Impact Factor
ISSN: 1837-9664
J Cancer 2023; 14(1):24-34. doi:10.7150/jca.78634 This issue Cite
Research Paper
Clinicopathological Significances and Prognostic Value of PPFIA4 in Colorectal Cancer
1. Department of Pathology, Tianjin Union Medical Center, Nankai University, Tianjin, 300121, P.R. China.
2. Graduate School, Tianjin University of Traditional Chinese Medicine, Tianjin, 301617, P.R. China.
3. Nankai University School of Medicine, Nankai University, Tianjin, 300071, P.R. China.
4. Department of Colorectal Surgery, Tianjin Union Medical Center, Tianjin, P.R. China.
# These authors equally contributed to the paper.
Received 2022-9-4; Accepted 2022-11-6; Published 2023-1-1
Abstract
Purpose: The PPFIA gene family (PPFIA1, PPFIA2, PPFIA3, and PPFIA4) is associated with multiple human diseases, particularly malignant tumors. However, the expression and prognostic value of the PPFIA family in human colorectal cancers (CRCs) have not been reported.
Materials and methods: In this study, several databases, including Oncomine, UALCAN, and the cancer cell line encyclopedia, were used to compare differences in PPFIA1, PPFIA2, PPFIA3, and PPFIA4 expression between normal colon samples and CRCs. The expression levels of these four proteins were used to evaluate the survival of patients with CRC, as determined by the Cancer Genome Atlas Program (TCGA) portal and gene expression profiling interactive analysis (GEPIA) databases. Western blotting and reverse transcription-polymerase chain reaction were performed to detect protein and mRNA levels of PPFIA1, PPFIA3, and PPFIA4, respectively. Immunohistochemical (IHC) staining was used to detect the correlation between PPFIA4 expression and the degree of CRC malignancy. Furthermore, potential miRNAs targeting PPFIA4 in CRCs were studied and confirmed.
Results: Bioinformatic analysis showed that the mRNA levels of PPFIA1, PPFIA3, and PPFIA4 were higher in CRC tissue samples than in normal colon tissue. Both mRNA and protein expression of PPFIA1, PPFIA3, and PPFIA4 were increased in the CRC cell lines LoVo and Hct116 compared with the normal colon epithelial cell line. Only PPFIA4 was associated with the prognosis of patients with CRC, which was confirmed by TCGA portal and GEPIA. IHC staining confirmed that the expression of PPFIA4 was higher in CRC tissues than in normal colon tissues and also increased in poorly differentiated CRC tissues and lymph node metastatic foci in comparison with well-differentiated CRC tissues and moderately differentiated CRC tissues. Functional annotation enrichment analysis indicated that the top 100 genes co-expressed with PPFIA4 were enriched in the G-protein coupled peptide receptor activity, leukotrience B4 receptor activity, and peroxisome proliferator-activated receptors and hypoxia-inducible factor-1 signaling pathways. In addition, miR-485-5p negatively regulates the expression of PPFIA4.
Conclusion: PPFIA4 expression is associated with the development of CRCs and may be a novel potential prognostic marker for human CRCs.
Keywords: Colorectal cancer, PPFIA4, MiR-485-5p, Bioinformatics Analysis, Prognosis
Introduction
Colorectal cancer (CRC) is the second most common cause of death and the third most common cause of malignant tumors [1, 2]. The analysis of differentially expressed genes between CRC and normal colon tissues may provide new targets for cancer research. Substantial biological information was identified in the post-genome era by biological information scientists, who also established a good deal of software and databases for systematic management and sufficient sequence data to study the molecular mechanisms of tumor initiation and progression. Additionally, the development of bioinformatics contributes to the search for appropriate biomarkers for tumor prognosis.
The PPFIA genes, which include PPFIA1, PPFIA2, PPFIA3 and PPFIA4, were discovered by encoding liprin-α family member proteins, which were discovered by interacting with the leukocyte common antigen-related gene (LAR) family [3, 4]. These four genes play important roles in the initiation and progression of tumors. Liprin-α1, encoded by PPFIA1, has the highest degree of alternative splicing in this family and is overexpressed in several epithelial cancers, including head and neck, breast, and ovarian cancers [5-8]. PPFIA2 expression is increased in the urinary sediments of patients with prostate cancer [9]. PPFIA3 is a significant marker of gastric cancer and has independent prognostic value in patients with pancreatic cancer [10-12]. Liprin-α4, encoded by PPFIA4, is located on chromosome 1q32. Overexpression of this protein promotes pancreatic cancer cells' proliferative and invasive abilities, whereas inhibition of this protein's expression improves the therapeutic effect in small cell lung cancer [13-15]. Liprin-α4 may be a novel therapeutic target for malignant tumors.
In this study, the expression of different PPFIA genes in normal colon samples and CRC tissues were analyzed using several databases, including Oncomine, UALCAN, and the cancer cell line encyclopedia (CCLE). Our results showed that PPFIA1, PPFIA3, and PPFIA4 expression were higher in colon cancer cell lines (LoVo and Hct116) than in the normal colon epithelial cell line. Among the four genes, PPFIA4 was associated with the prognosis of patients with CRC based on TCGAportal and gene expression profiling interactive analysis (GEPIA). Functional annotation enrichment analysis indicated that PPFIA4 is involved in the peroxisome proliferator-activated receptors (PPAR) and hypoxia-inducible factor-1 (HIF-1) signaling pathways, and miR-485-5p negatively regulates the expression of PPFIA4.
Materials and methods
1.PPFIAs expression levels analysis
UALCAN (http://ualcan.path.uab.edu) and Oncomine (https://www.oncomine.org/) databases were used to investigate the mRNA expression of PPFIA in normal colon and CRC samples. The criteria for the inclusion of patients downloaded from the database included: (1) patients did not receive chemotherapy and/or radiotherapy before radical operation; (2) patients did not accompany any other tumors; (3) patients' age was at least 18 years. The criteria for exclusion included: (1) The clinicopathologic data of patients was incomplete; (2) patients' age was less than 18 years. CCLE datasets were used to verify the mRNA expression levels of PPFIA genes in different cancer cell lines (https://sites.broadinstitute.org/ccle/). The Human Protein Atlas database was used to detect protein expression of PPFIA4 in normal colon and CRC tissues.
2. Cancer cell lines and culture
The normal human colon cell line NCM460 and colon cancer cell lines LoVo and Hct116 were obtained from the American Type Culture Collection (Manassas, VA, USA) and cultured at 37 °C in a 5% carbon dioxide (CO2) incubator. The cultural conditions were as previously described [16].
3. Real-time polymerase chain reaction (RT-PCR) analysis
Detailed information on the real-time polymerase chain reaction (RT-PCR) analysis is provided in the Supplementary Materials and Methods. The sequences of the RT-PCR primers are listed in Supplementary Table 1.
4. Western blot assay
PPFIA1, PPFIA3, and PPFIA4 protein expression in NCM460, LoVo, and Hct116 cells and PPFIA4 protein expression in LoVo and Hct116 cells transfected with and without miRNA mimics and inhibitors were analyzed by western blotting. Cells were collected and lysed in cold lysis buffer (RIPA, Solarbio, China) for 30 minutes. Electrophoresis was performed to separate the samples on 8% sodium dodecyl sulfate-polyacrylamide gels. The separated proteins were transferred to polyvinylidene fluoride or polyvinylidene difluoride (PVDF) membranes (Millipore, USA). At room temperature, 5% defatted milk (BD Biosciences) was used to block the membranes containing proteins for two hours. The PVDF membranes were then incubated with the primary antibodies (Supplementary Table 2) at 4 °C for 16 hours. The next day, secondary antibodies reacted with the primary antibodies for 1.5 hours at room temperature. A ChemiDoc imaging system (Bio-Rad) was used to assess protein expression.
5. Relapse and survival analyses
GEPIA (http://gepia.cancer-pku.cn/detail.php) and the TCGA portal (http://www.tcgaportal.org) were used to analyze the correlation between PPFIA mRNA expression and survival time of patients with CRC.
6. CRC tissue samples
Paraffin-embedded human CRC samples (n=219) and non-tumor colorectal tissue samples (n=5) were obtained from the Department of Pathology of Tianjin Union Medical Center. The diagnosis of CRC was confirmed by pathological examination. Based on the differentiation of CRCs and metastasis, these samples were divided into five groups: group I, five cases of colorectal epithelial tissue; group II, 51 cases of well-differentiated CRCs; group III, 54 cases of moderately differentiated CRCs; group IV, 51 cases of poorly differentiated CRCs; and group V, 63 cases of lymph node metastatic foci. The clinical parameters of the 224 cases including 219 cases CRC samples and 5 cases of non-tumor colorectal tissue samples were provided in the Supplementary Table 3. The experiments were approved by the hospital review board of the Tianjin Union Medical Center and the confidentiality of patient information was maintained.
7. Immunohistochemical (IHC) staining, scoring and quantification
IHC staining, scoring, and quantification were performed as previously described [16]. Detailed information is provided in the Supplementary Materials and Methods.
8. Functional enrichment analysis of correlated genes
The top 100 genes that were positively correlated with PPFIA4 in the CRC were selected from the UALCAN database. R software was employed to perform the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses.
9. Protein-protein interaction (PPI) networks and identification of candidate miRNAs (micro-RNA) and miRNA-mRNA regulation networks
A PPI network of 100 correlated genes was visualized using the STRING database. The Encyclopedia of RNA Interactomes (ENCORI; http://starbase.sysu.edu.cn/) was used to analyze the miRNA-ncRNA, miRNA-mRNA, ncRNA-RNA, RNA-RNA, RBP-ncRNA, and RBP-mRNA interactions from CLIP-seq, degradome-seq, and RNA-RNA interactome data. In our study, ENCORI was used to predict miRNAs that regulate PPFIA4. Cytoscape (version 3.7.2) was used to analyze the miRNA-PPFIA4 network [17].
10. Transient transfection
Inhibitors and mimics targeting miR-485-5P were synthesized by Gene-Pharma (Shanghai, China). Detailed information on the sequences is provided in Supplementary Table 4.
Results
1. PPFIA mRNA and protein expression levels were associated with the development of CRC
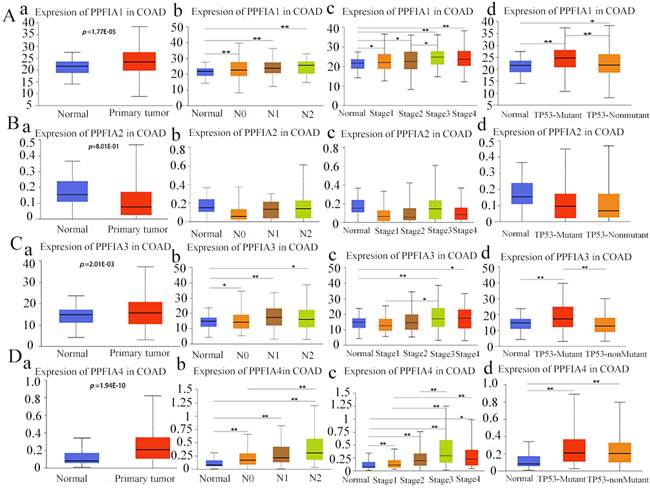
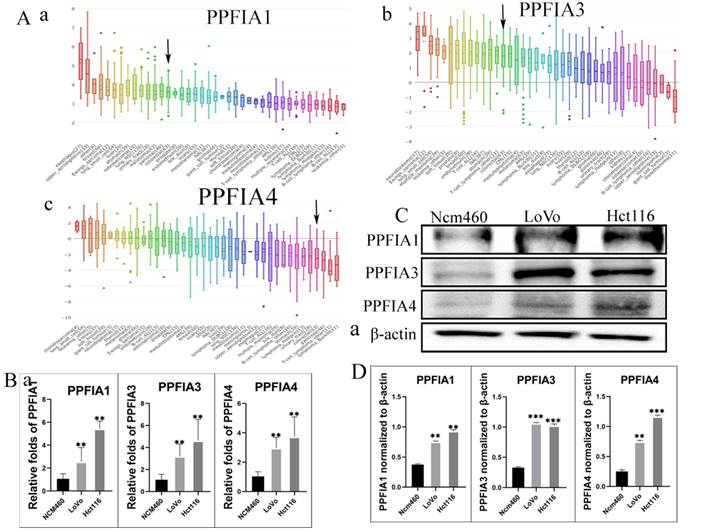
The Oncomine and UALCAN databases were used to compare the expression of the mRNA level expression of PPFIA in human CRC samples and normal colon epithelium samples. The results from the two databases demonstrated that the mRNA expression levels of PPFIA1, PPFIA3, and PPFIA4 were significantly higher in CRC samples than in normal colon epithelium. PPFIA2 mRNA expression was not significantly different between normal colon epithelial samples and CRC tissues (Figures 1A-a to d, 1B, 2A-a, 2B-a, 2C-a, 2D-a). In addition, the mRNA expression levels of PPFIA1, PPFIA3, and PPFIA4 were associated with tumor stage, lymph node metastasis, and TP53-mutation status (Figures 2A-b to d, 2B-b to d, 2C-b to d, and 2D-b to d). Results of CCLE database analysis also confirmed that the mRNA expression levels of PPFIA1, PPFIA3, and PPFIA4 in CRC cell lines were higher than those in normal colon cell lines (Figure 3A - a to c). Subsequently, RT-PCR and western blotting were performed to detect the mRNA and protein levels of PPFIA1, PPFIA3, and PPFIA4 in normal colon (NCM460) and CRC (LoVo and Hct116) cell lines. The results confirmed that both mRNA (Figure 3B - a to c) and protein (Figures 3C and 3D- a to c) expression of PPFIA1, PPFIA3, and PPFIA4 were increased in CRC cell lines compared to the normal colon cell line, and the differences were statistically significant (**P < 0.01, ***P < 0.001).
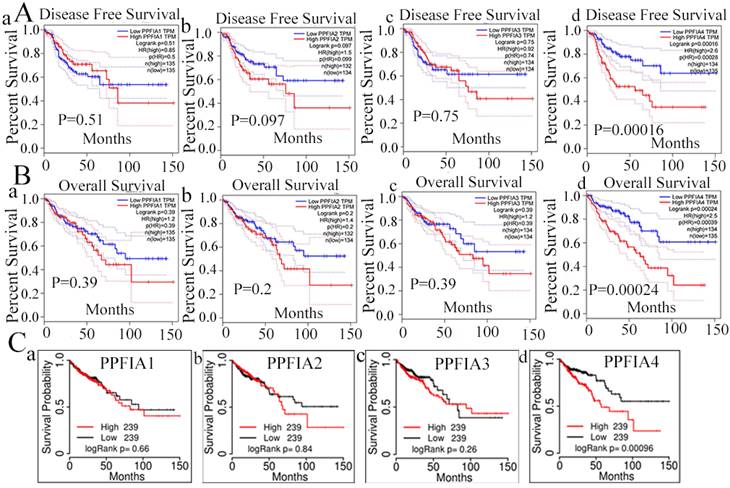
2. Prognostic value of PPFIA mRNA expression levels in CRCs
To evaluate the prognostic value of PPFIA1, PPFIA2, PPFIA3, and PPFIA4 in CRC patients, the GEPIA server and TCGAportal database were used to analyze the survival of CRC patients. The results indicated that the mRNA expression levels of PPFIA1, PPFIA2, and PPFIA3 were not significantly correlated with overall survival (OS) or disease-free survival (DFS) in CRC patients (Figures 4A-a to c, 4B-a to c). However, patients with high PPFIA4 mRNA expression levels of CRCs had shorter DFS and OS than those with low PPFIA4 mRNA expression levels, and the difference was statistically significant (Figure 4A-d, P = 0.00028; Figure 4C, P = 0.00039). These results are consistent with the TCGA portal database (Figure 4C). The results from the two databases demonstrated that high PPFIA4 mRNA expression levels were associated with poor prognosis in patients with CRC.
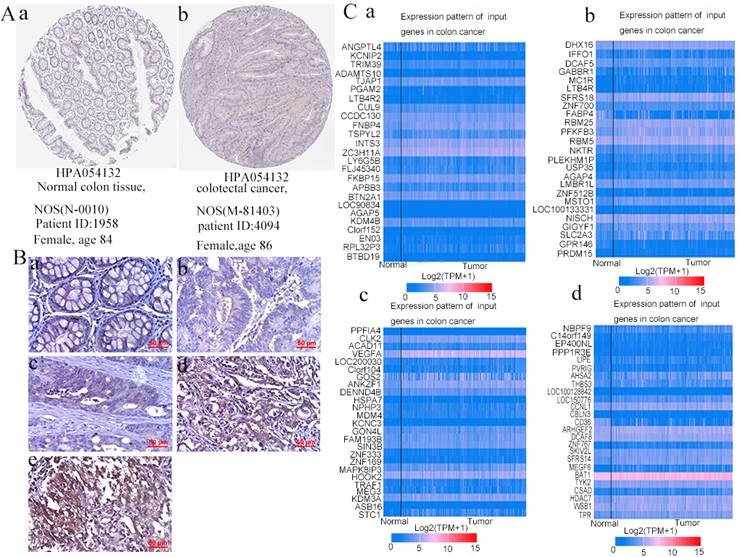
3. PPFIA4 IHC staining in human CRC tissues
PPFIA4 was selected for further study based on the results of the bioinformatics analysis described above. According to the Human Protein Atlas website, IHC analysis showed that the expression of PPFIA4 protein was higher in CRC tissues than in normal tissues (Figure 5A). In this study, IHC analysis was performed to detect the protein expression of PPFIA4 in five normal colon tissue samples and 219 cases. The results showed that the protein expression of PPFIA4 was higher in CRC tissues than in normal colon tissues (Figures 5B-a to e and Table 1). The expression of PPFIA4 was significantly lower in Group I than in Groups II (P = 0.009), III (P = 0.029), IV (P = 0.000), and V (P = 0.000). PPFIA4 expression was higher in groups IV (P = 0.000) and V (P = 0.000) than in group III. In addition, PPFIA4 expression was also higher in groups V (P = 0.004) and IV (P = 0.000) than in group II. The differences were not statistically significant between groups II and III (P = 0.299) or between groups IV and V (P = 0.128).
Comparison of the PPFIA4 staining index among different groups of human CRCs
| Group | n | Staining index | Value of statistics | P* | |
|---|---|---|---|---|---|
| Normal colon tissue | I | 5 | 2.2±0.83666 | χ2 = 53.837 | 0.000 |
| Well-differentiated CRCs | II | 51 | 5.5882±3.11882 | ||
| Moderately differentiated CRCs | III | 54 | 4.8519±2.17540 | ||
| Poorly differentiated CRCs | IV | 51 | 7.6471±3.56552 | ||
| Metastatic lymph node foci | V | 63 | 8.6349 ±3.27897 |
*P<0.05: statistically significant. (P: difference between the three groups; P1 (difference between groups I and II) = 0.009; P2 (difference between groups I and III) = 0.029; P3(difference between groups I and IV) = 0.000; P4(difference between groups I and V) = 0.000. P5 (difference between groups II and III) = 0.299. P6 (difference between Groups III and IV) = 0.000. P7 (difference between Groups III and V) = 0.000. P8 (difference between Groups II and IV) = 0.004. P9 (difference between groups II and V) = 0.000. P10 (difference between Groups IV and V) = 0.128.
CRC, colorectal cancer.
mRNA expression levels of PPFIA1, PPFIA2, PPFIA3, and PPFIA4 in different types of cancers and CRC. A. mRNA expression levels of PPFIA1 (a), PPFIA2 (b), PPFIA3 (c) and PPFIA4 (d) in normal colon and CRC samples as determined by Oncomine databases. B. mRNA expression levels of PPFIA1, PPFIA2, PPFIA3, and PPFIA4 in various types of cancers based on Oncomine database analysis. CRC: colorectal cancer.
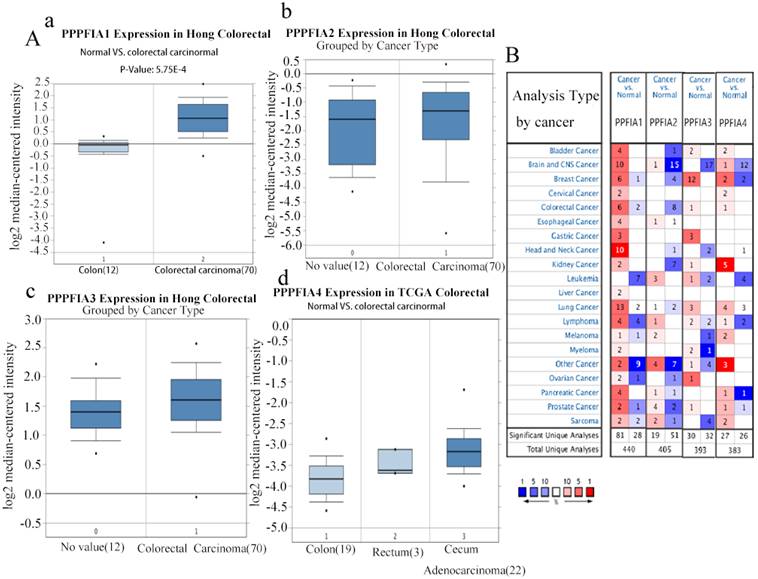
PPFIA1 (a), PPFIA2 (b), PPFIA3 (c) and PPFIA4 (d) mRNA expression levels were associated with lymphatic metastasis, pathological stage, and TP53-muation status in CRCs from the UALCAN database (*P < 0.05, ** P < 0.01).
A. mRNA expression levels of PPFIA1 (a), PPFIA3 (b), and PPFIA4 (c) in CRC cell lines. Black arrow points to the CRC cell lines. B. Statistical chart of the mRNA expression levels of PPFIA1 (a), PPFIA3 (b), and PPFIA4 (c) in LoVo and Hct116 compared with normal colon cell line (NCM460) determined by RT-PCR. C. The results of the western blot showed the expression of PPFIA1, PPFIA3 and PPFIA4 in LoVo and Hct116, and NCM460. D. The histogram revealed that the expression of PPFIA1 (a), PPFIA3 (c) and PPFIA4 (d) is statistically significant between LoVo, Hct116 and NCM460 (**P < 0.01, ***P < 0.001). CRC: colorectal cancer, RT-PCR: reverse transcription polymerase chain reaction.
PPFIA1, PPFIA2, PPFIA3, and PPFIA4 prognostic value in CRC patients. A. Based on GEPIA, disease free survival of PPFIA1 (a), PPFIA2 (b), PPFIA3 (c) and PPFIA4 (d) in CRC patients. B. Overall survival of PPFIA1 (a), PPFIA2 (b), PPFIA3 (c) and PPFIA4 (d) in CRC patients based on GEPIA. C. TCGA portal-based survival curves for PPFIA1 (a), PPFIA2 (b), PPFIA3 (c) and PPFIA4 (d) in CRC patients. CRC: colorectal cancer, GEPIA: gene expression profiling interactive analysis.
Micro-RNAs targeted PPFIA4 in ENCORI
| Number | miRNA | Coefficient-R | p-value |
|---|---|---|---|
| 1 | hsa-miR-101-3p | -0.051 | 2.76E-01 |
| 2 | hsa-miR-144-3p | -0.087 | 6.41E-02 |
| 3 | hsa-miR-329-3p | -0.019 | 6.86E-01 |
| 4 | hsa-miR-485-5p | -0.043 | 3.64E-01 |
| 5 | hsa-miR-580-3p | -0.022 | 6.42E-01 |
| 6 | hsa-miR-6884-5p | -0.097 | 4.06E-02 |
| 7 | hsa-miR-650 | -0.046 | 3.28E-01 |
| 8 | hsa-miR-660-5p | -0.032 | 4.94E-01 |
| 9 | hsa-miR-502-3p | -0.001 | 9.85E-01 |
| 10 | hsa-miR-362-3p | -0.131 | 5.41E-03 |
| 11 | hsa-miR-501-3p | -0.004 | 9.25E-01 |
| 12 | hsa-miR-138-5p | 0.021 | 6.59E-01 |
| 13 | hsa-miR-668-3p | 0.058 | 2.19E-01 |
| 14 | hsa-miR-134-5p | 0.076 | 1.06E-01 |
| 15 | hsa-miR-342-3p | 0.114 | 1.52E-02 |
| 16 | hsa-miR-423-3p | 0.09 | 5.54E-02 |
| 17 | hsa-miR-582-5p | 0.012 | 7.92E-01 |
4. PPFIA4 gene co-expression and functional enrichment analysis
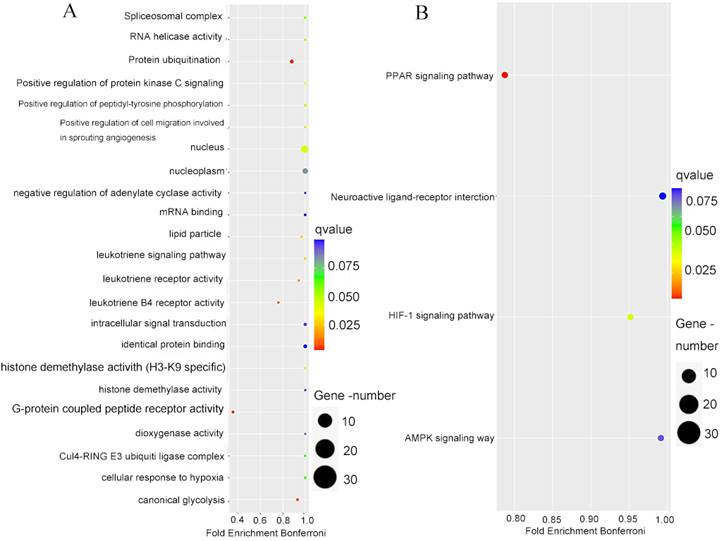
To better understand the biological processes and signaling pathways that are involved in PPFIA4, co-expressed genes with PPFIA4 were analyzed. The UALCAN database was used to identify the top 100 genes that were positively correlated with PPFIA4 expression in CRC (Figures 5C-a to d). Consequently, the PPI network was generated in the STRING protein interaction database to show the interaction among the 100 genes (Supplementary Figure 1), and then the 100 genes were imported into the Cytoscape platform (Version 3.7.1) to clarify the mutual relations of the 100 top genes co-expressed with PPFIA4 in CRC (Supplementary Figure 2). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses showed that the co-expressed genes of PPFIA4 were significantly enriched in G-protein coupled peptide receptor activity and leukotrience B4 receptor activity (Figure 6A). The most enriched KEGG pathways were the PPAR and HIF-1a signaling pathways (Figure 6B).
5. miRNAs and the expression of PPFIA4
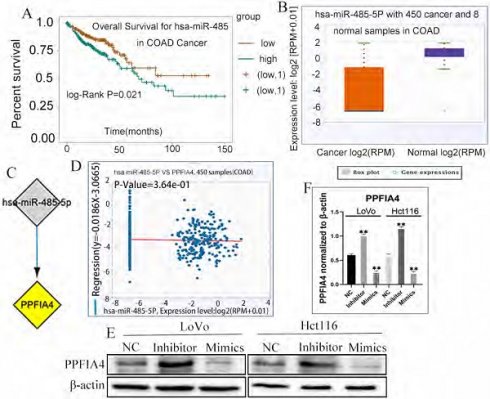
miRNAs are a class of small non-coding RNAs that play a critical role in the initiation and progression of diverse tumors [18, 19]. To determine the correlation between miRNAs and PPFIA4, the ENCORI platform was used to predict the miRNAs involved in the regulation of PPFIA4. Eventually, it was demonstrated that a total of 17 miRNAs could regulate the expression of PPFIA4 (Table 2). Among these, 14 miRNAs were negatively correlated with PPFIA4 expression. Four miRNAs among the 14 miRNAs showed decreased expression in CRCs, and only miR-485-5P was significantly associated with the prognosis of CRC patients (Figure 7A, P = 0.021). The results showed that miR-485-5P expression was decreased in CRC samples compared to that in normal colon tissue samples and negatively regulated the expression of PPFIA4, as determined by the ENCORI database (Figures 7B to 7D).
A. PPFIA4 expression in normal colon tissues and CRC tissues was determined using the human protein atlas. B. Immunohistochemical staining was performed to detect the expression of PPFIA4 in human normal colon and CRC tissues (400×). (a) Normal human colon tissues; (b) well-differentiated CRC; (c) moderately differentiated CRC; (d) poorly differentiated CRC; (e) Lymph node foci. C. Top 100 genes co-expressed with PPFIA4 in CRC based on UALCAN. a. Top 25 genes were co-expressed with PPFIA4 in CRC based on UALCAN. b. From 26 to 50 genes co-expressed with PPFIA4 in CRC based on UALCAN. c. From 51 to 75 genes were co-expressed with PPFIA4 in CRC based on UALCAN. d. From 76 to 100 genes were co-expressed with PPFIA4 in CRC based on UALCAN. CRC: colorectal cancer.
To further verify whether miR-485-5P can regulate the expression of PPFIA4, the expression of PPFIA4 was detected in LoVo and Hct116 cells after transfection with miR-485-5P inhibitor and miR-485-5P mimics. Intriguingly, the results indicated that PPFIA4 expression was decreased in LoVo and Hct116 cells after treatment with miR-485-5P mimics and increased after treatment with miR-485-5P inhibitors. These differences were statistically significant (Figure 7E and 7F, **P < 0.01). The results showed that miR-485-5P negatively modulated the expression of PPFIA4.
Discussion
High expression of PPFIA1 has been demonstrated in several cancers, including breast cancer, head and neck squamous cell carcinomas, and oropharyngeal carcinomas [20-28]. In studies, liprin-α1 has been shown to promote tumor cell migration and invasion [29, 30]. Inhibition of liprin-α1 resulted in increased expression of the transmembrane protein CD82, which plays an essential role in suppressing metastasis in several solid tumors [31]. Liprin-α2, encoded by PPFIA2, is located in the mature hippocampal presynapses. Liprin-α2 plays an important role in regulating neuronal activity and is highly expressed in the urinary sediments of patients with prostate cancer [9, 32-34]. PPFIA3 can be methylated in gastric cancer but is rarely methylated in normal gastric tissues. Furthermore, it has an independent prognostic value in patients with pancreatic cancer [10-12]. A study indicated that PPFIA4 was highly expressed in clear renal cell cancer compared to that in normal kidney tissue [35]. Xu et al. [38] showed that PPFIA4 was upregulated in human thyroid cancer tissues compared to nodular goitre tissues [36]. Additionally, PPFIA4 expression was significantly increased in castration-resistant prostate cancer tissues compared to that in prostate cancer [37]. Wang et al. [40] showed that PPFIA4 might be a potential biomarker for the diagnosis of pilocytic astrocytoma [38]. Results of Huang J, et al confirmed that PPFIA4 upregulation correlated with poor prognosis and higher clinical stages of CRC patients. Overexpression of PPFIA4 could increase the expression of EMT-related proteins and promote the proliferation, migration and invasion of colorectal cancer cells [39]. Bioinformatics analysis from Oncomine and UALCAN databases revealed that mRNA expression levels of PPFIA1, PPFIA3, and PPFIA4 were higher in CRC samples than in normal colon tissue. Evidence from CCLE databases and RT-PCR results also indicated that the mRNA of PPFIA1, PPFIA3, and PPFIA4 was overexpressed in the CRC cell lines LoVo and Hct116. The mRNA levels of PPFIA1, PPFIA3, and PPFIA4 were associated with tumor stage, lymph node metastasis, and TP53-mutation status. More importantly, survival analysis determined by TCGA portal and GEPIA illustrated that PPFIA4 expression was associated with the prognosis of patients with CRC. IHC staining analysis showed that PPFIA4 was closely associated with the degree of malignancy in CRCs.
To identify the biological processes and signal pathways of PPFIA4 involving in CRCs, the top 100 genes co-expressed with PPFIA4 were entered into the STRING database and Cytoscape software to obtain the PPI network. A gene set enrichment analysis was performed to study the role of genes co-expressed with PPFIA4 in CRCs. The GO enrichment analysis results indicated that these genes mainly participated in G-protein coupled peptide receptor activity and leukotriene B4 receptor activity. Furthermore, KEGG pathway enrichment analysis revealed that the co-expressed genes were enriched in PPAR and HIF-1 signaling pathways. Under hypoxia, PPFIA4 can promote the proliferation of cancer cells via mitogen-activated protein kinase (MAPK) or phosphoinositide 3-kinase (PI3K) signaling pathways. It enhances invasion ability through the epithelial-mesenchymal transition in pancreatic cancer. Moreover, PPFIA4 can promote chemotherapy resistance through MAPK pathways via HIF-1α expression in small cell lung cancer [14, 15]. Mattauch et al. revealed that PPFIA4 could be directly modulated by HIF-1α and by stabilizing E-cadherin and β-catenin to regulate cell junctions in renal cell carcinoma and breast cancer [35]. PPFIA4 also protects against nickel-induced cytotoxicity and modulates receptor protein tyrosine phosphatase-leukocyte antigen related receptor F (RPTP-LAR) activity [40]. PPFIA4 promotes castration-resistant prostate cancer progression via methylenetetrahydrofolate dehydrogenase 2 through mitochondrial metabolism [37].
The enrichment analysis of the top 100 genes co-expressed with PPFIA4. A. GO analysis of the top 100 genes co-expressed with PPFIA4. B. KEGG analysis of the top 100 genes co-expressed with PPFIA4. KEGG: Kyoto Encyclopedia of Genes and Genomes, GO: Gene Ontology.
A. Overall survival of miR-485-5p in CRC patients based on the ENCORI database. B. ENCORI determined the expression of miR-485-5p in normal colon and CRC samples. C. miRNA-PPFIA4 regulation network (Cytoscape). D. The negative correlation between miR-485-5p and PPFIA4 expression was determined by ENCORI. E. The expression of PPFIA4 in LoVo and Hct116 after treatment with inhibitors and mimics targeting miR-485-5p. F. The statistical chat of PPFIA4 expression in LoVo and Hct116 after treatment with inhibitors and mimics targeting miR-485-5p (**P < 0.01). CRC: colorectal cancer, ENCORI, encyclopedia of RNA Interactomes.
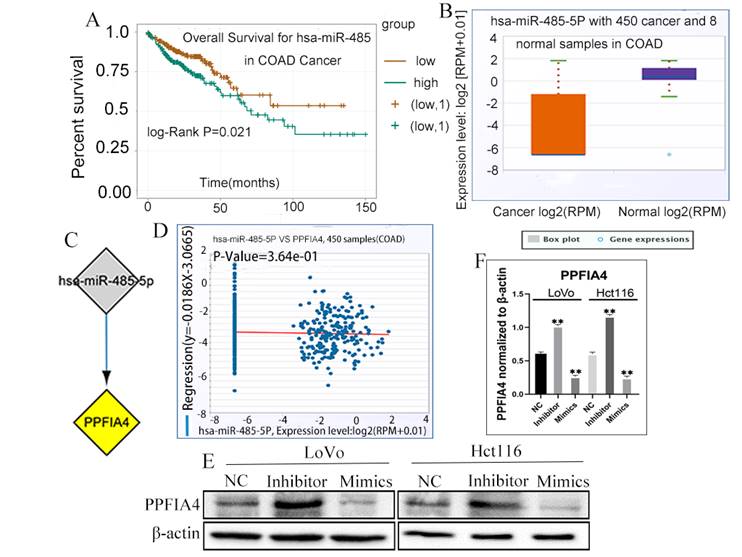
miRNAs are single-stranded, evolutionarily conserved molecules that modulate a wide range of target genes at the post-transcriptional level in physiological and pathological processes in humans [41, 42]. A recent study showed that miR-485-5p expression was decreased in CRC cell lines in comparison with the normal colon epithelial cell line, and in CRC tissues compared to paired para-cancerous tissues. MiR-485-5p overexpression suppressed the proliferation, migration, and invasion of CRC cells. CD147 was negatively regulated by miR-485-5p through binding a conserved sequence specifically within the CD147 [43]. miR-485-5p, which serves as a tumor suppressor, has been shown to be a potential prognostic biomarker for CRCs [19]. The expression of miR-485-5p could be sponged by LINC01224 and LINC01224 knockdown could increase the apoptosis of CRC cells [44]. In CRC, miR-485-5p plays a key role in targeting multiple glioma genes. In addition, HIF-1α can target the miR-485-5p promoter region to inhibit transcription [45]. Bioinformatic analysis showed that the expression of miR-485-5P was closely associated with the prognosis of CRC patients. The expression of PPFIA4 increased in LoVo and Hct116 cells when treated with miR-485-5P mimics and decreased when treated with miR-485-5P inhibitors. These results proved that miR-485-5P negatively regulates the expression of PPFIA4.
Our research indicated that PPFIA1, PPFIA3, and PPFIA4 expression increased in the CRC cell lines LoVo and Hct116. PPFIA4, which is involved in the PPAR and HIF-1 signaling pathways, is associated with the degree of malignancy of CRCs and could be used to evaluate the prognosis of patients with CRC. Thus, PPFIA4 may be a candidate biomarker and therapeutic target for CRCs. More studies on the potential mechanisms by which PPFIA4 regulates the development of CRC are needed in the future.
Abbreviations
CRC: colorectal cancer; GO: gene ontology; KEGG: kyoto encyclopedia of genes and genomes; CCLE: cancer cell line encyclopedia; GEPIA: gene expression profiling interactive analysis; ENCCRCORI: encyclopedia of RNA interactomes; OS: overall survival; DFS: disease free survival; IHC: immunohistochemical; miRNAs: micro-RNA; MAPK: mitogen-activated protein kinases; PI3K: phosphoinositide 3-kinase; RT-PCR: reverse transcription polymerase chain reaction; PPI: Protein-protein interaction; LAR: leukocyte common antigen-related gene.
Supplementary Material
Supplementary material and methods, figures, and tables.
Acknowledgements
This work was supported in part by grants from the National Science Foundation of China (#82173283 and #82103088), and Foundation of committee on science and technology of Tianjin (#20JCYBJC01230 and 21JCYBJC00190). The funders had no roles in the design of the study, data collection, analysis and interpretation, or decision to write and publish the work.
Author contributions
Conceptualization and supervision: Shiwu Zhang, Ming Gao; Methodology: Fangmei Fu, Xiaohui Yang, Linlin Fan; Validation: Fangmei Fu and Wenzheng Fu; Formal analysis: Fangmei Fu and Minying Zheng; Review and editing, Shiwu Zhang, Ming Gao; Funding acquisition: Shiwu Zhang, Minying Zheng. All authors have read and agreed to the published version of the manuscript.
Ethical approval
The study was approved by the Ethical Committee of Tianjin Union Medical Center.
Consent to participate
Written informed consents for the use of CRC tissue samples for scientific purposes were obtained for all individual participants and the use of human tissue samples was approved by the Hospital Review Board and the confidentiality of patients' information was maintained.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683-91
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-49
3. Stryker E, Johnson KG. LAR, liprin alpha and the regulation of active zone morphogenesis. J Cell Sci. 2007;120:3723-8
4. Serra-Pages C, Medley QG, Tang M, Hart A, Streuli M. Liprins, a family of LAR transmembrane protein-tyrosine phosphatase-interacting proteins. J Biol Chem. 1998;273:15611-20
5. Brown LA, Kalloger SE, Miller MA, Shih I-M, McKinney SE, Santos JL. et al. Amplification of 11q13 in ovarian carcinoma. Genes, Chromosomes and Cancer. 2008;47:481-9
6. Brown LA, Irving J, Parker R, Kim H, Press JZ, Longacre TA. et al. Amplification of EMSY, a novel oncogene on 11q13, in high grade ovarian surface epithelial carcinomas. Gynecologic Oncology. 2006;100:264-70
7. Schuuring E, Verhoeven E. et al. Amplification of genes within the chromosome 11q13 region is indicative of poor prognosis in patients with operable breast cancer. Cancer Research. 1992;52:5229-34
8. Chin SF, Wang Y, Thorne NP, Teschendorff AE, Pinder SE, Vias M. et al. Using array-comparative genomic hybridization to define molecular portraits of primary breast cancers. Oncogene. 2007;26:1959-70
9. Leyten GH, Hessels D, Smit FP, Jannink SA, de Jong H, Melchers WJ. et al. Identification of a Candidate Gene Panel for the Early Diagnosis of Prostate Cancer. Clin Cancer Res. 2015;21:3061-70
10. Li WH, Zhou ZJ, Huang TH, Guo K, Chen W, Wang Y. et al. Detection of OSR2, VAV3, and PPFIA3 Methylation in the Serum of Patients with Gastric Cancer. Dis Markers. 2016;2016:5780538
11. Meng Z, Yuan Q, Zhao J, Wang B, Li S, Offringa R. et al. The m(6)A-Related mRNA Signature Predicts the Prognosis of Pancreatic Cancer Patients. Mol Ther Oncolytics. 2020;17:460-70
12. Zong L, Hattori N, Yoda Y, Yamashita S, Takeshima H, Takahashi T. et al. Establishment of a DNA methylation marker to evaluate cancer cell fraction in gastric cancer. Gastric Cancer. 2016;19:361-9
13. Lee JY, Kim TH, Yang PS, Lim HE, Choi EK, Shim J. et al. Korean atrial fibrillation network genome-wide association study for early-onset atrial fibrillation identifies novel susceptibility loci. Eur Heart J. 2017;38:2586-94
14. Onishi H, Yamasaki A, Nakamura K, Ichimiya S, Yanai K, Umebayashi M. et al. Liprin-alpha4 as a New Therapeutic Target for SCLC as an Upstream Mediator of HIF1alpha. Anticancer Res. 2019;39:1179-84
15. Yamasaki A, Nakayama K, Imaizumi A, Kawamoto M, Fujimura A, Oyama Y. et al. Liprin-alpha4 as a Possible New Therapeutic Target for Pancreatic Cancer. Anticancer Res. 2017;37:6649-54
16. Li Z, Zheng M, Zhang H, Yang X, Fan L, Fu F. et al. Arsenic Trioxide Promotes Tumor Progression by Inducing the Formation of PGCCs and Embryonic Hemoglobin in Colon Cancer Cells. Front Oncol. 2021;11:720814
17. Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2010;27:431-2
18. Diaz G, Zamboni F, Tice A, Farci P. Integrated ordination of miRNA and mRNA expression profiles. BMC Genomics. 2015;16:767
19. Pan Y, Qin J, Sun H, Xu T, Wang S, He B. MiR-485-5p as a potential biomarker and tumor suppressor in human colorectal cancer. Biomark Med. 2020;14:239-48
20. Tan KD, Zhu Y, Tan HK, Rajasegaran V, Aggarwal A, Wu J. et al. Amplification and overexpression of PPFIA1, a putative 11q13 invasion suppressor gene, in head and neck squamous cell carcinoma. Genes Chromosomes Cancer. 2008;47:353-62
21. Dancau AM, Wuth L, Waschow M, Holst F, Krohn A, Choschzick M. et al. PPFIA1 and CCND1 are frequently coamplified in breast cancer. Genes Chromosomes Cancer. 2010;49:1-8
22. Blessmann M, Al-Dam A, Hanken H, Assaf AT, Riecke B, Klatt J. et al. Amplification of the PPFIA1 gene region on 11q13 in oral squamous cell carcinomas (OSCC). J Craniomaxillofac Surg. 2013;41:845-9
23. Choi EJ, Yun JA, Jabeen S, Jeon EK, Won HS, Ko YH. et al. Prognostic significance of TMEM16A, PPFIA1, and FADD expression in invasive ductal carcinoma of the breast. World J Surg Oncol. 2014;12:137
24. Chiaretti S, Astro V, Chiricozzi E, de Curtis I. Effects of the scaffold proteins liprin-alpha1, beta1 and beta2 on invasion by breast cancer cells. Biol Cell. 2016;108:65-75
25. Yang J, Wu NN, Huang DJ, Luo YC, Huang JZ, He HY. et al. PPFIA1 is upregulated in liver metastasis of breast cancer and is a potential poor prognostic indicator of metastatic relapse. Tumour Biol. 2017;39:1010428317713492
26. Barros-Filho MC, Reis-Rosa LA, Hatakeyama M, Marchi FA, Chulam T, Scapulatempo-Neto C. et al. Oncogenic drivers in 11q13 associated with prognosis and response to therapy in advanced oropharyngeal carcinomas. Oral Oncol. 2018;83:81-90
27. Zhou H, Cao T, Li WP, Wu G. Combined expression and prognostic significance of PPFIA1 and ALG3 in head and neck squamous cell carcinoma. Mol Biol Rep. 2019;46:2693-701
28. Alfarsi LH, El Ansari R, Craze ML, Masisi BK, Ellis IO, Rakha EA. et al. PPFIA1 expression associates with poor response to endocrine treatment in luminal breast cancer. BMC Cancer. 2020;20:425
29. Shen JC, Unoki M, Ythier D, Duperray A, Varticovski L, Kumamoto K. et al. Inhibitor of growth 4 suppresses cell spreading and cell migration by interacting with a novel binding partner, liprin alpha1. Cancer Res. 2007;67:2552-8
30. Asperti C, Astro V, Totaro A, Paris S, de Curtis I. Liprin-alpha1 promotes cell spreading on the extracellular matrix by affecting the distribution of activated integrins. J Cell Sci. 2009;122:3225-32
31. Pehkonen H, Lento M, von Nandelstadh P, Filippou A, Grenman R, Lehti K. et al. Liprin-alpha1 modulates cancer cell signaling by transmembrane protein CD82 in adhesive membrane domains linked to cytoskeleton. Cell Commun Signal. 2018;16:41
32. Spangler SA, Jaarsma D, De Graaff E, Wulf PS, Akhmanova A, Hoogenraad CC. Differential expression of liprin-alpha family proteins in the brain suggests functional diversification. J Comp Neurol. 2011;519:3040-60
33. Maurizy C, Quinternet M, Abel Y, Verheggen C, Santo PE, Bourguet M. et al. The RPAP3-Cterminal domain identifies R2TP-like quaternary chaperones. Nat Commun. 2018;9:2093
34. Torres VI, Inestrosa NC. Vertebrate Presynaptic Active Zone Assembly: a Role Accomplished by Diverse Molecular and Cellular Mechanisms. Mol Neurobiol. 2018;55:4513-28
35. Mattauch S, Sachs M, Behrens J. Liprin-alpha4 is a new hypoxia-inducible target gene required for maintenance of cell-cell contacts. Exp Cell Res. 2010;316:2883-92
36. Xu F, Xu H, Li Z, Huang Y, Huang X, Li Y. et al. Glycolysis-Based Genes Are Potential Biomarkers in Thyroid Cancer. Front Oncol. 2021;11:534838
37. Zhao R, Feng T, Gao L, Sun F, Zhou Q, Wang X. et al. PPFIA4 promotes castration-resistant prostate cancer by enhancing mitochondrial metabolism through MTHFD2. J Exp Clin Cancer Res. 2022;41:125
38. Wang G, Jia Y, Ye Y, Kang E, Chen H, Wang J. et al. Clinical and Epidemiological Study of Intracranial Tumors in Children and Identification of Diagnostic Biomarkers for the Most Common Tumor Subtype and Their Relationship with the Immune Microenvironment Through Bioinformatics Analysis. J Mol Neurosci. 2022;72:1208-23
39. Huang J, Yang M, Liu Z, Li X, Wang J, Fu N. et al. PPFIA4 Promotes Colon Cancer Cell Proliferation and Migration by Enhancing Tumor Glycolysis. Front Oncol. 2021;11:653200
40. Kiok K, Sun H, Clancy H, Bose S, Kluz T, Wu F. et al. Liprin-alpha4 is required for nickel induced receptor protein tyrosine phosphatase-leukocyte antigen related receptor F (RPTP-LAR) activity. PLoS One. 2011;6:e22764
41. Brower JV, Clark PA, Lyon W, Kuo JS. MicroRNAs in cancer: Glioblastoma and glioblastoma cancer stem cells. Neurochemistry International. 2014;77:68-77
42. Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nature Reviews Cancer. 2015;15:321-33
43. Hu XX, Xu XN, He BS, Sun HL, Xu T, Liu XX. et al. microRNA-485-5p Functions as a Tumor Suppressor in Colorectal Cancer Cells by Targeting CD147. J Cancer. 2018;9:2603-11
44. Gu J, Dong L, Wang Y, Nie W, Liu W, Zhao JA. LINC01224 promotes colorectal cancer progression through targeting miR-485-5p/MYO6 axis. World J Surg Oncol. 2021;19:281
45. Cheng L, Peng R, Guo P, Zhang H, Liu D, Liao X. et al. A HIF1A/miR-485-5p/SRPK1 axis modulates the aggressiveness of glioma cells upon hypoxia. Exp Cell Res. 2021;402:112547
Author contact
![]() Corresponding author: Shiwu Zhang, M.D., Ph.D., Department of Pathology, Tianjin Union Medical Center, Tianjin, 300121, China; Tel: (086)13652136865; Fax: (86)022-87721989; E-mail: zhangshiwu666com.
Corresponding author: Shiwu Zhang, M.D., Ph.D., Department of Pathology, Tianjin Union Medical Center, Tianjin, 300121, China; Tel: (086)13652136865; Fax: (86)022-87721989; E-mail: zhangshiwu666com.

 Global reach, higher impact
Global reach, higher impact