Impact Factor
ISSN: 1837-9664
J Cancer 2023; 14(12):2274-2288. doi:10.7150/jca.85811 This issue Cite
Research Paper
The NT5DC family: expression profile and prognostic value in pancreatic adenocarcinoma
1. Department of Pathology, Xiangya Hospital, Central South University, Changsha 410008, Hunan, China.
2. Department of blood transfusion, Affiliated Hospital of North Sichuan Medical College, Nanchong 637000, Sichuan, China.
3. Departments of Ultrasound Imaging, Xiangya Hospital, Central South University, Changsha 410008, Hunan, China.
4. Department of Orthopedics, Xiangya Hospital, Central South University, Changsha 410008, Hunan, China.
5. National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, China.
† These authors have contributed equally to this work.
* These authors jointly supervised this work.
Received 2023-5-3; Accepted 2023-7-12; Published 2023-7-16
Abstract
Pancreatic adenocarcinoma (PAAD) is a malignant tumor with high morbidity and mortality rates. The NT5DC family is an evolutionarily-conserved family of 5'-nucleosidases that catalyze the intracellular hydrolysis of nucleotides. Although the NT5DC family has been linked to the initiation and growth of several cancers, its function in PAAD remains unclear. A series of bioinformatic analyses was used to ascertain the expression, prognosis, gene changes, functional enrichment, and immune regulatory functions of the NT5DC family in PAAD. NT5C2 and NT5DC1/2 mRNA and protein levels are increased in PAAD. Furthermore, the high mRNA expressions of NT5C2, NT5DC2, and NT5DC4 indicate a poor prognosis in patients with PAAD. The enrichment of biological processes and gene expression in the NT5DC family in PAAD were investigated using Kyoto Encyclopedia of Genes and Genomes and Gene Ontology analyses. Further investigations into immune infiltration revealed a close relationship between NT5DC gene expression and immune cell infiltration. These findings provide new insights into the biological function and prognostic value of the NT5DC gene family in PAAD.
Keywords: NT5DC gene family, pancreatic adenocarcinoma, immune infiltration, prognosis, biomarkers
Introduction
Pancreatic adenocarcinoma (PAAD) is one of the most prevalent cancers worldwide [1, 2]. Due to its poor prognosis, PAAD accounts for almost as many deaths as cases and is the fourth most common cause of cancer death in both sexes [3]. Both the incidence and mortality of pancreatic cancer have increased over the past decade compared with those of most other solid tumors [4]. The management of PAAD has evolved with the introduction of new surgical and therapeutic techniques, such as laparoscopic techniques and neoadjuvant chemoradiotherapy. However, only modest improvements in outcomes have occurred [5]. To move toward an era of precision medicine, the identification of new biomarkers and therapeutic targets for PAAD are necessary [6, 7].
The NT5DC gene family consists of NT5C2, NT5DC1, NT5DC2, NT5DC3, and NT5DC4. The NT5DC family is an evolutionarily-conserved family of 5′-nucleotidases that catalyze the hydrolysis of nucleotides in the cell [8]. Dysfunctions of members of the NT5DC family have been associated with disorders of the immune system, sensitivity to cancer therapy, and metabolic disorders [9]. NT5C2 is responsible for the final dephosphorylation of 6-hydroxypurine monophosphates, including IMP, dIMP, GMP, dGMP, and XMP, before their extracellular export and plays a key role in the maintenance of the balance of nucleotides, nucleosides, and free groups in the purine library of the brain and spinal cord [10]. Single-nucleotide polymorphisms in NT5DC1 have been associated with susceptibility to chronic obstructive pulmonary disease [11]. Guo et al. [12] reported that NT5DC2 promotes the tumorigenicity of glioma stem cell-like cells via the upregulation of Fyn. Li et al. reported that NT5DC3 can be used as a biomarker to discriminate patients with type 2 diabetes (T2D) and T2D-induced colon cancer from healthy volunteers by detecting the expression level of NT5DC3 in the blood [13]. In addition, NT5DC3 may be used as target of a novel cancer preventive treatment strategy, especially in patients with T2D who are prone to colorectal cancer (CRC) [14]. Jin et al. [15] reported that suppressing NT5DC2 inhibits the metastatic progression of non-small cell lung carcinoma (NSCLC) by regulating the p53 signaling pathway. The reduced RNA binding protein IGF2BP2 down-regulates NT5DC2, which suppresses cell proliferation and induces cell cycle arrest and apoptosis in diffuse large B-cell lymphoma cells via the regulation of the p53 pathway [16]. NT5C2 methylation regulates the interaction between DNMT1 and the insulin receptor in patients with T2D [17]. Kulkarni et al. found that NT5C2 and AMPK activity in patients with T2D and obesity may be important for controlling insulin action and lipid metabolism in skeletal muscles [18]. Previous studies have shown that NT5DC family members are associated with tumor progression [19, 20] and can serve as tumor markers [21, 22]. However, the expression, prognosis, and immune properties of the NT5DC family in PAAD have not been comprehensively analyzed. In this study, the expression of NT5DC family genes and their association with clinical characteristics of PAAD were analyzed using research databases and bioinformatics tools. The results of this study provide new insights into the prognostic value and biological role of the NT5DC family in PAAD.
Materials and methods
Gene Expression Profiling Interactive Analysis 2
Gene Expression Profiling Interactive Analysis 2 (GEPIA2) has been used to study various genes in different cancer types and identify potential biomarkers and therapeutic targets using differential expression analysis, survival analysis, and similar gene identification [23]. The GEPIA2 database was used to analyze the expression of members of the NT5DC family of genes in 171 samples of normal pancreas and 181 samples of PAAD.
University of Alabama at Birmingham Cancer Data Analysis Portal
The University of Alabama at Birmingham Cancer Data Analysis Portal (UALCAN) is an in silico validation platform for target genes that can help identify relevant genes for biomarker analysis, expression profile analysis, and survival analysis [24]. In this study, the "individual cancer stage" model was used to analyze the relationships between the mRNA and protein expressions of different NT5DC family members and clinicopathological parameters in PAAD. A student's t-test was used to compare the results, and p < 0.05 was considered to be statistically significant.
cBioPortal
The cBioPortal (http://cbioportal.org) is an online portal for the exploration, visualization, and analysis of cancer genomic data [25]. A map from The Cancer Genome Atlas (TCGA) and 186 cBioPortal pancreatic cancer datasets of NT5DC gene families, which included mutations and mRNA expression information, were used in this study. The threshold for the absolute value of the log2 fold-change was 1.5. The p-value cutoff was set at 0.05.
Cytoscape
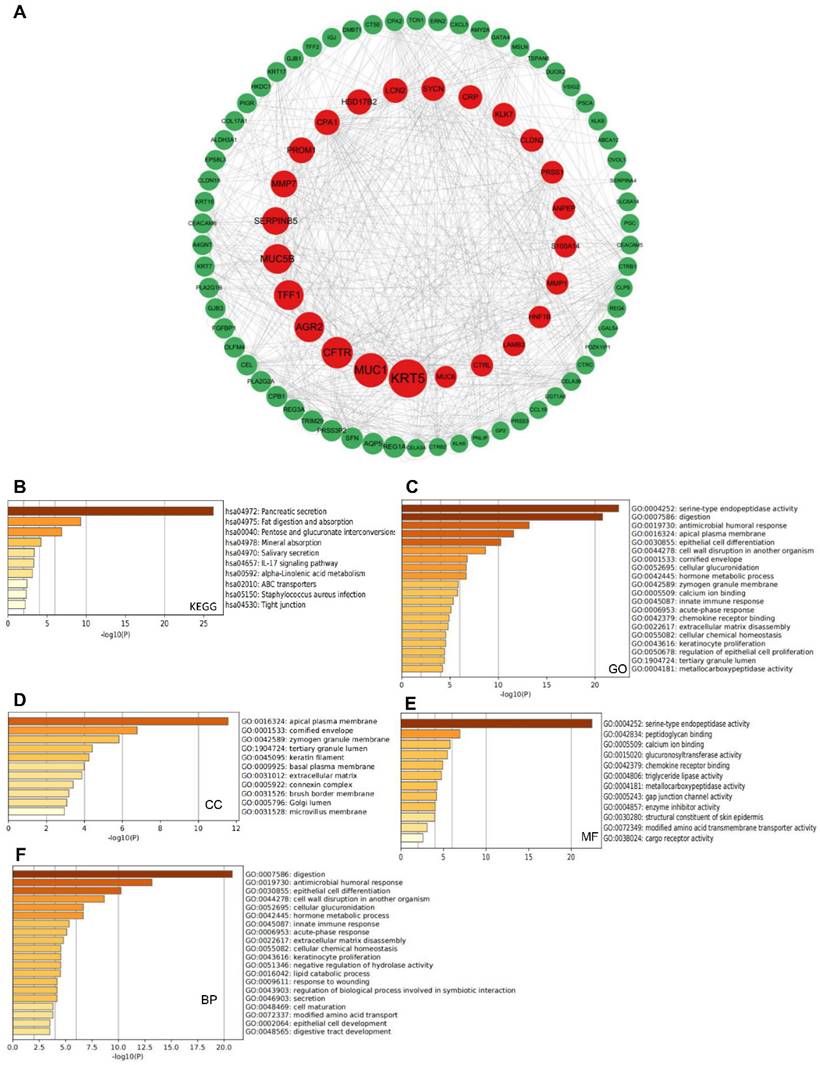
Cytoscape (http://cbioportal.org) allows for the integration of molecular interaction networks with expression data and other molecular states into one framework [26]. Cytoscape was used to selectively integrate 208 genes that were functionally co-expressed with members of the NT5DC family. The degrees to which proteins interact with each other are represented by the sizes of the nodes.
Metascape
Metascape combines functional enrichment, interactome analysis, gene annotation and membership search. It uses more than 40 independent knowledge bases within one integrated portal [27]. Metascape was used for Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) pathway enrichment analyses of the NT5DC family.
Tumor Immune Estimation Resource
The Tumor Immune Estimation Resource (TIMER2.0, http://timer.cistrome.org/) is a more robust estimation of immune infiltration levels for TCGA or user-provided tumor profiles that uses six state-of-the-art algorithms [28]. TIMER2.0 provides four modules with which to explore associations between immune infiltrates and genetic or clinical features and cancer-related associations in the TCGA cohorts [29]. The TIMER2.0 database was used to investigate the relationship between NT5DC family members and immune cell infiltration and the associations between the NT5DC family and B cells, CD8+ T cells, neutrophils, macrophages, dendritic NK-cells, Th1-cells, Treg-cells, and monocytic markers.
Kaplan-Meier Plotter Database
To integrate gene expression information and clinical prognostic data for the meta-analysis and to discover and validate survival-related molecular markers, the Kaplan-Meier Plotter database (https://kmplot.com) was used. Using an automatic selection of the best cutoff value, the 177 PAAD samples were divided into high- and low-expression groups, and the expressions of members of the NT5DC family were analyzed in terms of overall survival (OS), relapse-free survival (RFS), disease-specific survival, and progression-free survival from PAAD. Kaplan-Meier analysis and the log-rank test were used to analyze the results, and p < 0.05 was considered statistically significant.
Statistical analysis
Survival analysis statistics were obtained using the log-rank test, and associations of the NT5DC family with immune infiltration and immune cell type markers were evaluated using Spearman's correlation. The Student's t-test was used to compare two independent samples. Statistical significance was set at p < 0.05.
Results
Aberrant Expression of the NT5DC Family Members in PAAD
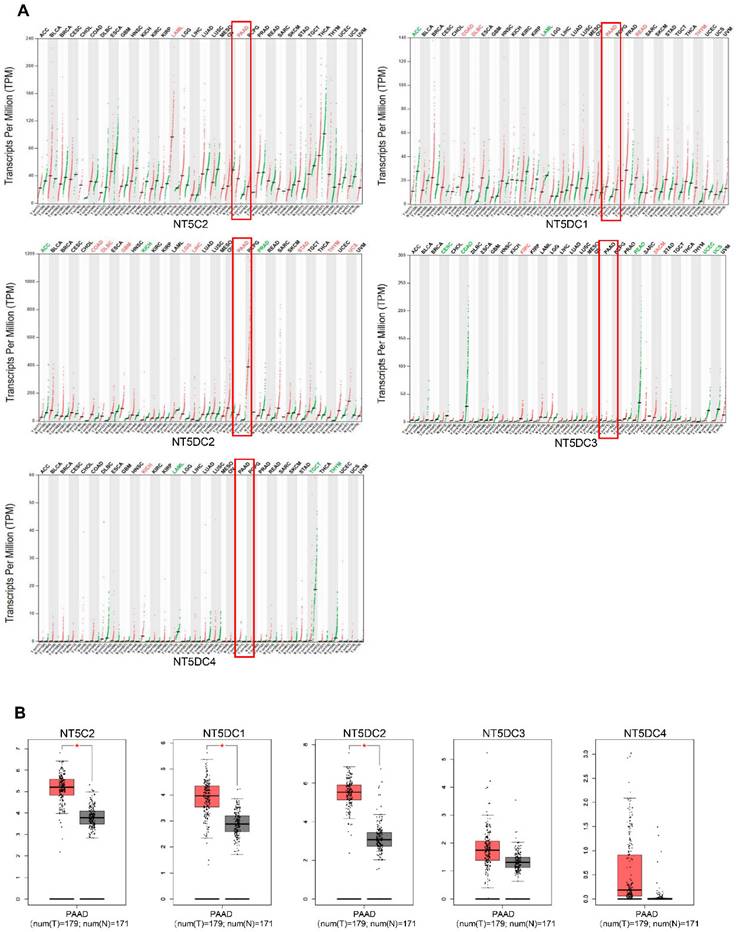
The mRNA levels of NT5DC family members were analyzed using the GEPIA database to compare the expression of NT5DC in different tumors with that in normal tissues (Figure 1A). The mRNA expression levels of NT5C2, NT5DC1, and NT5DC2 were upregulated in PAAD tissues compared with those in normal pancreatic tissues (Figure 1B).
Correlation of NT5DC Family Expressions with Clinicopathologic Features of PAAD
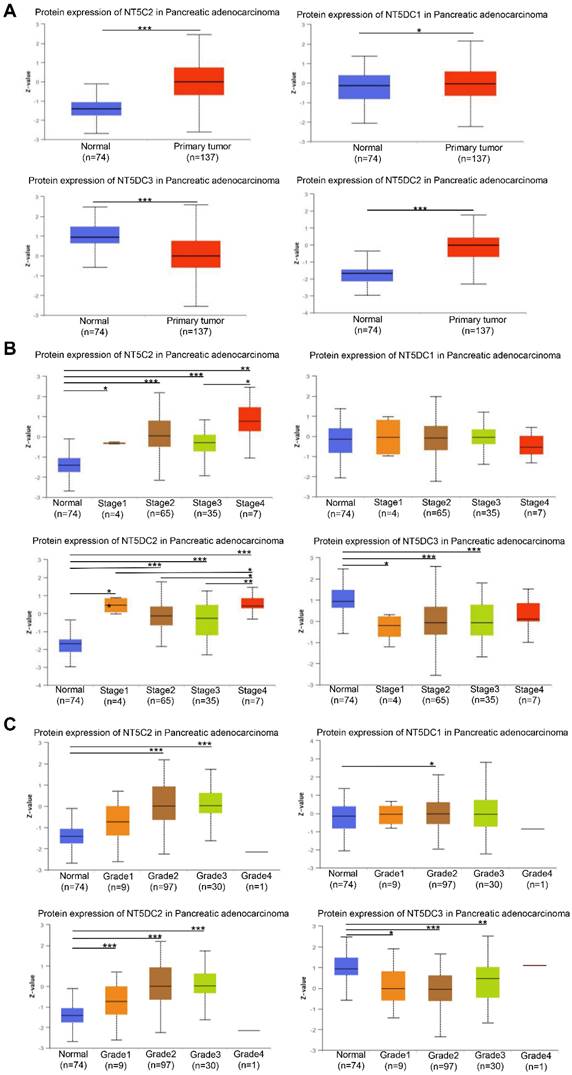
The expression levels of NT5DC family members were examined to determine whether they correlated with the stage of PAAD. The expressions of NT5C2, NT5DC1, and NT5DC2 proteins were higher in PAAD samples than in normal samples, though the expression of NT5DC3 protein was lower in PAAD samples (Figure 2A). The expression levels of NT5DC2/3 proteins were significantly correlated with the cancer stages of PAAD (Figure 2B). The expressions of NT5C2 and NT5DC1/2/3 proteins were closely correlated with the grades of cancer (Figure 2C).
The relationships between the expression levels of NT5DC family members and the clinicopathological features of PAAD were investigated. The clinicopathological features of 160 patients were downloaded from TCGA database. The mRNA expressions were ranked from lowest to highest and categorized into high and low expression groups based on the median value. NT5DC1 expression correlated with patient sex, NT5DC3 expression was significantly associated with cancer grade, and NT5DC4 expression was associated with tumor diameter (Table 1).
Genetic changes and functional enrichment analysis of NT5DC family members
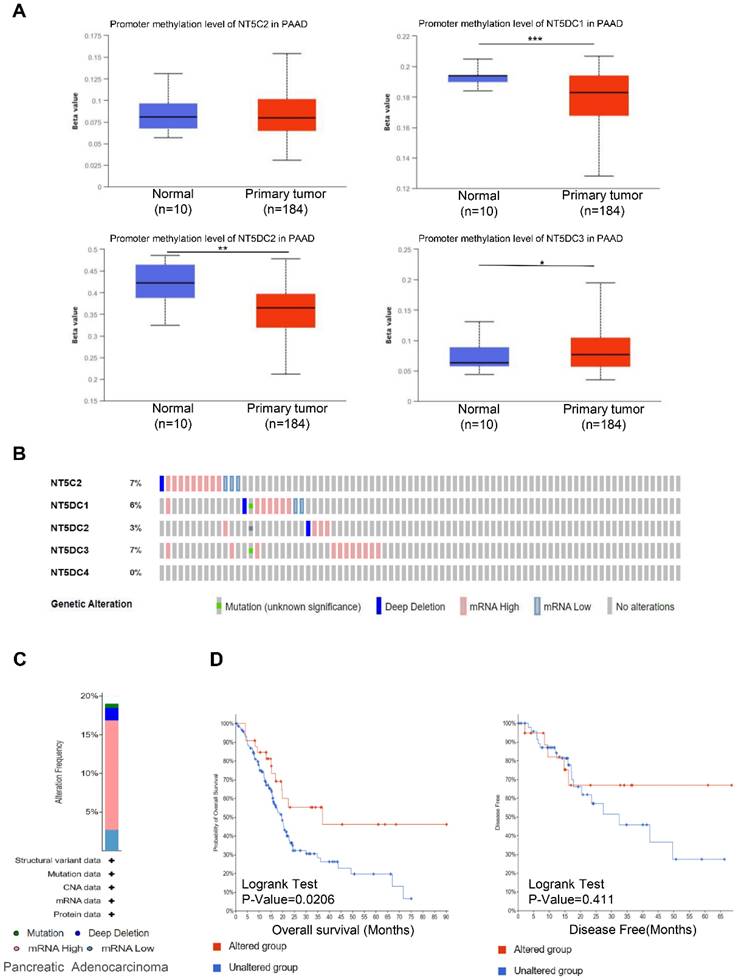
Using the UALCAN database, the methylation levels of NT5DC in patients with PAAD were identified. DNA methylation levels of NT5DC1/2 were lower in PAAD samples than in healthy individuals, while NT5DC3 had significantly higher levels in PAAD tissue (Figure 3A). The DNA methylation of NT5C2 was not significantly different between normal and PAAD tissues. Genetic variations in the NT5DC family were also investigated. The proportion of genetic variation in NT5DC family members increased from 0% to 7% in patients with PAAD. Mutations in NT5C2, NT5DC1, NT5DC2, NT5DC3, and NT5DC4 were found in 7%, 6%, 3%, 7%, and 0% of the samples, respectively (Figure 3B). The most common abnormalities of the NT5DC family in patients with PAAD were mRNA alterations (Figure 3C). The associations between NT5DC family member mutations and patient prognosis were also evaluated. Mutations in the NT5DC family members were associated with better OS (Figure 3D). However, NT5DC family gene mutations were not significantly correlated with disease-free survival (DFS). These results indicate that mutations in NT5DC family members may serve as potential therapeutic targets, which may significantly improve the prognosis of patients with PAAD.
A total of 208 co-expressed genes were identified from the cBioportal database and a map of co-expression networks of key genes associated with the NT5DC family members was created using Cytoscape_v.3.7.2 (Figure 4A). GO annotation and KEGG pathway analyses were used to analyze the biological functions of NT5DC members and their co-expressed genes using the Metascape database. Pancreatic secretion, fat digestion and absorption, pentose and glucuronate interconversion, and the IL-17 signaling pathway were explored for co-expressed genes (Figure 4B). The co-expressed genes were mainly correlated with serine-type endopeptidase activity, digestion, antimicrobial humoral responses, and innate immune responses (Figure 4C). Bioprocess analysis indicated that these genes were mainly involved in metabolism, humoral antimicrobial response, epithelial cell differentiation, and innate immune response (Figure 4D). Molecular function analysis revealed that the genes were mainly involved in serine-type endopeptidase activity, peptidoglycan binding, and chemokine receptor binding (Figure 4E). Analysis of the cellular components revealed that these genes were frequently associated with the apical plasma membrane and cornified envelope (Figure 4F).
NT5DC family member expressions in PAAD. (A) The pan-cancer expression of the mRNAs of NT5DCs is shown. (B) The expressions of the NT5DC family mRNA in PAAD are shown. *p < 0.01 compared with control. PAAD, pancreatic adenocarcinoma.
Relationships between stage and grade of PAAD and NT5DC family member expressions. (A) The protein expression levels of NT5DC family members in patients with PAAD are shown. (B) The correlations between NT5CD family member expressions and stages of PAAD are shown. (C) The correlations between NT5CD family member expressions and grades of PAAD are shown. *p < 0.05, **p < 0.01, ***p < 0.001 compared with control. UALCAN, University of Alabama at Birmingham Cancer Data Analysis Portal; PAAD, pancreatic adenocarcinoma.
Clinicopathologic parameters and the expressions of NT5DC family members in PAAD.
| Characteristics | N | NT5C2 | NT5DC1 | NT5DC2 | NT5DC3 | NT5DC4 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | High | p | Low | High | p | Low | High | p | Low | High | p | Low | High | p | ||
| Gender | 0.405 | 0.761 | 0.089 | 0.127 | 0.114 | |||||||||||
| Male | 89 | 51 | 38 | 48 | 41 | 57 | 32 | 55 | 34 | 56 | 33 | |||||
| Female | 71 | 36 | 35 | 40 | 31 | 36 | 35 | 52 | 19 | 53 | 18 | |||||
| Age | 0.177 | 0.032 | 0.522 | 0.065 | 0.932 | |||||||||||
| ≤55 | 32 | 14 | 18 | 23 | 9 | 17 | 15 | 17 | 15 | 22 | 10 | |||||
| >55 | 128 | 73 | 55 | 65 | 63 | 76 | 52 | 90 | 38 | 87 | 41 | |||||
| T Stage | 0.246 | 0.558 | 0.76 | 0.446 | 0.39 | |||||||||||
| T1 + T2 | 28 | 18 | 10 | 14 | 14 | 17 | 11 | 17 | 11 | 21 | 7 | |||||
| T3 + T4 | 132 | 69 | 63 | 74 | 58 | 76 | 56 | 90 | 42 | 88 | 44 | |||||
| N Stage | 0.641 | 0.519 | 0.911 | 0.102 | 0.452 | |||||||||||
| NX + N0 | 47 | 22 | 25 | 24 | 23 | 27 | 20 | 27 | 20 | 30 | 17 | |||||
| N1 | 113 | 65 | 48 | 64 | 49 | 66 | 47 | 80 | 33 | 79 | 34 | |||||
| M Stage | 0.215 | 0.215 | 0.164 | 0.956 | 0.963 | |||||||||||
| Mx | 82 | 47 | 35 | 49 | 33 | 52 | 30 | 55 | 27 | 56 | 26 | |||||
| M0 + M1 | 78 | 40 | 38 | 39 | 39 | 41 | 37 | 52 | 26 | 53 | 25 | |||||
| Stage | 0.743 | 0.825 | 0.636 | 0.16 | 0.58 | |||||||||||
| StageI | 19 | 11 | 8 | 10 | 9 | 12 | 7 | 10 | 9 | 14 | 5 | |||||
| StageII+III+IV | 141 | 76 | 65 | 78 | 63 | 81 | 60 | 97 | 44 | 95 | 46 | |||||
| Grade | 0.757 | 0.61 | 0.336 | 0.035 | 0.543 | |||||||||||
| Gx + G1 | 26 | 15 | 11 | 12 | 14 | 18 | 8 | 19 | 7 | 20 | 6 | |||||
| G2 | 86 | 48 | 38 | 49 | 37 | 46 | 40 | 50 | 36 | 58 | 28 | |||||
| G3 + G4 | 48 | 24 | 24 | 27 | 21 | 29 | 19 | 38 | 10 | 31 | 17 | |||||
| Diameter(cm) | 0.951 | 0.774 | 0.784 | 0.383 | 0.028 | |||||||||||
| ≤4 | 107 | 58 | 49 | 58 | 49 | 63 | 44 | 74 | 33 | 79 | 28 | |||||
| >4 | 53 | 29 | 24 | 30 | 23 | 30 | 23 | 33 | 20 | 30 | 23 | |||||
Bold font indicates significant difference.
Association Between the Expressions of NT5DC Family Members and Immune Infiltration in patients with PAAD
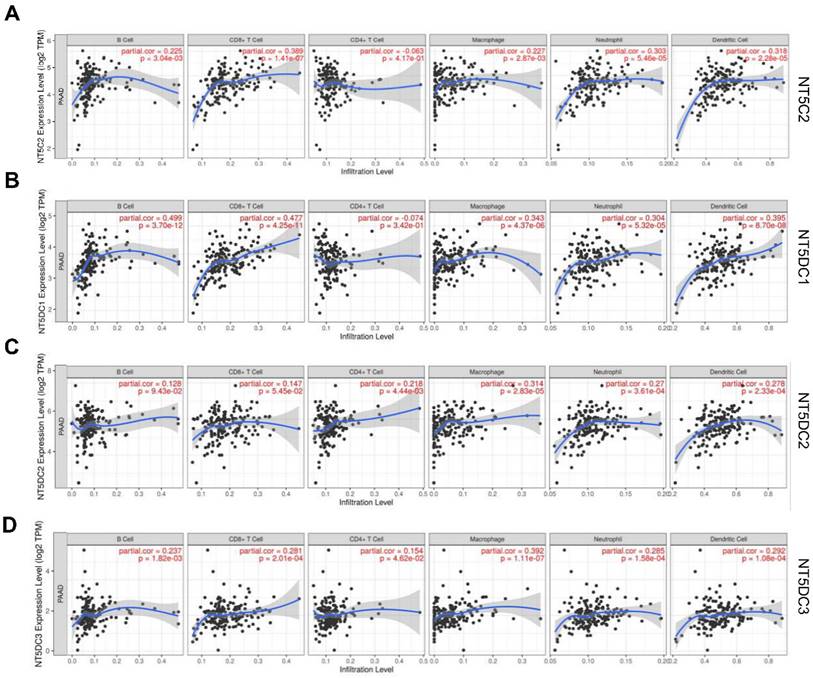
TIMER2.0 was used to investigate the correlation between the NT5DC gene family members and immune cell infiltration (Figure 5). The expression of NT5C2 mRNA was significantly associated with the infiltration of B cells (correlation coefficient (cor) = 0.225, p < 0.05), CD8+T cells (cor = 0.389, p < 0.05), and macrophages (cor = 0.227, p < 0.05). NT5DC1 expression was significantly associated with B cell (cor = 0.499, p < 0.05), CD8+T cell (cor = 0.477, p < 0.05), and macrophage (cor = 0.343, p < 0.05) infiltration. NT5DC2 expression was significantly associated with CD4+ T cell (cor = 0.218, p < 0.05) and macrophage (cor = 0.314, p < 0.05) infiltration. NT5DC3 expression was significantly associated with the infiltration of B cells (cor = 0.237, p < 0.05), CD8+ T cells (cor = 0.281, p < 0.05), CD4+ T cells (cor = 0.154, p < 0.05), and macrophages (cor = 0.392, p < 0.05).
To further explore the relationship between NT5DC family member expression and different immune cells, the marker types of dendritic cells, CD8+ T cells, neutrophils, and tumor-associated macrophages were analyzed in PAAD using the TIMER database. NT5C2 levels were strongly associated with Th2 and Tregs. NT5DC1 was strongly associated with M2 macrophages, neutrophils, Th17, Tregs, and monocytes and moderately associated with B cells and T cells. There was a moderate or weak correlation between NT5DC2 and B cells, M1 macrophages, M2 macrophages, and Treg markers. The NT5DC3 level was strongly associated with M2 macrophages and the exhaustion of T cells and monocytes (Table 2). The NT5DC gene family was implicated in the regulation of macrophage polarization in patients with PAAD, as a clear relationship was found between some components of the NT5DC gene family and M2 macrophage markers.
Prognostic Values of NT5DC Gene Family Members in Patients with PAAD
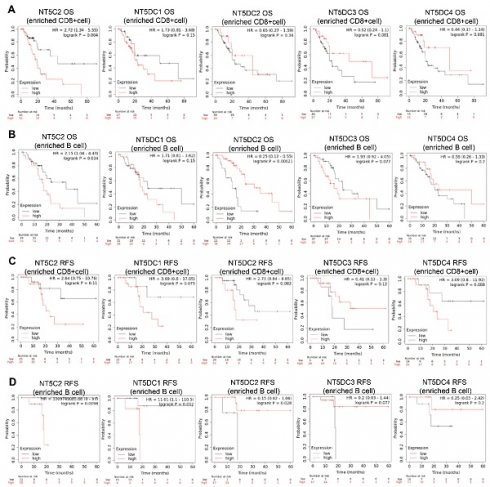
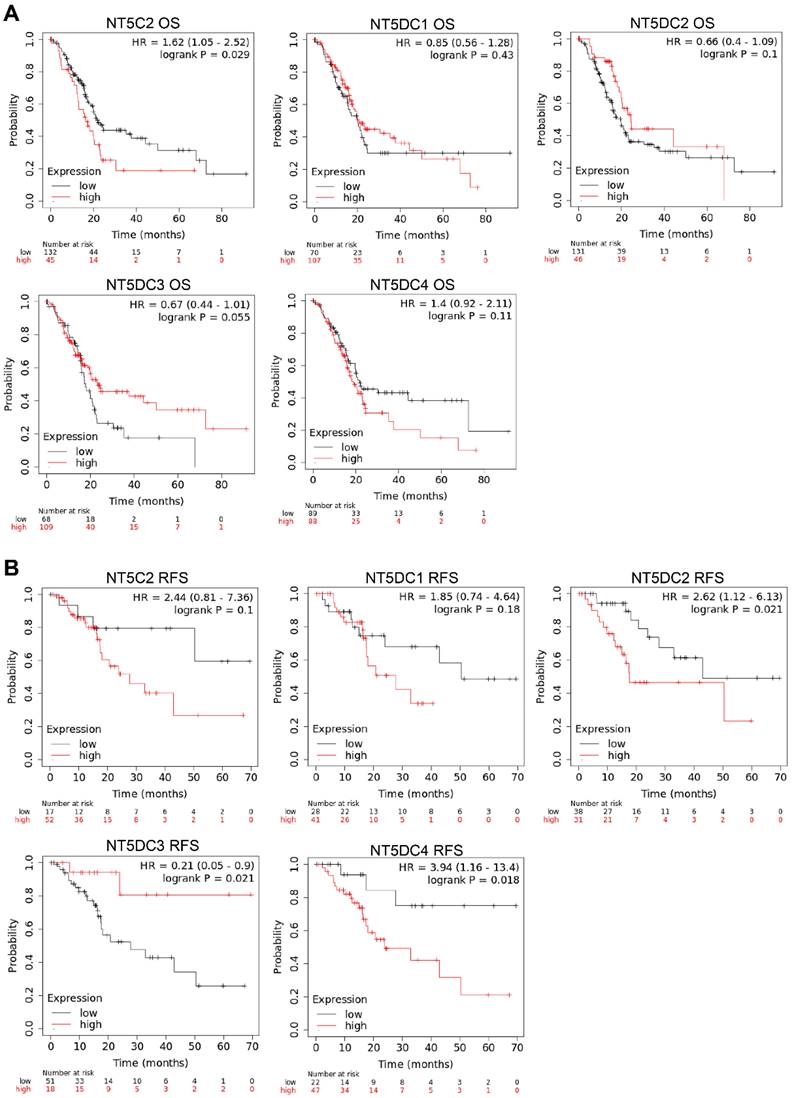
The Kaplan-Meier plotter database was used to investigate the prognostic values of NT5DC family member mRNA expression in patients with PAAD. Higher NT5C2 mRNA expression (OS hazard ratio (HR) = 1.62 (1.05-2.52), p = 0.024) was associated with poor OS in patients with PAAD (Figure 6A). Increased expressions of NT5DC2 (RFS HR = 2.62 (1.12-6.13), p = 0.033) and NT5DC4 (RFS: HR = 3.94 (1.16-13.4), p = 0.018) were significantly associated with poorer RFS in patients with PAAD. However, increased expression of NT5DC3 (RFS: HR = 0.21 (0.05-0.9), p = 0.021) was associated with better RFS (Figure 6B).
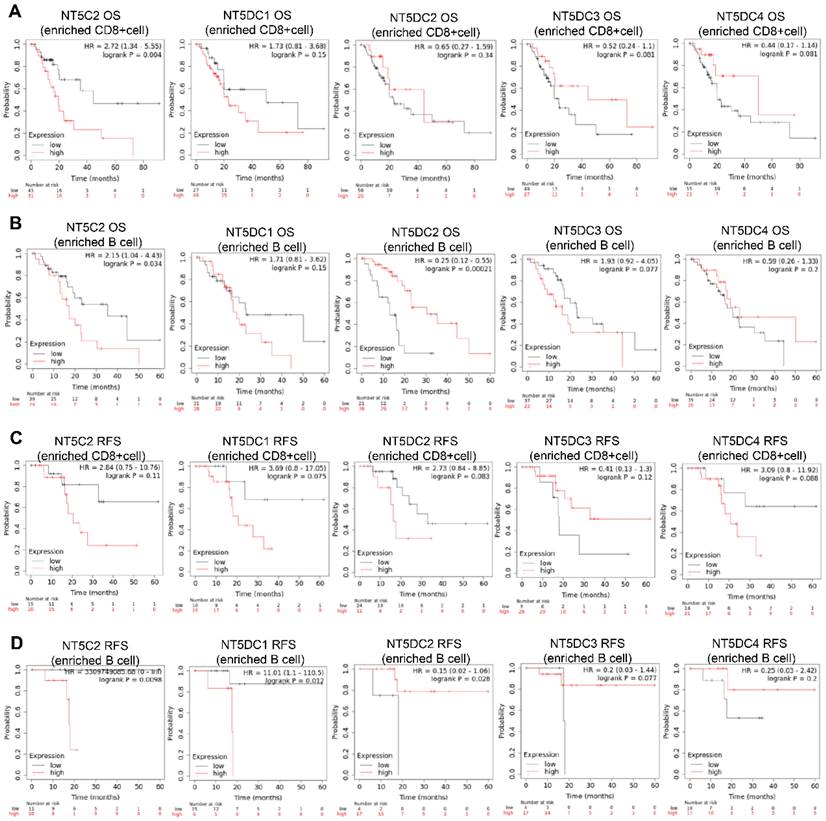
The prognostic values of the NT5DC family members in relation to immune cell enrichment in patients with PAAD were also investigated. NT5C2 upregulation was associated with poor OS when patients with PAAD were CD8+ T cell-enriched (Figure 7A). When patients with PAAD were enriched with B cells, NT5DC1 expression was significantly and negatively correlated with OS and RFS. In contrast, upregulated NT5DC2 expression was associated with favorable OS and RFS in patients with PAAD, and upregulated NT5C2 expression was associated with poor RFS in patients with PAAD with B cell enrichment (Figure 7B-D).
Genetic alterations and DNA methylation levels of the NT5DC family members in PAAD tissue. (A) DNA methylation changes in the NT5DC family members in PAAD tissue are shown, as assessed using the UALCAN database. (B-C) Summaries of the rate of alteration of the NT5DC family members in PAAD tissue are shown. These were determined using cBioPortal. (D) The associations between NT5DC family member mutations and the prognosis of patients with PAAD are shown. *p<0.05, **p<0.01, ***p<0.001 compared with control. PAAD, pancreatic adenocarcinoma; OS, overall survival; DFS, Disease-free survival; UALCAN, University of Alabama at Birmingham Cancer Data Analysis Portal.
Predicted functions and signaling pathways of NT5DCs and NT5DC-associated molecules co-expressed in patients with PAAD. (A) A total of 208 NT5DC-associated co-expressed molecules that are frequently altered in patients with PAAD are identified. The cBioPortal database was used to identify these molecules. The protein-protein interaction network of the NT5DC family members and their associated co-expressed genes, which was constructed using the Cytoscape database, is shown. (B-F) A functional enrichment analysis used to analyze the biological functions of NT5DC members and their co-expressed genes is shown. PAAD, pancreatic adenocarcinoma.
Association of the expression levels of NT5DC mRNA with the infiltration of immune cells. (A-D) The associations of NT5C2 and NT5DC 1-3 with immune cell infiltration, determined using the TIMER2.0 database, are shown. TIMER, Tumor Immune Estimation Resource.
Discussion
PAAD is a major cause of tumor-related death, with high mortality and metastasis rates. Despite improvements in the detection and treatment of PAAD, the prognosis remains poor, with a five-year survival rate < 10% [1, 30]. There is an urgent need to identify novel and effective diagnostic and prognostic biomarkers to significantly improve the prognosis of patients with PAAD [31, 32]. Intracellular nucleotide hydrolysis is performed by the evolutionarily-conserved 5'-nucleotide enzyme family NT5DC [8]. Several studies have reported that members of the NT5DC family are aberrantly expressed in various malignancies and are essential for cancer proliferation and development [13, 33, 34]. Unfortunately, the functions of the NT5DC family members in PAAD have not been thoroughly investigated. The biological effects of each NT5DC family member in relation to PAAD were elucidated from five different perspectives in this study: mRNA and protein expression levels, clinical features and disease prognosis, gene mutation, pathway analysis, and immune infiltration. The mRNA and protein levels of NT5C2, NT5DC1, and NT5DC2 were overexpressed in PAAD cells when compared to the expression in non-tumoral cells, whereas the protein levels of NT5DC3 were downregulated in PAAD cells. NT5DC family members can be used for clinicopathological cancer diagnosis, as reported in recent studies. Arik et al. [21] reported that the high expression of NT5C2 could be used as a prognostic marker in patients with lung cancer with poor prognoses. Guo et al. [12] discovered that NT5DC2 promotes the carcinogenesis of glioma stem cell-like cells by upregulating Fyn, and can be used as a potential therapeutic target for glioblastoma.
Prognostic values of mRNA expression levels of NT5DC family members in patients with PAAD. (A-B) Kaplan-Meier plots showing the OS and RFS of the NT5DC family members in patients with PAAD are shown. PAAD, pancreatic adenocarcinoma; OS, overall survival; RFS, relapse-free survival.
Correlations between the expressions of NT5DC family members and the markers of immune cells.
| NT5C2 | NT5DC1 | NT5DC2 | NT5DC3 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Cor | p | Cor | p | Cor | p | Cor | p | ||
| CD8 + Tcell | CD8A | 0.05 | 0.518 | 0.251 | 0.012 | 0.047 | 0.545 | 0.217 | 0.004 |
| CD8B | 0.058 | 0.450 | 0.185 | 0.011 | 0.131 | 0.087 | 0.08 | 0.297 | |
| GZMA | 0.04 | 0.602 | 0.179 | 0.019 | 0.117 | 0.128 | 0.176 | 0.022 | |
| B cell | CD19 | 0.007 | 0.925 | 0.155 | 0.043 | 0.256 | 0.001 | 0.108 | 0.160 |
| CD79A | -0.007 | 0.932 | 0.18 | 0.018 | 0.239 | 0.002 | 0.082 | 0.288 | |
| MS4A1 | 0.013 | 0.871 | 0.181 | 0.018 | 0.174 | 0.023 | 0.084 | 0.277 | |
| T cell | CD3D | 0.038 | 0.623 | 0.197 | 0.010 | 0.14 | 0.069 | 0.19 | 0.013 |
| CD3E | 0.064 | 0.409 | 0.236 | 0.002 | 0.12 | 0.119 | 0.208 | 0.006 | |
| CD2 | 0.064 | 0.407 | 0.259 | 0.001 | 0.104 | 0.176 | 0.175 | 0.022 | |
| TAM | CCL2 | -0.004 | 0.963 | 0.026 | 0.735 | 0.13 | 0.089 | 0.134 | 0.081 |
| CD68 | 0.313 | 0.000 | 0.361 | 0.000 | 0.174 | 0.023 | 0.209 | 0.006 | |
| IL10 | 0.097 | 0.209 | 0.125 | 0.103 | 0.21 | 0.006 | 0.254 | 0.001 | |
| M1 | IRF5 | 0.298 | 0.000 | 0.201 | 0.008 | 0.197 | 0.010 | 0.321 | 0.000 |
| PTGS2 | 0.297 | 0.000 | 0.142 | 0.064 | 0.315 | 0.000 | 0.19 | 0.013 | |
| NOS2 | 0.149 | 0.051 | 0.14 | 0.068 | 0.226 | 0.003 | 0.131 | 0.087 | |
| M2 | MS4A4A | 0.086 | 0.262 | 0.284 | 0.000 | 0.191 | 0.012 | 0.312 | 0.000 |
| CD163 | 0.117 | 0.128 | 0.316 | 0.000 | 0.193 | 0.011 | 0.365 | 0.000 | |
| VSIG4 | 0.104 | 0.176 | 0.255 | 0.001 | 0.21 | 0.006 | 0.308 | 0.000 | |
| Neutrophils | ITGAM | 0.184 | 0.016 | 0.265 | 0.000 | 0.227 | 0.003 | 0.225 | 0.003 |
| CCR7 | 0.068 | 0.376 | 0.244 | 0.002 | 0.141 | 0.066 | 0.18 | 0.019 | |
| SIGLEC5 | 0.143 | 0.061 | 0.288 | 0.000 | 0.236 | 0.002 | 0.228 | 0.003 | |
| DC | HLA-DQB1 | 0.07 | 0.364 | 0.241 | 0.002 | 0.07 | 0.360 | 0.229 | 0.003 |
| HLA-DPB1 | 0.035 | 0.646 | 0.219 | 0.004 | 0.205 | 0.007 | 0.197 | 0.010 | |
| HLA-DRA | 0.152 | 0.047 | 0.318 | 0.000 | 0.188 | 0.014 | 0.22 | 0.004 | |
| HLA-DPA1 | 0.117 | 0.128 | 0.327 | 0.000 | 0.184 | 0.016 | 0.236 | 0.002 | |
| ITGAX | 0.068 | 0.375 | 0.134 | 0.081 | 0.263 | 0.001 | 0.247 | 0.001 | |
| CD1C | 0.142 | 0.065 | 0.285 | 0.000 | 0.145 | 0.059 | 0.13 | 0.091 | |
| NRP1 | 0.294 | 0.000 | 0.3 | 0.000 | 0.249 | 0.001 | 0.29 | 0.000 | |
| NK cell | KIR2DL1 | 0.035 | 0.651 | 0.119 | 0.120 | -0.038 | 0.620 | 0.099 | 0.199 |
| KIR2DL3 | 0.044 | 0.565 | 0.018 | 0.816 | 0.064 | 0.407 | 0.088 | 0.252 | |
| KIR2DL4 | 0.163 | 0.033 | 0.103 | 0.178 | -0.054 | 0.480 | 0.148 | 0.054 | |
| KIR3DL1 | -0.048 | 0.535 | 0.043 | 0.575 | -0.018 | 0.814 | 0.117 | 0.129 | |
| KIR3DL2 | 0.0324 | 0.674 | 0.164 | 0.032 | 0.098 | 0.204 | 0.137 | 0.073 | |
| KIR3DL3 | 0.123 | 0.108 | 0.066 | 0.393 | 0.067 | 0.381 | -0.02 | 0.792 | |
| KIR2DS4 | -0.068 | 0.378 | 0.132 | 0.086 | -0.076 | 0.325 | 0.055 | 0.478 | |
| Th1 | TBX21 | -0.002 | 0.974 | 0.174 | 0.023 | 0.002 | 0.974 | 0.212 | 0.005 |
| STAT1 | 0.433 | 0.000 | 0.363 | 0.000 | 0.06 | 0.436 | 0.208 | 0.006 | |
| STAT4 | -0.055 | 0.474 | 0.165 | 0.031 | 0.074 | 0.336 | 0.271 | 0.000 | |
| IFNG | 0.043 | 0.574 | 0.099 | 0.199 | -0.025 | 0.743 | 0.149 | 0.052 | |
| Th2 | STAT6 | 0.561 | 0.000 | 0.39 | 0.000 | 0.023 | 0.767 | 0.246 | 0.001 |
| GATA3 | 0.215 | 0.004 | 0.107 | 0.163 | 0.263 | 0.001 | 0.079 | 0.302 | |
| STAT5A | 0.266 | 0.000 | 0.233 | 0.002 | 0.219 | 0.004 | 0.337 | 0.000 | |
| Tfh | BCL6 | 0.392 | 0.000 | 0.282 | 0.000 | 0.185 | 0.015 | 0.356 | 0.000 |
| IL21 | 0.035 | 0.654 | 0.077 | 0.316 | 0.051 | 0.504 | 0.079 | 0.305 | |
| Th17 | STAT3 | 0.396 | 0.000 | 0.43 | 0.000 | 0.189 | 0.014 | 0.449 | 0.000 |
| IL17A | 0.031 | 0.688 | 0.213 | 0.005 | -0.123 | 0.109 | 0.071 | 0.353 | |
| Treg | FOXP3 | 0.164 | 0.032 | 0.238 | 0.002 | 0.257 | 0.001 | 0.194 | 0.011 |
| STAT5B | 0.317 | 0.000 | 0.403 | 0.000 | 0.212 | 0.014 | 0.462 | 0.000 | |
| CCR8 | 0.284 | 0.000 | 0.363 | 0.000 | 0.187 | 0.015 | 0.231 | 0.002 | |
| T exhaustion-cell | PDCD1 | -0.053 | 0.492 | 0.119 | 0.122 | 0.141 | 0.065 | 0.217 | 0.004 |
| CTLA4 | 0.082 | 0.286 | 0.197 | 0.010 | 0.188 | 0.014 | 0.285 | 0.000 | |
| HAVCR2 | 0.13 | 0.089 | 0.247 | 0.001 | 0.218 | 0.004 | 0.272 | 0.000 | |
| LAG3 | -0.061 | 0.429 | 0.109 | 0.157 | 0.018 | 0.819 | 0.31 | 0.000 | |
| Monocyte | CD86 | 0.14 | 0.068 | 0.256 | 0.001 | 0.206 | 0.007 | 0.257 | 0.001 |
| C3AR1 | 0.113 | 0.143 | 0.281 | 0.000 | 0.186 | 0.015 | 0.27 | 0.000 | |
| CSF1R | 0.087 | 0.257 | 0.285 | 0.000 | 0.169 | 0.028 | 0.302 | 0.000 | |
Bold font indicates significant difference.
Therefore, the prognostic significance of aberrantly expressed members of the NT5DC family in patients with PAAD and the clinicopathological associations of these genes were further investigated in this study. The protein expression of NT5DC family members was significantly correlated with the stage and grade of PAAD. In patients with PAAD, overexpression of NT5C2 mRNA was associated with poor OS; overexpression of NT5DC2/4 mRNA was significantly associated with poor RFS; and overexpression of NT5DC3 mRNA was correlated with better RFS. These results suggest that NT5C2 and NT5DC2 may have important prognostic values and great potential as diagnostic markers in patients with PAAD.
Prognostic values of the NT5DC family members in relation to immune cell enrichment in patients with PAAD. (A-D) The prognostic values of the NT5DC family members in relation to CD8+ T cell and B cell enrichment in patients with PAAD are shown. PAAD, pancreatic adenocarcinoma; OS, overall survival; RFS, relapse-free survival.
Genetic mutations that alter the biochemistry of tissue cells can lead to the development of cancer. Previous studies have reported that variants of the NT5DC family members are involved in tumor development. NT5C2 is associated with chemoresistance and a poor prognosis in patients with relapsed acute lymphoblastic leukemia [35]. NT5DC1 polymorphisms are associated with susceptibility to chronic obstructive pulmonary disease [11]. The NT5DC gene alteration group (which mainly includes mutations and mRNA changes) is associated with a better OS than the unaltered group. These results are in accordance with the results of the current study regarding NT5DC family gene alterations in PAAD. NT5DC1/2 DNA methylation levels in tumor tissue were lower than those in normal tissue, while the DNA methylation levels of NT5DC3 were significantly increased in tumor tissue, suggesting that DNA methylation-targeting drugs may have applications in cancer treatment [36] as they implicate epigenetic regulation in the biological process of PAAD induced by the NT5DC family. The co-expressed genes of the NT5DC family, including UGT2B7, MT1G, ACADL, MT1H, and others, were found to be associated with pancreatic cancer in previous studies [37-40]. Enrichment analyses of co-expressed genes revealed strong correlations between members of the NT5DC family and the antimicrobial humoral response, innate immune response, IL-17 signaling pathway, serine-type endopeptidase activity, and other immune-related signaling pathways. Therefore, the NT5DC family members interact with key molecules in tumor immune-related pathways and should be considered attractive therapeutic targets for the treatment of PAAD.
Pancreatic cancer is a tumor of the digestive tract with a very poor prognosis. The clinical characteristics of pancreatic cancer, including the difficulty of early detection, low surgical resection rate, and high recurrence and metastasis rates after surgery, make it particularly difficult to diagnose [1]. Surgical resection is currently the primary treatment for pancreatic cancer; however, patient survival rates are low [41, 42]. Pancreatic cancer mortality is largely due to an immunosuppressive environment, inadequate T cell infiltration, and a low mutation burden. However, immunotherapy appears to have a synergistic effect with other treatment modalities and increases response rates. Therefore, the development of multimodal treatments that target the mechanism of resistance to immunotherapy is necessary [43, 44]. Li et al. reported that NT5DC2 upregulated Epidermal growth factor receptor (EGFR) expression by downregulating EGFR ubiquitination and preventing ubiquitin-proteasome degradation but not transcription. Upregulating EGFR activated downstream signaling, playing a critical role in the protumourigenic effects of NT5DC2. Furthermore, NT5DC2 expression was associated with larger tumor size and microvascular invasion, and was independently associated with RFS. These findings suggest that NT5DC2 may be a potential molecular target for the treatment of hepatocellular carcinoma [34]. Hu et al. [19] reported that TEAD4 binds to the NT5DC2 promoter to activate its transcription. The overexpression of TEAD4 was associated with poor survival, suggesting that NT5DC2 plays an important role in the development of leiomyosarcomas and may serve as a potential therapeutic target. In addition, NT5C2 is involved in metabolizing cladribine, an immunomodulatory drug used to treat multiple sclerosis [45]. Low-intensity maintenance therapy with 6-mercaptopurine (6-MP) limits the incidence of acute lymphoblastic leukemia (ALL) relapse, though activating mutations in the NT5C2 gene confer resistance to 6-MP in patients with early relapse of ALL and directly contribute to ALL relapse [46]. NT5C2 has been identified as an important therapeutic target in hematological cancers. Zsuzsanna et al. used a combined approach based on fragment-based drug design and in silico methods to design potential inhibitors of NT5C2 that act synergistically with known antitumor agents [47]. The results of these studies indicate that the NT5DC family has great potential in immunotherapies. This study aimed to identify potential targets for PAAD treatment by investigating the associations between NT5DC family members and immune cell infiltration. The results of this study indicate that immune cells are correlated with the NT5DC family to different degrees. NT5C2 and NT5DC1/2/3 were associated with neutrophils, dendritic cells, and macrophages, suggesting that the NT5DC family may be a good indicator of the immunological health of patients with PAAD. Furthermore, the infiltration of antitumor immune cells, such as CD8+ T and B cells, improves the prognosis of patients with PAAD. The results of the current study demonstrate that NT5C2 and NT5DC2 may be useful immunotherapeutic targets for the treatment of PAAD. However, the current study is limited by the fact that the information used to analyze the data was collected from internet databases. To verify the function and potential molecular mechanisms of NT5C2 and NT5DC2 in PAAD, in vitro and in vivo experiments and clinical studies are necessary.
Conclusion
Bioinformatics tools were used to comprehensively evaluate the expression and prognosis of NT5DC family members in patients with PAAD to understand the biological and immunological effects of the NT5DC family in PAAD. The findings suggest that NT5C2 and NT5DC2 may be targets and valuable biomarkers to establish personalized treatments for patients with PAAD, which may help with the development of improved diagnostic and therapeutic modalities to improve patient prognoses. Further research is needed to identify new pharmacological therapies and to evaluate the mechanisms of their effects on carcinogenesis and development.
Supplementary Material
Supplementary table.
Acknowledgements
Funding
This study was supported by the National Natural Science Foundation of China (81903032), the Outstanding Youth Foundation of Hunan Provincial Natural Science Foundation of China (2022JJ20098), the Natural Science Foundation of Hunan Province (2021JJ41013 and 2022JJ40784), the Changsha Municipal Natural Science Foundation (kq2202374), the Scientific Research Program of North Sichuan Medical College (CBY22-QNA32), the Central South University Innovation-Driven Research Programme (2023CXQD075) and the Student Innovation Project of Central South University (2022ZZTS0938).
Author contributions
Xiaoqian Yu and Ru Sun conducted experimental operations, sample processing, data analysis, and performed the experiments. All authors participated in writing the paper. Chunlin Ou and Hongbin Guo conceived and designed the experiments. All authors read and approved the final manuscript.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Competing Interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Vincent A, Herman J, Schulick R. et al. Pancreatic cancer. Lancet. 2011;378(9791):607-620
2. Nie H, Wu Y, Ou C. et al. Comprehensive Analysis of SMC Gene Family Prognostic Value and Immune Infiltration in Patients With Pancreatic Adenocarcinoma. Front Med (Lausanne). 2022;9:832312
3. Sung H, Ferlay J, Siegel RL. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209-249
4. Grossberg AJ, Chu LC, Deig CR. et al. Multidisciplinary standards of care and recent progress in pancreatic ductal adenocarcinoma. CA Cancer J Clin. 2020;70(5):375-403
5. McGuigan A, Kelly P, Turkington RC. et al. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J Gastroenterol. 2018;24(43):4846-4861
6. Nie H, Liao Z, Wang Y. et al. Exosomal long non-coding RNAs: Emerging players in cancer metastasis and potential diagnostic biomarkers for personalized oncology. Genes Dis. 2021;8(6):769-780
7. Han Y, Wang D, Peng L. et al. Single-cell sequencing: a promising approach for uncovering the mechanisms of tumor metastasis. J Hematol Oncol. 2022;15(1):59
8. Singgih EL, van der Voet M, Schimmel-Naber M. et al. Investigating cytosolic 5'-nucleotidase II family genes as candidates for neuropsychiatric disorders in Drosophila (114/150 chr). Transl Psychiatry. 2021;11(1):55
9. Jordheim LP. Expanding the clinical relevance of the 5'-nucleotidase cN-II/NT5C2. Purinergic Signal. 2018;14(4):321-329
10. Oka J, Matsumoto A, Hosokawa Y. et al. Molecular cloning of human cytosolic purine 5'-nucleotidase. Biochem Biophys Res Commun. 1994;205(1):917-922
11. Guo Y, Gong Y, Shi G. et al. Single-nucleotide polymorphisms in the TSPYL-4 and NT5DC1 genes are associated with susceptibility to chronic obstructive pulmonary disease. Mol Med Rep. 2012;6(3):631-638
12. Guo S, Ran H, Xiao D. et al. NT5DC2 promotes tumorigenicity of glioma stem-like cells by upregulating fyn. Cancer Lett. 2019;454:98-107
13. Li H, Li C, Zhang B. et al. Lactoferrin suppresses the progression of colon cancer under hyperglycemia by targeting WTAP/m(6)A/NT5DC3/HKDC1 axis. J Transl Med. 2023;21(1):156
14. Li H, Yao Q, Li C. et al. Lactoferrin Inhibits the Development of T2D-Induced Colon Tumors by Regulating the NT5DC3/PI3K/AKT/mTOR Signaling Pathway. Foods. 2022 11(24)
15. Jin X, Liu X, Zhang Z. et al. NT5DC2 suppression restrains progression towards metastasis of non-small-cell lung cancer through regulation p53 signaling. Biochem Biophys Res Commun. 2020;533(3):354-361
16. Cui Y, Wen Y, Lv C. et al. Decreased RNA-binding protein IGF2BP2 downregulates NT5DC2, which suppresses cell proliferation, and induces cell cycle arrest and apoptosis in diffuse large B-cell lymphoma cells by regulating the p53 signaling pathway. Mol Med Rep. 2022 26(3)
17. Chen YT, Lin WD, Liao WL. et al. NT5C2 methylation regulatory interplay between DNMT1 and insulin receptor in type 2 diabetes. Sci Rep. 2020;10(1):16087
18. Kulkarni SS, Karlsson HK, Szekeres F. et al. Suppression of 5'-nucleotidase enzymes promotes AMP-activated protein kinase (AMPK) phosphorylation and metabolism in human and mouse skeletal muscle. J Biol Chem. 2011;286(40):34567-34574
19. Hu B, Zhou S, Hu X. et al. NT5DC2 promotes leiomyosarcoma tumour cell growth via stabilizing unpalmitoylated TEAD4 and generating a positive feedback loop. J Cell Mol Med. 2021;25(13):5976-5987
20. Zhu Z, Hou Q, Guo H. NT5DC2 knockdown inhibits colorectal carcinoma progression by repressing metastasis, angiogenesis and tumor-associated macrophage recruitment: A mechanism involving VEGF signaling. Exp Cell Res. 2020;397(1):112311
21. Schulze AB, Kuntze A, Schmidt LH. et al. High Expression of NT5DC2 Is a Negative Prognostic Marker in Pulmonary Adenocarcinoma. Cancers (Basel). 2022 14(6)
22. Li R, Liu R, Zheng S. et al. Comprehensive Analysis of Prognostic Value and Immune Infiltration of the NT5DC Family in Hepatocellular Carcinoma. J Oncol. 2022;2022:2607878
23. Tang Z, Kang B, Li C. et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556-w560
24. Chandrashekar DS, Bashel B, Balasubramanya SAH. et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19(8):649-658
25. Gao J, Aksoy BA, Dogrusoz U. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1
26. Shannon P, Markiel A, Ozier O. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498-2504
27. Zhou Y, Zhou B, Pache L. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523
28. Li T, Fan J, Wang B. et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017;77(21):e108-e110
29. Li T, Fu J, Zeng Z. et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509-w514
30. Kleeff J, Korc M, Apte M. et al. Pancreatic cancer. Nat Rev Dis Primers. 2016;2:16022
31. Sun R, He XY, Mei C. et al. Role of exosomal long non-coding RNAs in colorectal cancer. World J Gastrointest Oncol. 2021;13(8):867-878
32. Zhou H, He X, He Y. et al. Exosomal circRNAs: Emerging Players in Tumor Metastasis. Front Cell Dev Biol. 2021;9:786224
33. Dieck CL, Ferrando A. Genetics and mechanisms of NT5C2-driven chemotherapy resistance in relapsed ALL. Blood. 2019;133(21):2263-2268
34. Li KS, Zhu XD, Liu HD. et al. NT5DC2 promotes tumor cell proliferation by stabilizing EGFR in hepatocellular carcinoma. Cell Death Dis. 2020;11(5):335
35. Tzoneva G, Dieck CL, Oshima K. et al. Clonal evolution mechanisms in NT5C2 mutant-relapsed acute lymphoblastic leukaemia. Nature. 2018;553(7689):511-514
36. Kunz JB, Rausch T, Bandapalli OR. et al. Pediatric T-cell lymphoblastic leukemia evolves into relapse by clonal selection, acquisition of mutations and promoter hypomethylation. Haematologica. 2015;100(11):1442-1450
37. Jones NR, Lazarus P. UGT2B gene expression analysis in multiple tobacco carcinogen-targeted tissues. Drug Metab Dispos. 2014;42(4):529-536
38. Li K, Zhang Z, Mei Y. et al. Metallothionein-1G suppresses pancreatic cancer cell stemness by limiting activin A secretion via NF-κB inhibition. Theranostics. 2021;11(7):3196-3212
39. Hamilton-Williams EE, Cheung J, Rainbow DB. et al. Cellular mechanisms of restored β-cell tolerance mediated by protective alleles of Idd3 and Idd5. Diabetes. 2012;61(1):166-174
40. Zhu B, Huo R, Zhi Q. et al. Increased expression of zinc transporter ZIP4, ZIP11, ZnT1, and ZnT6 predicts poor prognosis in pancreatic cancer. J Trace Elem Med Biol. 2021;65:126734
41. Schizas D, Charalampakis N, Kole C. et al. Immunotherapy for pancreatic cancer: A 2020 update. Cancer Treat Rev. 2020;86:102016
42. Padoan A, Plebani M, Basso D. Inflammation and Pancreatic Cancer: Focus on Metabolism, Cytokines, and Immunity. Int J Mol Sci. 2019 20(3)
43. Morrison AH, Byrne KT, Vonderheide RH. Immunotherapy and Prevention of Pancreatic Cancer. Trends Cancer. 2018;4(6):418-428
44. Bear AS, Vonderheide RH, O'Hara MH. Challenges and Opportunities for Pancreatic Cancer Immunotherapy. Cancer Cell. 2020;38(6):788-802
45. Carlini F, Ivaldi F, Gualandi F. et al. Different Susceptibility of T and B Cells to Cladribine Depends On Their Levels of Deoxycytidine Kinase Activity Linked to Activation Status. J Neuroimmune Pharmacol. 2022;17(1-2):195-205
46. Reglero C, Dieck CL, Zask A. et al. Pharmacologic Inhibition of NT5C2 Reverses Genetic and Nongenetic Drivers of 6-MP Resistance in Acute Lymphoblastic Leukemia. Cancer Discov. 2022;12(11):2646-2665
47. Marton Z, Guillon R, Krimm I. et al. Identification of Noncompetitive Inhibitors of Cytosolic 5'-Nucleotidase II Using a Fragment-Based Approach. J Med Chem. 2015;58(24):9680-9696
Author contact
![]() Corresponding authors: Chunlin Ou. Department of Pathology, Xiangya Hospital, Central South University, Changsha 410008, Hunan, China. Email: ouchunlinedu.cn; Hongbin Guo, Department of Orthopedics, Xiangya Hospital, Central South University, Changsha 410008, Hunan, China. Email: ghb_0113com.
Corresponding authors: Chunlin Ou. Department of Pathology, Xiangya Hospital, Central South University, Changsha 410008, Hunan, China. Email: ouchunlinedu.cn; Hongbin Guo, Department of Orthopedics, Xiangya Hospital, Central South University, Changsha 410008, Hunan, China. Email: ghb_0113com.

 Global reach, higher impact
Global reach, higher impact