | 0.01 |
| rs1318648 | ESPL1 | hsa-miR-149 | TG | 96.50 | 0.30 | 0.84 | 0.04 | 0.01 | 0.01 |
| rs2076345 | TCEB3 | hsa-miR-320b | CT | 93.63 | 0.25 | 0.17 | 0.04 | 0.76 | 0.10 |
| rs2228128 | POLR2A | has-miR-744 | TC | 93.63 | 0.06 | 0.28 | 0.05 | 0.97 | 0.02 |
| rs3217933 | CCND2 | hsa-miR-17 | TC | 99.68 | 0.09 | 0.68 | 0.59 | 0.004 | 0.39 |
| rs7654 | TPM3 | hsa-miR-615-3p | CA | 99.68 | 0.08 | 0.15 | 0.62 | 0.01 | 0.35 |
| rs2306409 | GTPBP4 | hsa-miR-16 | TC | 98.41 | 0.39 | 0.84 | 0.86 | 0.004 | 0.15 |
| rs7091596 | PARD3 | hsa-miR-93* | AT | 98.09 | 0.24 | 0.63 | 0.09 | 0.02 | 0.03 |
| rs2261988 | UHRF1 | has-miR-615-3p | CA | 98.41 | 0.15 | 0.16 | 0.09 | 0.06 | 0.04 |
Abbreviation: CR, call rate; MAF, minor allele frequency; and HWE, Hardy-Weinberg equilibrium.
aInformation about SNPs and SNP ID were obtained from NCBI database (https://www.ncbi.nlm.nih.gov/). The transcription start site was counted as +1 in reference sequences.
bP values were calculated using multivariate Cox proportional hazard models, adjusted for age, sex, smoking status, stage, ECOG performance status, TKI benefit, maintenance therapy, and chemotherapy regimen.
Associations between EXO1 rs1047840 and CAMKK2 rs1653586 and clinical outcomes
| Polymorphism/Genotype | Target Gene | miRNA | No. of cases (%)a | Response | Overall Survival | ||||
|---|---|---|---|---|---|---|---|---|---|
| Responders (%)b | Non-responders (%)b | OR (95% CI)c | Pc | HR (95% CI)d | Pd | ||||
| rs1047840f | EXO1 | hsa-miR-30e | |||||||
| GG | (cds-non) | 191 (62.2) | 94 (49.2) | 97 (50.8) | 1.00 | 1.00 | |||
| GA | 97 (31.6) | 28 (28.9) | 69 (71.1) | 0.40 (0.23-0.69) | 0.001 | 1.35 (1.01-1.81) | 0.05 | ||
| AA | 19 (6.2) | 6 (31.6) | 13 (68.4) | 0.47 (0.16-1.37) | 0.17 | 1.28 (0.75-2.21) | 0.37 | ||
| Dominant | 0.41 (0.24-0.68) | 0.001 | 1.34 (1.01-1.77) | 0.04 | |||||
| Recessive | 0.63 (0.22-1.79) | 0.38 | 1.14 (0.67-1.94) | 0.62 | |||||
| Ptrend e | 0.52 (0.34-0.79) | 0.002 | 1.22 (0.98-1.51) | 0.07 | |||||
| rs1653586f | CAMKK2 | hsa-miR-185 | |||||||
| GG | (UTR-3) | 257 (82.1) | 118 (45.9) | 139 (54.1) | 1.00 | 1.00 | |||
| GT | 56 (17.9) | 12 (21.4) | 44 (78.6) | 0.33 (0.16-0.67) | 0.002 | 1.50 (1.06-2.13) | 0.02 | ||
| TT | 0 (0.0) | 0 (0.0) | 0 (0.0) | - | - | - | - | ||
| Dominant | 0.33 (0.16-0.67) | 0.002 | 1.50 (1.06-2.13) | 0.02 | |||||
| Recessive | - | - | - | - | |||||
| Ptrend e | 0.33 (0.16-0.67) | 0.002 | 1.50 (1.06-2.13) | 0.02 | |||||
Abbreviations: OR, odds ratio; CI, confidence interval; HR, hazard ratio.
aColumn percentage.
bRow percentage.
cOR, 95% CI, and their corresponding P values were calculated by multivariate regression analysis, adjusted for age, sex, smoking status, stage, ECOG performance status, and chemotherapy regimen.
dHRs, 95% CIs and their corresponding P values were calculated using multivariate Cox proportional hazard model, adjusted for age, sex, smoking status, stage, ECOG performance status, TKI benefit, maintenance therapy, and chemotherapy regimen.
ePtrend for the additive model.
fGenotype failure: 7 cases for rs1047840, 1 for rs1653586.
Functional analysis of EXO1 rs1047840G>A by dual luciferase reporter assay. (A) Schematic representation of reporter plasmids containing EXO1 rs1047840G>A, and the complementarity between miR-30e and EXO1 coding sequence targeted. (B) Renilla luciferase assay for the effect of miRNA binding on rs1047840G>A using H1299 cells. Renilla luciferase activity was normalized to firefly luciferase activity and data are presented relative to the Mock control. Each bar represents mean ± SE. P values by Student's t-test. R-luc, Renilla luciferase; F-luc, firefly luciferase; HSV-TK, Herpes simplex virus thymidine kinase.
Discussion
In this study, we investigated whether the treatment outcomes of pemetrexed chemotherapy in lung adenocarcinoma patients are different according to the genotypes of genetic polymorphisms in miRNA target sites. The study revealed that two SNPs in miRNA binding sites, EXO1 rs1047840G>A and CAMKK2 rs1653586G>T, could predict the response to pemetrexed chemotherapy and survival outcome. The luciferase assay suggested that rs1047840G>A alters the binding of miRNA and consequently increases the expression level of EXO1. These findings suggest that the two SNPs, especially EXO1 rs1047840G>A, could be used as potential biomarkers to predict therapeutic outcomes in lung adenocarcinoma patients treated with pemetrexed chemotherapy, which may help to establish an optimal personalized treatment strategy.
Human exonuclease 1 (EXO1) is a member of the Rad2/XPG family of nucleases which exhibits 5'→3' exonuclease, 5' flap endonuclease, and 5'→3' RNase H activity [24, 25]. This enzyme mainly contributes to the regulation of cell cycle checkpoints and post-replicative DNA repair pathways, such as mismatch repair (MMR), translesion DNA synthesis (TLS), nucleotide excision repair (NER), and double-strand break repair (DSBR) [26-30]. Dysfunction of EXO1 could impair DNA repair processes which can cause replication stress followed by genomic instability and development of cancer [31]. EXO1 genetic variants have been associated with the risk and prognosis of different types of cancers, including lung cancer [32-37]. In addition, EXO1 overexpression and its relation to the resistance to chemotherapy or radiotherapy and poor prognosis was reported in multiple types of cancers [38-41]. Pemetrexed interferes with folate metabolism leading to ineffective DNA synthesis and tumor cell growth failure [5]. Inhibition of TS by pemetrexed induces uracil misincorporation into DNA resulting in cell death, and base excision repair (BER) system is responsible for the removal of misincorporated uracil. Therefore, the upregulation of BER is associated with pemetrexed resistance and is found in small cell and squamous cell carcinoma which are known as the pemetrexed-resistant histological subtypes of lung cancer [42]. Likewise, given its role in DNA damage response and cell cycle checkpoint activation, the upregulation of EXO1 may lead to the resistance to pemetrexed chemotherapy. In this study, EXO1 rs1047840G>A was associated with worse clinical outcomes in lung adenocarcinoma patients after pemetrexed chemotherapy and linked to increased expression of EXO1. Based on our results, it is postulated that the rs1047840-induced change in miRNA binding efficiency may increase the expression level of EXO1, resulting in worse response to pemetrexed chemotherapy and survival outcomes. Although many previous studies have investigated EXO1 polymorphism, especially rs1047840, in lung cancer, most of those studies evaluated the relationship between EXO1 rs1047840 and the risk of lung cancer (33, 35, 44). Regarding the chemotherapy outcomes, one study by R.Li et al. showed the association between EXO1 rs9350 and survival of patients receiving platinum-based chemotherapy in NSCLC (37). To the best of our knowledge, this is the first study to suggest a predictive role of EXO1 rs1047840 in the therapeutic response to pemetrexed chemotherapy and also survival in lung adenocarcinoma patients. In addition, by the SNP selection using a database for miRNA binding site variants, we could suggest that the rs1047840-induced change in miRNA binding efficiency may be a mechanism of the association between EXO1 rs1047840 and lung cancer. Alternatively, the EXO1 rs1047840G>A is a non-synonymous SNP (E589K) with an amino acid change, which may affect EXO1 protein functions. Further studies are required to understand the role of EXO1 in the mechanism of pemetrexed resistance. On the other hand, potential oncogenic roles of EXO1 have also been suggested, which may be another possible mechanism of the association between EXO1 rs1047840G>A and worse clinical outcomes after pemetrexed chemotherapy. Expression of degradation-resistant EXO1 could result in DNA hyper-resection in homologous recombination, which severely compromised DSBR and lead to chromosomal instability [43]. A bioinformatic analysis study showed that EXO1 was identified as one of the hub genes which were markedly associated with poor prognosis in patients with lung cancer [44]. Recently, Zhou et al. showed that EXO1 was overexpressed in lung adenocarcinoma tissues and the high expression of EXO1 was associated with poor prognosis [45]. By bioinformatic analyses using public database, they revealed the correlation between increased EXO1 expression and decreased tumor infiltrating B cells and CD4+ T cells, suggesting that EXO1 may induce an immune-suppressive tumor microenvironment in lung adenocarcinoma. Given that immune checkpoint blockade combined with pemetrexed-platinum doublet chemotherapy has been approved as the standard first-line treatment for lung adenocarcinoma, it will be worth to further evaluate EXO1 rs1047840G>A as a biomarker to predict clinical outcomes of combined immunotherapy and pemetrexed chemotherapy.
In this study, CAMKK2 rs1653586G>T was significantly related with the worse response to pemetrexed and survival outcome in lung adenocarcinoma patients. Calmodulin (CaM) is an intracellular calcium receptor which controls the downstream calcium/calmodulin kinase (CaMK) cascade that has been associated with various metabolic diseases [46]. Calcium/calmodulin-dependent kinase kinase 2 (CAMKK2) belongs to the subfamily of calcium/calmodulin-dependent protein kinase and is involved in the maintenance of energy balance, adiposity, glucose homeostasis, inflammation, and cancer [47]. CAMKK2 is generally upregulated in several cancers, including prostate cancer, and high expression of CAMKK2 was correlated with poor overall survival in hepatocellular carcinoma and glioma [48-50]. It was reported that knockdown of CAMKK2 potentiated the effect of carboplatin in ovarian cancer by modulating Akt pathway [51]. However, the role of CAMKK2 in the pathogenesis and prognosis of lung cancer has yet to be clarified. Further study is required to understand the molecular mechanism of the association between the genetic variants and clinical outcomes.
There are some limitations in this study. First, because pemetrexed maintenance regimen was not approved in the early part of the enrollment period, maintenance therapy was not prescribed for some patients who are considered eligible for the maintenance therapy by the current treatment guidelines. Although maintenance therapy was adjusted for in the multivariate analysis, the prognostic differences between patients with and without pemetrexed maintenance therapy could not be ignored. Second, as a single center retrospective study, the treatment other than pemetrexed in the course of treatment including targeted agents could not be strictly controlled in this study, which should be considered in the interpretation of the survival analysis. Third, the study cohort is relatively small. Although stratified analyses may show the clinical significance of EXO1 rs1047840 and CAMKK2 rs1653586 in different subgroups of patients, i.e. stage III/IV and postsurgical recurrence groups, this study is underpowered for stratification analyses or propensity score matching. Therefore, a well-designed and properly powered study is warranted to validate our findings.
In conclusion, two SNPs in miRNA binding sites, especially EXO1 rs1047840G>A, were associated with the treatment response to pemetrexed chemotherapy and survival in lung adenocarcinoma patients. Evaluation of genetic variants in miRNA binding sites may be useful for refining therapeutic decisions in the treatment of lung adenocarcinoma by helping to identify subgroups of patients who will benefit from pemetrexed chemotherapy. To verify EXO1 rs1047840G>A and CAMKK2 rs1653586G>T as biomarkers for predicting clinical outcomes, the results of this study need to be further tested in a larger population with diverse ethnicity.
Supplementary Material
Supplementary table.
Acknowledgements
Funding Statement
This research was supported by Kyungpook National University Research Fund, 2021.
Ethical Approval Statement
The biospecimens for this study were provided by National Biobank of Korea-Kyungpook National University Hospital (KNUH), which is supported by the Ministry of Health, Welfare and affairs. All materials derived from the National Biobank of Korea-KNUH were obtained (with informed consent) under institutional review board (IRB)-approved protocols.
Data Availability Statement
The datasets and materials in this study are available on reasonable request from corresponding authors or first author.
Author Contributions
Conceived and designed the study: S.Y.L. and J.Y.P. Acquired clinical data: J.E.P., S.Y.L., Y.S., S.H.C., Y.H.L., H.S., J.Y.J., K.M.S., S.S.Y., J.L., S.I.C., C.H.K., and J.Y.P. Performed experiments: M.J.H., J.H.L., J.E.C., H.-G.K., and S.K.D. Analyzed and interpreted data: M.J.H., J.E.P., J.H.L., W.K.L., S.Y.L., and J.Y.P. Wrote main manuscript: M.J.H., J.E.P., and S.Y.L. Supervised the study: S.Y.L. and J.Y.P. All authors reviewed the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Thai AA, Solomon BJ, Sequist LV. et al. Lung cancer. Lancet. 2021;398:535-554
2. Yuan M, Huang LL, Chen JH. et al. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct Target Ther. 2019;4:61
3. Mamdani H, Matosevic S, Khalid AB. et al. Immunotherapy in Lung Cancer: Current Landscape and Future Directions. Front Immunol. 2022;13:823618
4. Gandhi L, Rodriguez-Abreu D, Gadgeel S. et al. Pembrolizumab plus Chemotherapy in Metastatic Non-Small-Cell Lung Cancer. N Engl J Med. 2018;378:2078-2092
5. Adjei AA. Pharmacology and mechanism of action of pemetrexed. Clin Lung Cancer. 2004;5(Suppl 2):S51-S55
6. Scagliotti G, Hanna N, Fossella F. et al. The differential efficacy of pemetrexed according to NSCLC histology: a review of two Phase III studies. Oncologist. 2009;14:253-263
7. Takezawa K, Okamoto I, Okamoto W. et al. Thymidylate synthase as a determinant of pemetrexed sensitivity in non-small cell lung cancer. Br J Cancer. 2011;104:1594-1601
8. Krawczyk P, Kucharczyk T, Kowalski DM. et al. Polymorphisms in TS, MTHFR and ERCC1 genes as predictive markers in first-line platinum and pemetrexed therapy in NSCLC patients. J Cancer Res Clin Oncol. 2014;140:2047-2057
9. Fennell DA, Myrand SP, Nguyen TS. et al. Association between gene expression profiles and clinical outcome of pemetrexed-based treatment in patients with advanced non-squamous non-small cell lung cancer: exploratory results from a phase II study. PLoS One. 2014;9:e107455
10. Bartel DP. Metazoan MicroRNAs. Cell. 2018;173:20-51
11. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297
12. Ng R, Song G, Roll GR. et al. A microRNA-21 surge facilitates rapid cyclin D1 translation and cell cycle progression in mouse liver regeneration. J Clin Invest. 2012;122:1097-1108
13. Rayner KJ, Esau CC, Hussain FN. et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478:404-407
14. Zhang P, Bill K, Liu J. et al. MiR-155 is a liposarcoma oncogene that targets casein kinase-1alpha and enhances beta-catenin signaling. Cancer Res. 2012;72:1751-1762
15. Taganov KD, Boldin MP, Chang KJ. et al. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481-12486
16. Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390-7394
17. Iqbal MA, Arora S, Prakasam G. et al. MicroRNA in lung cancer: role, mechanisms, pathways and therapeutic relevance. Mol Aspects Med. 2019;70:3-20
18. Preskill C, Weidhaas JB. SNPs in microRNA binding sites as prognostic and predictive cancer biomarkers. Critical Reviews™ in Oncogenesis. 2013;18:327-40
19. Helwak A, Kudla G, Dudnakova T. et al. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654-665
20. Ciuleanu T, Brodowicz T, Zielinski C. et al. Maintenance pemetrexed plus best supportive care versus placebo plus best supportive care for non-small-cell lung cancer: a randomised, double-blind, phase 3 study. The Lancet. 2009;374:1432-1440
21. Eisenhauer EA, Therasse P, Bogaerts J. et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228-247
22. Bhattacharya A, Ziebarth JD, Cui Y. PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res. 2014;42(Database issue):D86-91
23. Higgins ME, Claremont M, Major JE. et al. CancerGenes: a gene selection resource for cancer genome projects. Nucleic Acids Res. 2007;35(Database issue):D721-726
24. Wilson DM, Coleman MA, Adamson AW. et al. Hex1: a new human Rad2 nuclease family member with homology to yeast exonuclease 1. Nucleic acids research. 1998;26:3762-3768
25. Qiu J, Qian Y, Chen V. et al. Human exonuclease 1 functionally complements its yeast homologues in DNA recombination, RNA primer removal, and mutation avoidance. J Biol Chem. 1999;274:17893-17900
26. Szankasi P, Smith GR. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science. 1995;267:1166-1169
27. Sertic S, Mollica A, Campus I. et al. Coordinated Activity of Y Family TLS Polymerases and EXO1 Protects Non-S Phase Cells from UV-Induced Cytotoxic Lesions. Mol Cell. 2018;70:34-47 e34
28. Qiu J, Guan M-X, Bailis AM. et al. Saccharomyces cerevisiae exonuclease-1 plays a role in UV resistance that is distinct from nucleotide excision repair. Nucleic acids research. 1998;26:3077-3083
29. Bolderson E, Tomimatsu N, Richard DJ. et al. Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks. Nucleic Acids Res. 2010;38:1821-1831
30. Keijzers G, Liu D, Rasmussen LJ. Exonuclease 1 and its versatile roles in DNA repair. Crit Rev Biochem Mol Biol. 2016;51:440-451
31. Keijzers G, Bakula D, Petr MA. et al. Human Exonuclease 1 (EXO1) Regulatory Functions in DNA Replication with Putative Roles in Cancer. Int J Mol Sci. 2018;20:74
32. Wang H-C, Chiu C-F, Tsai R-Y. et al. Association of genetic polymorphisms of EXO1 gene with risk of breast cancer in Taiwan. Anticancer research. 2009;29:3897-3901
33. Hsu N-Y, Wang H-C, Wang C-H. et al. Lung cancer susceptibility and genetic polymorphisms of Exo1 gene in Taiwan. Anticancer research. 2009;29:725-730
34. Bayram S, Akkiz H, Bekar A. et al. The significance of Exonuclease 1 K589E polymorphism on hepatocellular carcinoma susceptibility in the Turkish population: a case-control study. Mol Biol Rep. 2012;39:5943-5951
35. Jin G, Wang H, Hu Z. et al. Potentially functional polymorphisms of EXO1 and risk of lung cancer in a Chinese population: A case-control analysis. Lung Cancer. 2008;60:340-346
36. Nogueira GA, Lourenco GJ, Oliveira CB. et al. Association between genetic polymorphisms in DNA mismatch repair-related genes with risk and prognosis of head and neck squamous cell carcinoma. Int J Cancer. 2015;137:810-818
37. Li R, Gu J, Heymach JV. et al. Hypoxia pathway genetic variants predict survival of non-small-cell lung cancer patients receiving platinum-based chemotherapy. Carcinogenesis. 2017;38:419-424
38. Dai Y, Tang Z, Yang Z. et al. EXO1 overexpression is associated with poor prognosis of hepatocellular carcinoma patients. Cell Cycle. 2018;17:2386-2397
39. de Sousa JF, Torrieri R, Serafim RB. et al. Expression signatures of DNA repair genes correlate with survival prognosis of astrocytoma patients. Tumour Biol. 2017;39:1010428317694552
40. He D, Li T, Sheng M. et al. Exonuclease 1 (Exo1) Participates in Mammalian Non-Homologous End Joining and Contributes to Drug Resistance in Ovarian Cancer. Med Sci Monit. 2020;26:e918751
41. Zhou J, Wang Y, Wang Y. et al. FOXM1 modulates cisplatin sensitivity by regulating EXO1 in ovarian cancer. PLoS One. 2014;9:e96989
42. Weeks LD, Fu P, Gerson SL. Uracil-DNA glycosylase expression determines human lung cancer cell sensitivity to pemetrexed. Mol Cancer Ther. 2013;12:2248-2260
43. Tomimatsu N, Mukherjee B, Harris JL. et al. DNA-damage-induced degradation of EXO1 exonuclease limits DNA end resection to ensure accurate DNA repair. J Biol Chem. 2017;292:10779-10790
44. Li Z, Sang M, Tian Z. et al. Identification of key biomarkers and potential molecular mechanisms in lung cancer by bioinformatics analysis. Oncol Lett. 2019;18:4429-4440
45. Zhou CS, Feng MT, Chen X. et al. Exonuclease 1 (EXO1) is a Potential Prognostic Biomarker and Correlates with Immune Infiltrates in Lung Adenocarcinoma. Onco Targets Ther. 2021;14:1033-1048
46. Marcelo KL, Means AR, York B. The Ca(2+)/Calmodulin/CaMKK2 Axis: Nature's Metabolic CaMshaft. Trends Endocrinol Metab. 2016;27:706-718
47. Racioppi L, Means AR. Calcium/calmodulin-dependent protein kinase kinase 2: roles in signaling and pathophysiology. J Biol Chem. 2012;287:31658-31665
48. Massie CE, Lynch A, Ramos-Montoya A. et al. The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. The EMBO journal. 2011;30:2719-2733
49. Lin F, Marcelo KL, Rajapakshe K. et al. The camKK2/camKIV relay is an essential regulator of hepatic cancer. Hepatology. 2015;62:505-520
50. Liu DM, Wang HJ, Han B. et al. CAMKK2, Regulated by Promoter Methylation, is a Prognostic Marker in Diffuse Gliomas. CNS Neurosci Ther. 2016;22:518-524
51. Gocher AM, Azabdaftari G, Euscher LM. et al. Akt activation by Ca(2+)/calmodulin-dependent protein kinase kinase 2 (CaMKK2) in ovarian cancer cells. J Biol Chem. 2017;292:14188-14204
Author contact
![]() Corresponding authors: Shin Yup Lee, MD, PhD, Lung Cancer Center, Kyungpook National University Chilgok Hospital, 807, Hoguk-ro, Buk-gu, Daegu 41404, Korea; Tel: +82-53-200-2632; Fax: +82-53-200-2027, E-mail: shinyupac.kr; Jae Yong Park, MD, PhD, Lung Cancer Center, Kyungpook National University Chilgok Hospital, 807, Hoguk-ro, Buk-gu, Daegu 41404, Korea; Tel: +82-53-200-2631; Fax: +82-53-200-2027, E-mail: jaeyongac.kr.
Corresponding authors: Shin Yup Lee, MD, PhD, Lung Cancer Center, Kyungpook National University Chilgok Hospital, 807, Hoguk-ro, Buk-gu, Daegu 41404, Korea; Tel: +82-53-200-2632; Fax: +82-53-200-2027, E-mail: shinyupac.kr; Jae Yong Park, MD, PhD, Lung Cancer Center, Kyungpook National University Chilgok Hospital, 807, Hoguk-ro, Buk-gu, Daegu 41404, Korea; Tel: +82-53-200-2631; Fax: +82-53-200-2027, E-mail: jaeyongac.kr.

 Global reach, higher impact
Global reach, higher impact