Impact Factor
ISSN: 1837-9664
J Cancer 2023; 14(12):2198-2208. doi:10.7150/jca.85681 This issue Cite
Research Paper
Diagnostic value of aberrant decreased 5-Methylcytosine RNA modification in leukocytes for non-small cell lung cancer
1. Department of Clinical Laboratory, Zhuhai People's Hospital (Zhuhai Hospital affiliated with Jinan University), Zhuhai, Guangdong, China.
2. Department of Clinical Laboratory, The Third Affiliated Hospital of Guangzhou Medical University, Guangzhou, Guangdong, China.
3. Department of Medical Laboratory, Shenzhen People's Hospital (The Second Clinical Medical College, Jinan University; The First Affiliated Hospital, Southern University of Science and Technology), Shenzhen, Guangdong, China.
4. Department of Clinical Medical Laboratory, Guangzhou First' People Hospital, School of Medicine, South China University of Technology, Guangzhou, Guangdong, China.
5. Department of Clinical Laboratory, The Seventh Affiliated Hospital of Sun Yat-sen University, Shenzhen, Guangdong, China.
# These authors contributed equally to this work.
Received 2023-4-27; Accepted 2023-7-6; Published 2023-7-16
Abstract
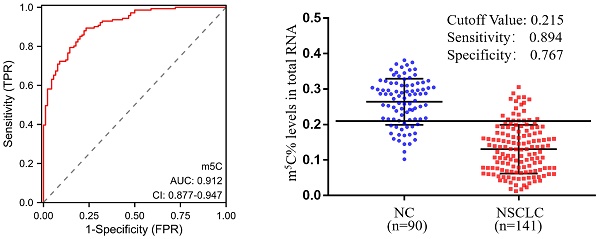
Background: Non-small cell lung cancer (NSCLC) was a disease with poor outcomes, partly because there were no high-efficiency non-invasive diagnostic biomarkers. The RNA modification status of 5-Methylcytosine (m5C) has been shown to be a biomarker for various diseases, but its potentiality to be a diagnostic biomarker for NSCLC remained inconclusive.
Methods: In this research, we collected peripheral leukocyte samples from 141 patients with NSCLC and 90 normal people as controls to evaluate the extent of m5C RNA modification.
Results: We found that the m5C modification levels in leukocytes of NSCLC patients were decreased dramatically, which were compared to the normal controls, and levels of m5C modification decreased progressively with tumor stage. Importantly, m5C modification exhibited superior diagnostic value compared to carcinoembryonic antigen (CEA), squamous cell carcinoma antigen (SCC), cytokeratin 19 fragment (Cyfra21-1), and carbohydrate antigen 125 (CA125), which demonstrated area under the curves (AUCs) of 0.912, 0.773, 0.669, 0.754, and 0.732, respectively. The combination of m5C modification with these serum tumor biomarkers further improved the AUC to 0.960. A nomogram model incorporating m5C modification also provided an effectively diagnostic tool for NSCLC.
Conclusion: Collectively, our findings suggested that m5C modification in leukocytes held promise as a prospective biomarker for NSCLC diagnosis.
Keywords: 5-Methylcytosine, non-small cell lung cancer, leukocytes, diagnosis, biomarker
Introduction
According to current statistics, lung cancer is the most prevalent tumor worldwide, accounting for the highest incidence and mortality rate of all malignancies in men and the second highest in women[1]. NSCLC was the main categories of lung cancer, and it remained a huge challenge in clinical management. Early detection of NSCLC was crucial for improving the five-year survival rates following surgical intervention. Unfortunately, the absence of early clinical symptoms and diagnostic techniques resulted in over 70% of NSCLC cases being identified at advanced stages[2]. Therefore, it was crucial to diagnose NSCLC accurately and at an early stage to improve patient prognosis.
Due to limited clinical resources, large-scale imaging screening and invasive pathology were currently not feasible for population-based screening of NSCLC[3]. As a result, serological tests had become increasingly popular for non-invasive and dynamic clinical surveillance of NSCLC patients[4]. However, currently available clinical diagnostic biomarkers, such as SCC, CEA, CA125, and Cyfra21-1, exhibited inadequate sensitivity and specificity for NSCLC screening, particularly in early-stage patients[5-7]. Consequently, a non-invasive and efficient method to improve the early diagnosis of NSCLC was urgently needed.
RNA methylation modifications, which included 5-Methylcytosine (m5C) and N6-methyladenosine (m6A), were regulated by a family of Writers, Erasers and Readers[8, 9]. These modifications were primarily concentrated within the CDS and 3'UTR regions of RNA, and could affect RNA transcriptional, post-transcriptional, and translational processes, making them a key regulator of various diseases[9, 10]. Our previous research, as well as other studies, had shown that m6A modification levels in leukocytes were markedly increased in NSCLC patients and closely associated with the key Erasers molecules FTO and ALKBH5, which is suggested that m6A methylation can be used as a tumor biomarker to modify the peripheral blood leukocytes[11, 12]. Additionally, our previous study has identified m5C methylation modification levels of white bold cells in colorectal cancer (CRC) patients as a novel tumor marker, with elevated levels seen in various primary CRC mouse models[13]. Therefore, we aimed to investigate whether m5C modification status in leukocytes could be a novel tumor biomarker in diagnosing NSCLC.
Surprisingly, our research found that, in contrast to CRC patients, the level of m5C RNA modification detected in leukocytes of NSCLC patients was apparently reduced. Furthermore, m5C modification provided exceptional diagnostic capability for NSCLC patients. The combination of m5C modification with other serum tumor biomarkers and the construction of a nomogram model incorporating m5C modification could further enhance diagnostic efficacy.
Materials and Methods
Human samples
The Institutional Review Board of The Third Affiliated Hospital of Guangzhou Medical University had approved the retrospective study (IRB number: S2021-114) on 20 Jan 2021. Between Jan 2021 and Dec 2022, we collected 231 peripheral blood samples using EDTA anticoagulation tubes at The Third Affiliated Hospital of Guangzhou Medical University. Blood samples were taken to test for tumor markers including SCC, CA125, CEA and Cyfra21-1 in the clinical laboratory. After the necessary tests, any remaining blood was collected with the patient's consent. Among those samples, 141 samples were from NSCLC patients and the other 90 samples were normal controls (NC) without a history of diabetes, hypertension, stroke, aneurysm, myocardial infarction or other serious diseases. 0.5 mL whole blood were mixed with 1 mL red blood cell lysis solution (TIANGEN, Beijing, China), after centrifuged, 1 mL triol was used to dissolve the precipitate to stabilize RNA. Then put the mixed samples at -80℃ to store. All the patients with NSCLC were selected according to the result of histopathology or cytopathology, and informed consent was obtained from all participants. 141 NSCLC patients were followed and had the blood samples both at time of first diagnosis before they got treatment (including surgery, chemotherapy, radiotherapy, targeted therapy, or immunotherapy) and samples from 11 NSCLC patients were collected at six months after they underwent therapy. Ethics approval was obtained from the Ethics Committee of The Third Affiliated Hospital of Guangzhou Medical University. All the patients' clinical and biological characteristics are displayed in Supplementary Table 1 and Supplementary Table 2.
RNA isolation and m5C quantification
TRIzol (Thermo Scientific, MA, USA) was used to extract the RNA. Levels of m5C in RNA were quantified using the MethyFlash 5-mC RNA Methylation ELISA Easy Kit (Fluorometric) (Epigentek, New York, USA). Briefly, added 200ng RNA into the assay wells which were coated with biding solution, and then incubated at 37℃ for 90minutes.
Subsequently, a serial diluted concentration of 5-mC antibody, signal indicator and enhancer solution were added consecutively, then incubated at room temperature for 1 hour. Finally, added fluorescent developer and incubated for another 3 minutes at room temperature. Using synergyH1 multi-mode readers (BioTek, Vermont, USA) to measure the fluorescence at wavelengths of 530 nm for excitation and 590 nm for emission, within 2 to 10 minutes after adding the development solution.
Statistical analysis
For normally distributed data between two groups, unpaired Student's t test was used to analyze the variability, which was shown as the SD (mean±SD). Otherwise, nonparametric Mann-Whitney test was used to analyze the data. For multiple groups, the significant differences were determined by one-way analysis of variance (ANOVA).
There was a statistical significance if P was less than 0.05. The receiver operating characteristic (ROC) curve with an area under the curve (AUC) was used to assess the diagnostic value of biomarkers to differentiate NSCLC from NC. A nomogram was constructed based on multivariate logistic regression analysis for predicting NSCLC.
Results
Levels of m5C modification in leukocytes of NSCLC patients
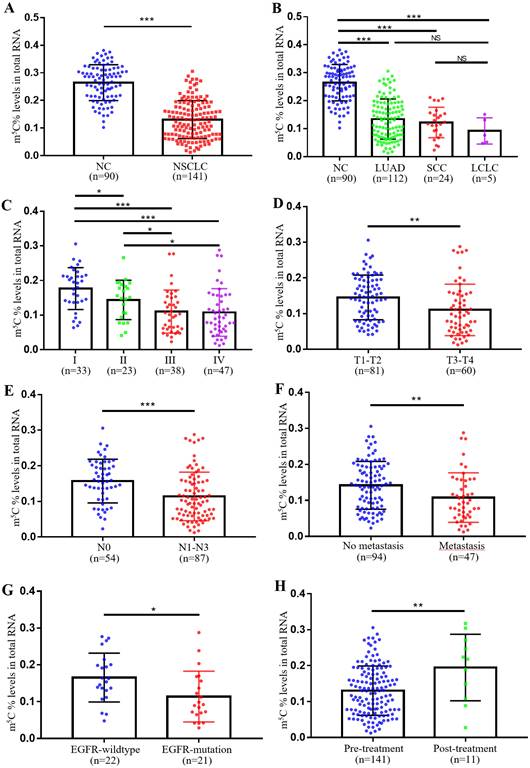
In our previous study, we observed remarkably higher levels of m5C RNA modification in leukocytes of CRC patients. Therefore, we further checked its expression in leukocytes of NSCLC patients. Supplementary Table 1 showed the baseline characteristics of NC individuals and NSCLC patients were well-matched. Surprisingly, unlike in CRC, NSCLC patients exhibited markedly lower levels of m5C modification in their leukocytes (Fig. 1A). Furthermore, no significant difference in m5C modification was found between different pathological classifications of NSCLC, which included adenocarcinoma, squamous cell carcinoma, and large cell lung carcinoma (Fig. 1B). Statistical analyses presented that the m5C levels were associated with gender, clinical stage, N classification, M classification, differentiation, and EGFR genotyping (Supplementary Table 2). Among them, levels of m5C RNA modification gradually decreased with tumor stage progression (Fig. 1C). As T classification and N classification increased, levels of m5C RNA modification were also reduced in leukocytes (Fig. 1D-E). NSCLC patients with distant-metastases had lower m5C levels than non-metastatic NSCLC patients (Fig. 1F). The results also indicated that poor prognosis NSCLC patients with EGFR mutations shared reduced m5C levels (Fig. 1G). Additionally, Leukocyte m5C levels rebounded in NSCLC patients after treatment (Fig. 1H). Taken together, our findings demonstrated that in contrast to CRC patients, levels of m5C RNA modification detected in leukocytes of NSCLC patients decreased apparently.
Comparison of m5C modification levels in leukocytes and tumor biomarker levels in serum
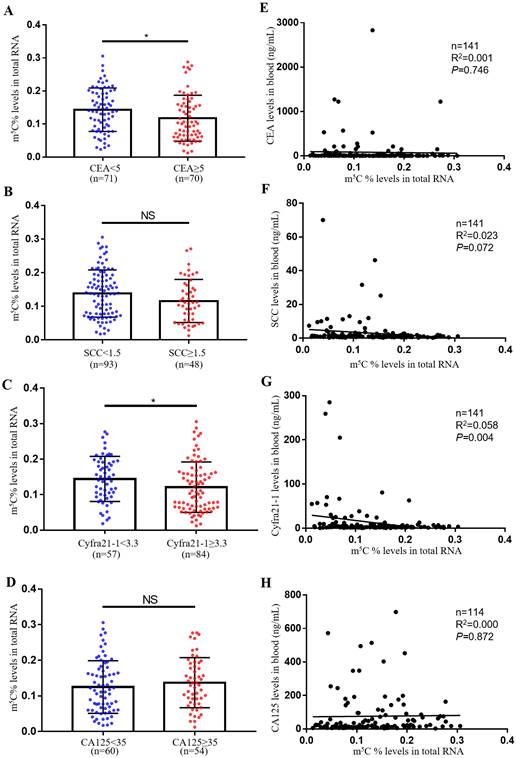
The most usually used serum biomarkers for NSCLC were SCC, CA125, CEA and Cyfra21-1. In this study, we investigated the relationship between the level of m5C modification and these tumor biomarkers. Notably, we observed that m5C modification levels were lower in the CEA and Cyfra21-1 abnormally increased groups compared to the control group (Fig. 2A-D). However, we only found a negative correlation between m5C modification levels and Cyfra21-1 levels, and no significant correlation with CEA, SCC, and CA125 levels (Fig. 2E-H). Therefore, Cyfra21-1 was identified as the serum biomarkers of NSCLC that was most associated with m5C modification of leukocytes.
Clinical utility of m5C modification levels in leukocytes for the diagnosis of NSCLC
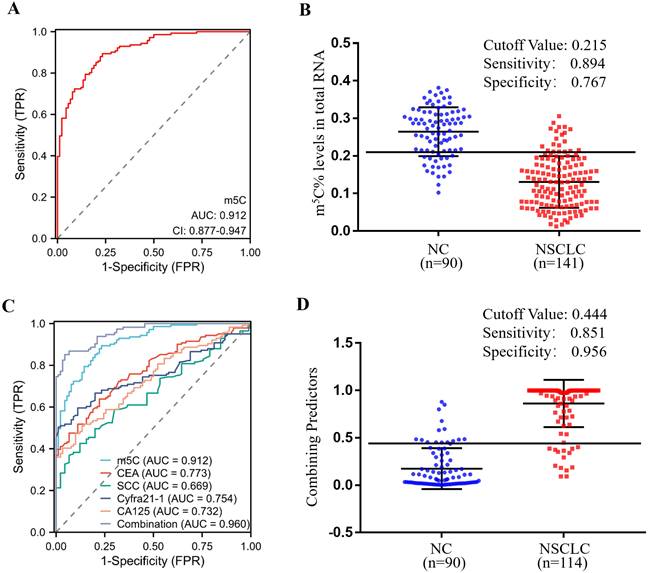
In order to estimate the diagnostic value of m5C modification in leukocytes for NSCLC furtherly, ROC curves were generated. As illustrated in Fig. 3A, m5C modification displayed a high ability to distinguish NSCLC patients from NC, with an AUC of 0.912 (95% CI 0.877 to 0.947). The optimal cutoff value for m5C modification level, based on the Youden index, was determined to be 0.215, with a sensitivity of 0.894 and specificity of 0.767 (Fig. 3B). Moreover, our findings suggested that m5C modification possessed superior diagnostic value compared to CEA, SCC, Cyfra21-1, and CA125, which demonstrated AUCs of 0.773, 0.669, 0.754, and 0.732, respectively (Fig. 3C; Table 1). While m5C modification was combined with these commonly used serum biomarkers, the AUC of the biomarker panel increased to 0.960 (Fig. 3C). The combined biomarker score for diagnosis was defined as: 0.539-23.816*m5C+0.084*CEA+0.682*SCC+0.701*Cyfra21-1+0.060*CA125. The optimal cutoff value for the combination biomarker was 0.444, with a sensitivity of 0.851 and specificity of 0.956 (Fig. 3D). Taken together, these results indicated that m5C modification levels in leukocytes RNA provided exceptional diagnostic capability for NSCLC patients.
Sensitivity and specificity of the diagnostic value of various biomarkers alone and in combination in NSCLC.
| Marker | Sensitivity | Specificity | AUC | 95% CI |
|---|---|---|---|---|
| m5C | 0.894 | 0.767 | 0.912 | 0.877-0.947 |
| CEA | 0.638 | 0.778 | 0.773 | 0.714-0.831 |
| SCC | 0.426 | 0.878 | 0.669 | 0.600-0.737 |
| Cyfra21-1 CA125 | 0.504 0.509 | 0.989 0.867 | 0.754 0.732 | 0.692-0.815 0.665-0.800 |
| Combination | 0.851 | 0.956 | 0.960 | 0.938-0.982 |
The diagnostic value of m5C modification levels in leukocytes at different stages of NSCLC
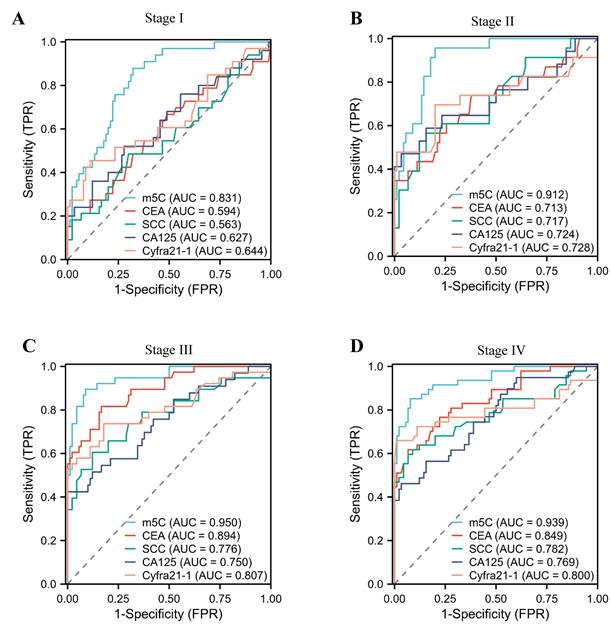
In addition, we assessed the diagnostic potential of m5C modification in leukocytes at various stages of NSCLC. Notably, stage I NSCLC patients could be accurately distinguished from healthy individuals by m5C modification, with AUCs of 0.831 (Fig. 4A). Conversely, conventional used serum biomarkers provided limited diagnostic capability for stage I NSCLC patients (Fig. 4A; Supplementary Table 3). Encouragingly, the AUC of m5C modification for NSCLC patients increased gradually with advancing stage (Fig. 4B-D). These results underscored the potential of m5C modification in leukocytes as a promising diagnostic biomarker for early NSCLC.
Levels of m5C modification in leukocytes of NSCLC patients. (A) Levels of m5C modification in RNA of peripheral blood leukocytes from NC (n=90) and NSCLC patients (n=141). (B) m5C modification levels of leukocytes in different pathological classifications of NSCLC, including LUAD (adenocarcinoma, n=112), SCC (squamous cell carcinoma, n=24), and LCLC (large cell lung carcinoma, n=5). (C) m5C modification levels of leukocytes in NSCLC patients at different clinical stages, including stage-I (n=33), stage-II (n=23), stage-III (n=38), and stage-IV (n=47). (D) Comparison of m5C modification levels of leukocytes in NSCLC patients with different T stages. (E) Comparison of m5C modification levels of leukocytes in NSCLC patients with different N stages. (F) Comparison of m5C modification levels of leukocytes in NSCLC patients with (n=47) and without (n=94) distant-metastasis. (G) Comparison of m5C modification levels of leukocytes in NSCLC patients with (n=21) and without (n=22) EGFR mutation. (H) Comparison of m5C modification levels of leukocytes in NSCLC patients with (n=11) and without (n=141) treatment. Data were shown as mean ± SD; * P <0.05, ** P <0.01, and *** P <0.001. NS, no significance.
Comparison of m5C modification levels in leukocytes and tumor biomarker levels in serum. (A) Comparison of m5C modification levels of leukocytes in NSCLC patients with CEA ≥ 5 ng/ml (n = 70) and CEA < 5 ng/ml (n = 71). (B) Comparison of m5C modification levels in leukocytes between NSCLC patients with SCC≥ 1.5 ng/ml (n = 48) and those with SCC < 1.5 ng/ml (n = 93). (C) Comparison of m5C modification levels in leukocytes between NSCLC patients with Cyfra21-1≥ 3.3 ng/ml (n = 84) and those with Cyfra21-1< 3.3 ng/ml (n = 57). (D) Comparison of m5C modification levels in leukocytes between NSCLC patients with CA125≥ 35 ng/ml (n = 54) and those with CA125< 35 ng/ml (n = 60). (E-H) Spearman correlation analysis of m5C modification levels with CEA levels (E), SCC levels (F), Cyfra21-1 levels (G), and CA125 levels (H) in NSCLC patients. Data were shown as mean ± SD; * P <0.05. NS, no significance.
Clinical utility of m5C modification levels in leukocytes for the diagnosis of NSCLC. (A, B) ROC curve (A) and corresponding cutoff value (B) for discriminating NSCLC from NC based on m5C modification levels in leukocytes. (C) ROC curve for m5C modification, CEA, SCC, Cyfra21-1, and CA125 alone or together. (D) Cutoff value for m5C modification, CEA, SCC, Cyfra21-1, and CA125 in combination for discriminated NSCLC from NC.
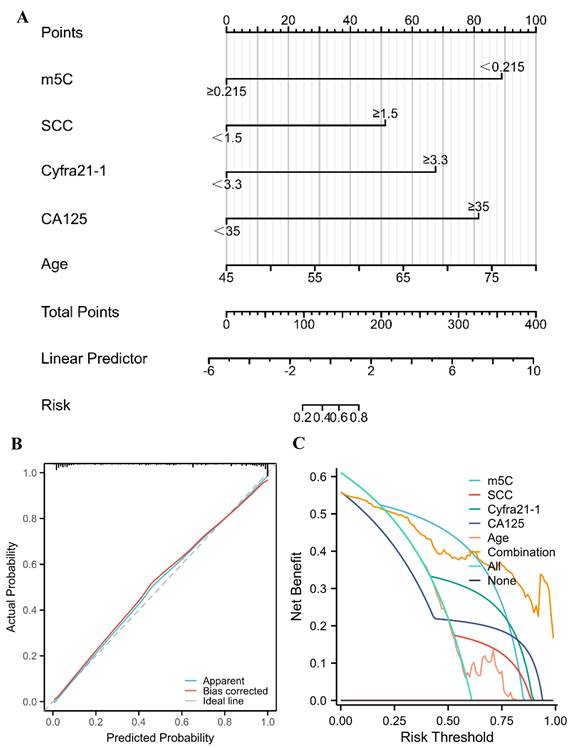
A nomogram for predicting the probability of NSCLC for healthy individuals
The risk of NSCLC that related to m5C modification, CEA, SCC, Cyfra21-1, CA125, and age was indicated by the results of univariate logistic regression analysis (Table 2). Furthermore, m5C modification, SCC, Cyfra21-1, CA125, and age were independent diagnostic factors for NSCLC, which were identified by multivariate logistic regression analyses (Table 2). According to the results of the multivariate logistic regression, a nomogram was constructed to differentiate NSCLC patients from healthy individuals (Fig. 5A). The concordance index (C-index) of the nomogram was 0.945 (95% CI 0.917 to 0.972), suggesting a strong capacity to distinguish NSCLC patients from healthy individuals (Fig. 5A). The calibration curves of the nomogram displayed high consistencies between observed and predicted values, with the Hosmer-Lemeshow test yielding a non-significant P value of 0.361 (Fig. 5B). Furthermore, the clinical validity of the nomogram model was confirmed by the decision curve analysis (DCA), which obtained a net benefit for almost all threshold probabilities (Fig. 5C). Collectively, a nomogram model incorporating m5C modification also provided an effectively diagnostic tool for NSCLC.
Univariate and multivariate logistic regression for NSCLC diagnosis.
| Variables | Univariate Analysis | Multivariate Analysis | ||||
|---|---|---|---|---|---|---|
| OR (95% CI) | P value | OR (95% CI) | P value | |||
| m5C % | ||||||
| <0.215 vs. ≥0.215 | 22.078 (21.340-22.815) | <0.001 | 26.310 (25.182-27.437) | <0.001 | ||
| Gender | ||||||
| Female vs. Male | 1.080 (0.520-1.640) | 0.788 | ||||
| CEA (ng/mL) | ||||||
| ≥5 vs. <5 | 8.286 (7.533-9.039) | <0.001 | 2.775 (1.670-3.881) | 0.070 | ||
| SCC (ng/mL) | ||||||
| ≥1.5 vs. <1.5 | 7.280 (6.366-8.194) | <0.001 | 6.799 (5.176-8.423) | 0.021 | ||
| Cyfra21-1 (ng/mL) | ||||||
| ≥3.3 vs. <3.3 | 12.267 (11.510-13.024) | <0.001 | 10.701 (9.526-11.876) | <0.001 | ||
| CA125 (ng/mL) | ||||||
| ≥35 vs. <35 | 20.343 (19.133-21.553) | <0.001 | 22.831 (21.181-24.480) | <0.001 | ||
| Age | 1.057(1.021-1.094) | 0.003 | 1.120 (1.040-1.199) | 0.006 | ||
The diagnostic value of m5C modification levels in leukocytes at different stages of NSCLC. (A-D) ROC curve for m5C modification compared with CEA, SCC, Cyfra21-1, and CA125 in NSCLC patients at stage I (A), stage II (B), stage III (C), and stage IV (D).
A nomogram for predicting the probability of NSCLC for healthy individuals. (A) A nomogram for differentiating between NSCLC patient and healthy individual was established based on multivariate logistic regression results. (B) Calibration curves of the nomogram. (C) DCA for the nomogram and various markers alone.
Discussion
Growing evidence suggested that m5C RNA modification and its regulators were abnormally expressed in different types of cancers[14]. Meanwhile, m5C RNA methylation status was closely linked to cancer pathogenesis, including cancer development, metastasis, recurrence, and drug resistance[15]. Previous study had identified abundant m5C modification in the lncRNA of CRC, which participated in the transcription of lncRNA by influencing the promoters and super-enhancers[16]. our recent study also found significantly increased levels of m5C RNA modification in leukocytes from CRC patients as a novel tumor biomarker[13]. Given the lack of effective non-invasive diagnostic biomarkers for NSCLC, we proposed to investigate whether the m5C modification status in leukocytes could also be used as a tumor biomarker for NSCLC.
The relationship between m5C modification and NSCLC has been controversial[15]. One of the earliest studies found that elevated levels of m5C modification could be detected in circulating tumor cells from NSCLC patients[17]. While recent study also demonstrated that NSUN3 and NSUN4, regulators of m5C modification, could promote NSCLC progression[18]. Meanwhile, NSUN3 was strictly linked to the infiltration of CD8+ T cells, whereas NSUN4 was associated with the infiltration of neutrophils[18]. However, in contradiction to these results, m5C methylation modification patterns were constructed by bioinformatic methods based on 11 m5C regulators, with higher scores suggesting a better prognosis[19]. In our study, the level of m5C modification of leukocytes in NSCLC patients was markedly lower than that in NC (Fig. 1), although the reasons for this reduction and the specific regulators that played a crucial role required further investigation. The pathological subtype of large cell lung cancer, which was associated with a poorer prognosis, exhibited relatively lower levels of m5C compared to the pathological subtypes of lung squamous carcinoma and lung adenocarcinoma, which were associated with a better prognosis. However, this difference did not reach statistical significance, likely due to the limited number of cases of large cell lung cancer in our study (Fig. 1B; Supplementary Table 2).
Nevertheless, abnormally reduced m5C modification exhibited superior diagnostic value compared to CEA, SCC, Cyfra21-1, and CA125, which demonstrated AUCs of 0.912, 0.773, 0.669, 0.754, and 0.732, respectively (Fig. 3A-B). The combined biomarker score of m5C modification in combination with these traditional biomarkers further increased AUC to 0.960 without considering the economic cost (Fig. 3C-D). Meanwhile, to optimize the diagnostic model for clinical application, we identified 5 variables (m5C modification, SCC, Cyfra21-1, CA125, and age) based on logistic regression analysis and constructed a nomogram model, which demonstrated excellent discrimination and reclassification performance in the diagnosis of NSCLC (Table 2; Fig. 5). Notably, our prediction model selected common clinical characteristics as predictors, making it ideal for replication in primary hospitals and areas with limited healthcare resources. As all the samples we used in this research were only collected from a single institution, further studies to verify the reliability of this model would require much larger sample sizes from other medical centers.
An accumulating body of research had indicated that RNA methylation modifications in peripheral blood cells were associated with disease diagnosis and prognosis. The concentrations of m6A in peripheral blood RNA had shown its potential as a biomarker to diagnose lung cancer, gastric cancer, and, breast cancer[12, 20, 21]. The demethyltransferase FTO has been found to exert a pro-carcinogenic effect in acute myeloid leukemia by regulating m6A levels on the mRNA of downstream target genes, such as ASB2 and RARA[22]. Additionally, regulated levels of m6A RNA modifications in peripheral blood following myocardial infarction might hold potential as a novel biomarker for the development of heart failure[23]. While previous studies had mainly focused on m6A in RNA methylation modification, little had been done on m5C. Our study found that the m5C modification was distinct from m6A modification in leukocytes, which was not abnormally high in all diseases.
To investigate the underlying reasons for the decreased m5C levels in leukocytes of NSCLC patients, we analyzed key regulatory factors previously implicated in m5C modification in NSCLC tumor tissues[15]. qRT-PCR analysis revealed a significant upregulation of the methylesterase NSUN3 and an apparent downregulation of the demethyltransferase TET2 in NSCLC patients compared to healthy individuals (Supplementary Figure. 1). However, no significant differences were observed in NOP2, ALYREF, and YBX1 (Supplementary Figure. 1). These findings suggested that NSUN3 and TET2 might contribute to the abnormal m5C modification levels in leukocytes of NSCLC patients, but further experimental data was needed to provide stronger evidence. Additionally, m5C modification leaded to the regulation of numerous downstream genes, and it remained crucial to investigate which specific leukocyte genes were affected by m5C modification and participated in tumor progression in NSCLC patients through further in vitro experiments.
With the development of molecular biology, liquid biopsy had become a hot research topic in tumor diagnosis[24]. Compared to traditional tissue biopsy, liquid biopsy was non-invasive and dynamic[25]. There were three main types of biomarkers of liquid biopsy for tumor patients: Circulating Tumor Cells (CTCs), Cell-Free DNA (cfDNA) and Exosomes (Exos). Epithelial-Mesenchymal Transition (EMT) was a biologic process that epithelial cells lost their adhesion and polarity to transform into mesenchymal cells, and CTCs were shed from the primary focus into the circulatory system[26]. CTCs were the first liquid biopsy indicator to be clinically tested, but tumor patients only contained 1-10 CTCs cells per mL of blood. Therefore, CTCs testing was more suitable for prognostic assessment of patients with advanced tumors, and its clinical application was limited[27]. Meanwhile, m5C modification could accurately distinguish early-stage NSCLC patients from healthy individuals with AUCs of 0.831 (Fig. 4). Continuous monitoring of cfDNA levels could also detect post-operative recurrence and metastases in gastric cancer patients up to 12 months earlier than imaging[28]. However, DNA fragments were also released during the death of normal cells and were more abundant, which might overwrite the cfDNA information of tumor cells and therefore were not suitable for early tumor diagnosis[29]. The method of isolation of Exos was not yet standardized, and different isolation methods could extract Exos with different characteristics, making them a long way from clinical application[30, 31]. In comparison, peripheral blood leukocyte samples were easy to obtain, and the m5C methylation detection method was inexpensive. While our research suggested that m5C methylation modification status of leukocytes held great promise as a superior diagnostic tool for NSCLC, there were certain challenges that must be addressed. For instance, storing RNA samples was difficult, and the process for detecting RNA methylation modifications was cumbersome.
While our findings hold promise, several limitations should be acknowledged in our study. Future investigations should aim to address these limitations, including the inclusion of larger sample sizes from multiple centers to mitigate potential biases such as NSCLC bias, control type bias, and selection bias. Additionally, long-term observation of lung cancer patients is warranted to determine whether m5C modification has a significant impact on prognosis. These efforts will provide a more comprehensive understanding of the implications of our findings and their clinical implications.
Conclusion
In summary, our research indicated firstly the clinical diagnosis values of m5C methylation modification status in leukocytes of NSCLC. Additionally, combining m5C modification with other serum tumor biomarkers and constructing the nomogram model incorporating m5C modification could further enhance diagnostic efficacy.
Abbreviations
ROC: Receiver operating characteristic; AUC: Area under the curve; NSCLC: Non-small cell lung cancer; m5C: 5-Methylcytosine; m6A: N6-methyladenosine; CEA: Carcinoembryonic antigen; SCC: squamous cell carcinoma antigen; Cyfra21-1: cytokeratin 19 fragment; CA125: carbohydrate antigen 125; CRC: colorectal cancer; CTCs: Circulating Tumor Cells; cfDNA: Cell-Free DNA; Exos: Exosomes.
Supplementary Material
Supplementary figure and tables.
Acknowledgements
Funding
This study was supported by the fund from the National Nature Science Foundation of China (81902693; 82103346; 82202829); Guangdong Basic and Applied Basic Research Foundation (2021A1515110094; 2022A1515111199).
Data Availability Statement
All data generated or analyzed during this study are included in this published article. Further inquiries can be directed to the corresponding authors.
Ethics Statement
Written informed consent was obtained from the individuals for the publication of any potentially identifiable images or data included in this article.
Author Contributions
HHH, HM, and JX initiated and designed this study. YHF and JHB performed the experiments. ML and JP contributed to the data analysis. YHF and ML were responsible for writing the manuscript. All authors contributed to the article and approved the submitted version.
Ethics Approval and Consent to Participate
All participants provided written informed consent. The study design was conducted in accordance with the principles of the Declaration of Helsinki and approved by the Ethics Committee of The Third Affiliated Hospital of Guangzhou Medical University had approved the retrospective study (IRB number: S2021-114).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Chen P, Liu Y, Wen Y, Zhou C. Non-small cell lung cancer in China. Cancer Commun (Lond). 2022;42:937-70
2. Lahiri A, Maji A, Potdar PD, Singh N, Parikh P, Bisht B. et al. Lung cancer immunotherapy: progress, pitfalls, and promises. Mol Cancer. 2023;22:40
3. Adams SJ, Stone E, Baldwin DR, Vliegenthart R, Lee P, Fintelmann FJ. Lung cancer screening. Lancet. 2023;401:390-408
4. Chen S, Huang H, Liu Y, Lai C, Peng S, Zhou L. et al. A multi-parametric prognostic model based on clinical features and serological markers predicts overall survival in non-small cell lung cancer patients with chronic hepatitis B viral infection. Cancer Cell Int. 2020;20:555
5. Dal Bello MG, Filiberti RA, Alama A, Orengo AM, Mussap M, Coco S. et al. The role of CEA, CYFRA21-1 and NSE in monitoring tumor response to Nivolumab in advanced non-small cell lung cancer (NSCLC) patients. J Transl Med. 2019;17:74
6. Zhou J, Diao X, Wang S, Yao Y. Diagnosis Value of Combined Detection of Serum SF, CEA and CRP in Non-Small Cell Lung Cancer. Cancer Manag Res. 2020;12:8813-9
7. Wang X, Wang M, Feng L, Song J, Dong X, Xiao T. et al. Four-protein model for predicting prognostic risk of lung cancer. Front Med. 2022;16:618-26
8. Huang H, Weng H, Chen J. m(6)A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell. 2020;37:270-88
9. Nombela P, Miguel-Lopez B, Blanco S. The role of m(6)A, m(5)C and Psi RNA modifications in cancer: Novel therapeutic opportunities. Mol Cancer. 2021;20:18
10. Boulias K, Greer EL. Biological roles of adenine methylation in RNA. Nat Rev Genet. 2023;24:143-60
11. Yin H, Hong H, Yin P, Lu W, Niu S, Chen X. et al. Increased levels of N6-methyladenosine in peripheral blood RNA: a perspective diagnostic biomarker and therapeutic target for non-small cell lung cancer. Clin Chem Lab Med. 2023;61:473-84
12. Pei Y, Lou X, Li K, Xu X, Guo Y, Xu D. et al. Peripheral Blood Leukocyte N6-methyladenosine is a Noninvasive Biomarker for Non-small-cell Lung Carcinoma. Onco Targets Ther. 2020;13:11913-21
13. Yin H, Huang Z, Niu S, Ming L, Jiang H, Gu L. et al. 5-Methylcytosine (m(5)C) modification in peripheral blood immune cells is a novel non-invasive biomarker for colorectal cancer diagnosis. Front Immunol. 2022;13:967921
14. Li M, Tao Z, Zhao Y, Li L, Zheng J, Li Z. et al. 5-methylcytosine RNA methyltransferases and their potential roles in cancer. J Transl Med. 2022;20:214
15. Song H, Zhang J, Liu B, Xu J, Cai B, Yang H. et al. Biological roles of RNA m(5)C modification and its implications in Cancer immunotherapy. Biomark Res. 2022;10:15
16. Hu H, Shu M, He L, Yu X, Liu X, Lu Y. et al. Epigenomic landscape of 5-hydroxymethylcytosine reveals its transcriptional regulation of lncRNAs in colorectal cancer. Br J Cancer. 2017;116:658-68
17. Mackenzie JW, Lewis RJ, Sisler GE, Lin W, Rogers J, Clark I. Urinary catabolites of ribonucleic acid as cancer markers: a preliminary report of their use in patients with lung cancer. Ann Thorac Surg. 1984;38:133-9
18. Pan J, Huang Z, Xu Y. m5C RNA Methylation Regulators Predict Prognosis and Regulate the Immune Microenvironment in Lung Squamous Cell Carcinoma. Front Oncol. 2021;11:657466
19. Chen H, Ge XL, Zhang ZY, Liu M, Wu RY, Zhang XF. et al. M(5)C regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in lung adenocarcinoma. Transl Lung Cancer Res. 2021;10:2172-92
20. Ge L, Zhang N, Chen Z, Song J, Wu Y, Li Z. et al. Level of N6-Methyladenosine in Peripheral Blood RNA: A Novel Predictive Biomarker for Gastric Cancer. Clin Chem. 2020;66:342-51
21. Xiao H, Fan X, Zhang R, Wu G. Upregulated N6-Methyladenosine RNA in Peripheral Blood: Potential Diagnostic Biomarker for Breast Cancer. Cancer Res Treat. 2021;53:399-408
22. Li Z, Weng H, Su R, Weng X, Zuo Z, Li C. et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N(6)-Methyladenosine RNA Demethylase. Cancer Cell. 2017;31:127-41
23. Vausort M, Niedolistek M, Lumley AI, Okninska M, Paterek A, Maczewski M. et al. Regulation of N6-Methyladenosine after Myocardial Infarction. Cells. 2022;11:2271
24. Visser E, Genet S, de Kock R, van den Borne B, Youssef-El Soud M, Belderbos HNA. et al. Liquid biopsy-based decision support algorithms for diagnosis and subtyping of lung cancer. Lung Cancer. 2023;178:28-36
25. Li W, Liu JB, Hou LK, Yu F, Zhang J, Wu W. et al. Liquid biopsy in lung cancer: significance in diagnostics, prediction, and treatment monitoring. Mol Cancer. 2022;21:25
26. Thanh Huong P, Gurshaney S, Thanh Binh N, Gia Pham A, Hoang Nguyen H, Thanh Nguyen X. et al. Emerging Role of Circulating Tumor Cells in Gastric Cancer. Cancers (Basel). 2020;12:695
27. Zhang WW, Rong Y, Liu Q, Luo CL, Zhang Y, Wang FB. Integrative diagnosis of cancer by combining CTCs and associated peripheral blood cells in liquid biopsy. Clin Transl Oncol. 2019;21:828-35
28. Ren J, Lu P, Zhou X, Liao Y, Liu X, Li J. et al. Genome-Scale Methylation Analysis of Circulating Cell-Free DNA in Gastric Cancer Patients. Clin Chem. 2022;68:354-64
29. Pellini B, Chaudhuri AA. Circulating Tumor DNA Minimal Residual Disease Detection of Non-Small-Cell Lung Cancer Treated With Curative Intent. J Clin Oncol. 2022;40:567-75
30. Meng W, He C, Hao Y, Wang L, Li L, Zhu G. Prospects and challenges of extracellular vesicle-based drug delivery system: considering cell source. Drug Deliv. 2020;27:585-98
31. Karimi N, Dalirfardouei R, Dias T, Lotvall J, Lasser C. Tetraspanins distinguish separate extracellular vesicle subpopulations in human serum and plasma - Contributions of platelet extracellular vesicles in plasma samples. J Extracell Vesicles. 2022;11:e12213
Author contact
![]() Corresponding authors: Honghai Hong, Email: gaolaosaocom; Haofan Yin, Email: yinhfsysu.edu.cn; Xi Jiang, Email: 1084942258com.
Corresponding authors: Honghai Hong, Email: gaolaosaocom; Haofan Yin, Email: yinhfsysu.edu.cn; Xi Jiang, Email: 1084942258com.

 Global reach, higher impact
Global reach, higher impact