Impact Factor
ISSN: 1837-9664
J Cancer 2023; 14(17):3203-3213. doi:10.7150/jca.86967 This issue Cite
Review
Mechanisms underlying the changes in acetaldehyde dehydrogenase 1 in cholangiocarcinoma
1. Department of Hepatobiliary Surgery, The First Affiliated Hospital of Hunan Normal University, Changsha, 410005 Hunan Province, China
2. Central Laboratory of Hunan Provincial People's Hospital (The First Affiliated Hospital of Hunan Normal University), Changsha, 410015, China.
3. Key Laboratory of Molecular Epidemiology of Hunan Province, School of Medicine, Hunan Normal University, Changsha, China.
*Contributed equally.
Received 2023-6-9; Accepted 2023-9-8; Published 2023-9-25
Abstract
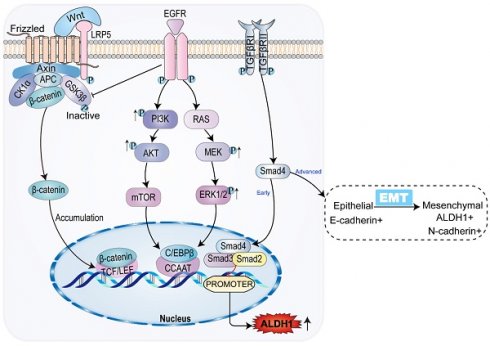
Cholangiocarcinoma (CCA) is the most recurrent malignant tumor found in the biliary system. It originates from the bile duct epithelial cells characterized by easy metastasis, high intermittent rate, and poor prognosis. Acetaldehyde dehydrogenase 1 (ALDH1), a marker of cancer stem cells, the levels of which are particularly elevated in various of malignant tumors. Additionally, the increased ALDH1 levels are closely related to the degree and prognosis of malignant tumors. This study reviewed the mechanisms underlying the changes in ALDH1 levels in CCA.
Keywords: Cholangiocarcinoma, ALDH1, prognosis
Introduction
Cholangiocarcinoma (CCA) is a common primary liver malignancy, accounting for approximately 15% of all primary liver tumors [1-3]. CCA is categorized into three subtypes according to its original anatomical sites: intrahepatic CCA, hilar CCA, and distal CCA [3, 4]. At present, CCA currently accounts for 3% of all gastrointestinal malignancies and 2% of cancer-related deaths [2, 3]. Radical resection (R0 resection) currently provides the most effective treatment therapy for CCA. Patients who undergo complete tumor removal can have a median survival time of over 40 months, with a 25% to 40% overall survival rate after five years. On the other hand, patients who did not go through surgical resection had a much lower median survival time of only 10-12 months [3, 5, 6]. Since patients with CCA are often asymptomatic in the early stages with a lack of specific tumor markers, when diagnosing, the disease is often at an advanced stage, resulting in limit treatment options and resulting in a poor prognosis [7].
Acetaldehyde dehydrogenase (ALDH) is an intracellular oxidase that alleviates intracellular acetaldehyde or drug toxicity, and also a significant cancer stem cell (CSC) marker that influences a series of important processes such as relief from ethanol or drug cytotoxicity [8-10]. ALDH1, an important member of the ALDH family, is mainly categorized into ALDH1A1, ALDH1A2, ALDH1A3, and other subtypes. The ALDH1A1 coding gene is located on autosomal chromosome 9 with 13 exons, while ALDH1A2 and ALDH1A3 are located on chromosome 15 [11, 12]. ALDH1, an enzyme that oxidize retinol to retinoic acid (RA), is involved in cell proliferation and RA-related metabolism [13, 14]. At the same time, it shows antioxidative activity and the ability to regulate osmotic pressure, participating in drug metabolism and cell differentiation. ALDH1 also serves as a marker significantly related to CSC, which can be used to extract tumor cell subsets from various cell lines and primary tumors [15]. In recent years, the expression of ALDH1 has been detected in CCA, therefore, we reviewed the significance of ALDH1 expression in CCA.
1. Tumor-associated signaling pathways and proteins affect the expression of ALDH1
1.1 Common regulatory proteins for the transcription of ALDH1 in tumor cells
The ALDH1 gene promoter contains a CCAAT box (71 to 67 bp) activated by the CCAAT/enhancer binding protein β (C/EBPβ), and the binding of C/EBPβ to the promoter initiates ALDH1 expression [16, 17] In breast cancer, mucin 1 (MUCI-1) induces phosphorylation of extracellular regulated protein kinase (ERK) and activation of C/EBPβ transcription factors (Figure 1). MUCI-1, phosphorylated ERK, and activated C/EBPβ transcription factors form a complex at the ALDH1 promoter and activate ALDH1 [18]. Aberrant activation of the signal transducer and transcription 3 activator (STAT3) significantly promotes DNA transcription of ALDH1 [19]. STAT3 forms a complex with the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), which interferes with the interaction between C/EBPβ and DDIT3/CHOP/GADD153 to form a complex avoiding C/EBPβ inactivation thus allowing C/EBPβ-dependent ALDH1 promoter expression [20]. In addition, the ALDH1 promoter contains androgen receptor (AR) binding sites. So, direct regulation of the AR pathway regulates ALDH1 expression in androgen-dependent prostate cancer cells (Figure 1). The androgen dihydrotestosterone directly upregulates ALDH1 by inducing AR transcriptional activity [21, 22]. Tumors associated with AR, such as prostate cancer, breast cancer, and endometrial cancer, all show high expression levels of ALDH1 [21].
Common molecules in tumors related to ALDH1, including polycomb family genes, androgen, NRF2, etc., promote the expression of ALDH1 by interacting with ALDH1.
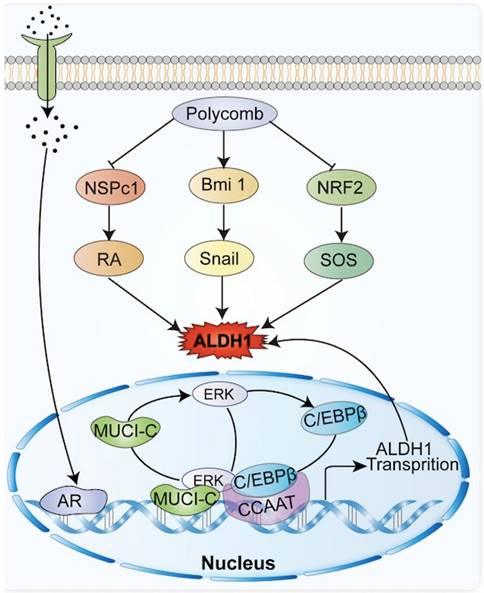
1.2 Interactions between ALDH1 and molecules in tumors
In breast cancer, Notch signal pathway induces deacetylation and activation of ALDH1Lys-353 through mammalian Sir2 homologue (SIRT2), controls the activity of ALDH1 (Figure 1), and is related to tumorigenesis and tumor growth [13]. The nuclear erythroid-associated factor 2 (NRF2) is a protein associated with erythroid and platelet development. NRF2 (Figure 1) is involved in antioxidant processes, and high levels of NRF2 interact with antioxidants and detoxifying enzymes to involve in tumor chemoresistance [23]. Increased expression of NRF2-mediated antioxidant enzymes inhibits reactive oxygen species (ROS) production, and reduced ROS production serves as a negative feedback signal promoting ALDH1 expression [24]. Polycomb proteins are a family of proteins, relating to CSC and tumor transformation, which could reshape chromatin and thereby express epigenetically silence genes (Figure 1). The Polycomb genes Bmi-1 and NSPc 1 can promote ALDH 1 expression [25, 26].
A high expression of ALDH1 could increase the risk of malignant tumors related to ethanol [27]. Mounting evidence suggests that patients with malignant tumors exhibiting a high expression of ALDH1 have an increased propensity for metastasis and a poor prognosis [21]. Other malignant tumors, such as acute lymphoblastic leukemia and prostate cancer, tumor cells exhibit higher ALDH1 activity levels and a worse tumor prognosis with increasing tumor invasiveness [28, 29]. ALDH1 interacts with the common genes and signal pathway proteins in tumors. Tumor-promoting factors can upregulate the expression of ALDH1, promoting occurrence, proliferation, and metastasis of tumor cells, subsequently, enhancing tumor resistance.
2. Mechanism of action of ALDH1 in CSC
CSC, known as tumor-initiating or tumor-proliferating cells [30], are also the underlying cause of tumor heterogeneity. CSC are a subdivision of tumor cells with characteristics of self-renewal, high tumorigenicity, and multidirectional differentiation [31]. An increasing number of studies have reported that ALDH1 shows high activity in a variety of CSC, and its expression is closely associated with tumor metastasis, drug resistance, and restenosis. In ALDH1 family, ALDH1A1 and ALDH1A3 are especially significant in CSC [32-34]. ALDH1 is involved in many signaling pathways that regulate CSC, including RA and transforming growth factor-β (TGF-β) [35].
2.1 Enzymatic functions of ALDH1 and CSC
In the presence of ALDH1, retinaldehyde is irreversibly metabolized to RA (all-trans, 9-cis, 13-cis) [36, 37]. RA is a signaling molecule that regulates the transcription of gene and is involved in the differentiation and proliferation of CSC [38]. In the classical RA pathway, the RA molecule enters the nucleus and binds with the heterodimer created by nuclear retinoic acid receptor (RARα) and retinol X receptor (RXR). Histone acetylation induced by RA activation promotes downstream AKT molecule activation, upregulation of ALDH1 expression, and promotes CSC differentiation and cell proliferation [39].
ALDH1 plays a role in acetaldehyde metabolism that facilitates CSC proliferation. Ethanol is metabolized into acetaldehyde through the actions of ethanol dehydrogenase (ADH), catalase, and cytochrome P4502E1 (CYP2E1), which disrupt antioxidant defense systems and produce ROS. ROS inhibit DNA repair and methylation, forming the DNA and protein complexes that inhibit the growth of CSC [31, 33, 40]. ALDH1 oxidizes toxic aldehydes while removing ROS, preventing the toxic effect of retinoids and ROS to CSC [41].
2.2 ALDH1 induces epithelial-mesenchymal transition associated with CSC
Epithelial-mesenchymal transition (EMT) is not a single stereotyped process, but a multistep process that involves a transition from epithelial to mesenchymal cells through an intermediate mixed state (partial EMT state, P-EMT) [42-44]. While the TGF-β-induced transition from epithelial to P-EMT is reversible, the transition from P-EMT to mesenchymal cells may not be reversible [45]. In the case of CCA, the TGF-β pathway induces EMT. The acquisition of mesenchymal stroma, assisted by upregulation of ALDH1 expression, is a key event in increasing tumor aggressiveness and CSC properties [46, 47]. EMT allows tumors to acquire a pluripotent stem cell-like phenotype of tumor cells and to express CSC-specific transcription factors. SOX and others are involved in maintaining the multiple differentiation potential of tumors. Meanwhile, Notch1 and Wnt2 regulate CSC self-renewal [48].
CSC have been demonstrated to be crucial in tumor growth. ALDH1, as a practical marker of CSC, indicates the acquisition of tumor stemness and helps the discrimination of CSC in tumors [49].
3. ALDH1 in the prognosis of CCA
3.1 ALDH1 expression indicates a poor prognosis in CCA
In patients with CCA, high expression of ALDH1 is consistently associated with poor tumor differentiation and a poor prognosis, which agree with the findings from studies on other types of tumors [50, 51]. In breast cancer, ovarian cancer, and many other tumors, high expression of CD274 (PD-L1) can activate the immunosuppressive response and lead to tumor progression [52-54]. In contrast, low expression of CD274 in CCA is associated with an increase in ALDH1 activity and a reduction in the production of active aldehyde and ROS, resulting in the expression of the CSC characteristics of CCA cells. Meanwhile, the levels of the CSC markers Oct4, Sox2, and Nanog are also elevated [55]. As a negative tumor regulator of CCA, CD274 downregulates the expression of the CSC marker ALDH1 and decreases the production of ROS and active aldehydes.
LIN28B is a type of RNA-binding protein [56]. The activity and expression level of the CSC marker ALDH1 are especially increased in CCA cell lines overexpressing LIN28B. Furthermore, LIN28B overexpression in CCA cells activates the carcinogenic signaling pathway and leads to increased STAT3 expression [57]. STAT3 has the ability to participate in ALDH1 transcription and reduce the sensitivity of chemotherapeutic drugs. The LIN28B/STAT3/ALDH1 pathway play a complicated role in bile duct cell proliferation and differentiation differentiation and is associated with the occurrence of CCA. These results show that ALDH1 is a CSC marker of CCA, and a prognostic factor for CCA [57].
3.2 Signal pathways related to ALDH1 and CCA
3.2.1 ALDH1 and the classical tumor activation pathway
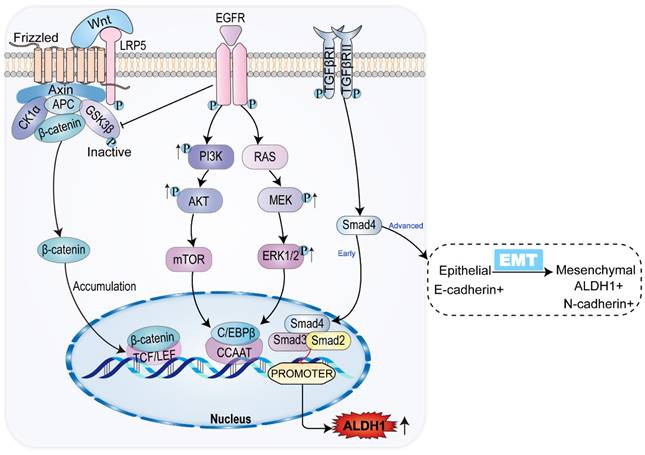
Interleukin-6 (IL-6), vascular endothelial growth factor (VEGF), epidermal growth factor (EGF), and hepatic liver growth factor (HGF) participate in classical oncogenic pathways, including the RAS-MAPK-ERK-MTOR and PI3K-AKT-MTOR pathways (Figure 2). Expression of ALDH1 can be upregulated through the classical carcinogenic pathway [58]. The mammalian target of rapamycin (mTOR), a 289-kDa serine/threonine protein kinase, regulates cell proliferation, survival, and angiogenesis [12, 59]. IL-6 and other agents can increase the expression of granulin precursor proteins, which activate the AKT-mTOR pathway of cell mitosis, cell migration, and angiogenesis. Meanwhile, mTOR activates downstream ALDH1A1 expression [47, 60].
IL-6 and others activate p38MAPK, a group of protein kinases responsible for cell differentiation and proliferation, promoting ALDH1 expression and leading to reduced expression of p21, a mediator of cell senescence, as well as to promotion of mitosis, and enhancement of cell proliferation [61]. MAPK can enable ERK phosphorylation and activation, which activates mTOR. Activation of the RAS-MAPK-ERK-MTOR pathway subsequently activates downstream ALDH1A3 [12]. The expression of mTOR is common in gallbladder cancer and CCA, suggesting a tumor prognosis. Inhibition of mTOR exhibits antitumor effects in gallbladder cancer and CCA [62]. Additionally, mTOR can be activated by phosphorylation of C/EBPβ2. Activation and proliferation of C/EBPβ2 upregulates the expression of ALDH1 by recognizing and binding to the ALDH1 promoter located in the CCAAT box of the cis-acting element [59]. ALDH1 expression is upregulated in the classical tumor activation pathway, which is closely related to the growth and progression of CCA, indicating a poor prognosis for CCA.
3.2.2 ALDH1 and TGF-β-SMAD pathway
TGF-β plays a dual role in cancer. In addition to controlling CCA progression by inducing apoptosis, TDF-β can also increase the progression of CCA by promoting EMT, tumor migration and invasion, and suppressing the immune system [63].
In the early stage of CCA, TGF-β inhibits the expression of ALDH1 via SMAD, while the TGF-β signaling pathway is associated with a decrease in tumor-initiating cells in CCA. The decreased expression of ALDH1 suggests that tumor activation is regulated in CCA [64, 65]. However, in advanced CCA, activation of the TGF-β signaling pathway can contribute to tumor growth, with tumor progression [15, 66]. The percentage of ALDH1-positive cells significantly increases in CCA cells treated with TGF-β. Meanwhile, ALDH1-positive cells exhibit stem cell characteristics and EMT features [47, 67-69]. TGF-β induces the level and activity of SMAD, promoting EMT by inhibiting the transcription of E-calmodulin, the epithelial morphological marker [70]. EMT is the key to acquiring CSC properties [71]. In the TGF-β pathway, ALDH1 expression is upregulated and the invasive activity of CCA is elevated [46]. The results of immunohistochemical analysis showed that both TGF-β and ALDH1 expression were enhanced in advanced CCA (Figure 2). Consequently, TGF-β and ALDH1 could be independent prognostic factors in CCA, where the upregulation of ALDH1 and TGF-β expression indicated shortened overall survival of patients [46]. ALDH1 acts as a prognostic marker for CCA, indicating a poor prognosis of CCA in both in the early and late stages of the tumor.
3.2.3 ALDH1 and WNT-β-catenin pathway
When the bile duct epithelium is damaged, inflammatory macrophages produce WNT ligands, which usually play an epithelial repair role [72]. Macrophages upregulate the expression and transcription of WNT7b and WNT10a, inhibiting the degradation of intracellular β-catenin by binding to the FZD receptor and its co-receptor LRP5/LRP6 on bile duct cells, thus leading to β-catenin accumulation [73]. In the nucleus family, β-catenin interacts with the TCF/LEF transcription factor, causing the expression of ALDH1. This leads to increased cell viability and resistance to apoptosis [74]. SOX17, an oncogene, encodes a protein that can forms a complex with other proteins and act as a transcriptional regulator. It antagonizes the WNT-β-catenin signaling pathway, negatively regulates the expression of ALDH1, regulates cholangiocyte differentiation, and acts as a tumor suppressor in CCA (Figure 2). Studies have shown that the SOX17 promoter is highly methylated, resulting in decreased expression of SOX17. This reduces the inhibition of the WNT-β-catenin pathway, leading to upregulation of ALDH1 expression, and worsening the prognosis of CCA [75, 76].
Common signaling pathways and ALDH1 expression in CCA. In CCA, WNT/β-catenin signaling pathway promotes ALDH1 expression by binding to transcriptional regulatory factor TCF, while classical tumor signaling pathways PI3K/AKT/mTOR and RAS/MEK/ERK promote ALDH1 expression by promoting transcriptional enhancer CCAAT. TGF-β/Smad signaling pathway has the opposite effect on the expression of ALDH1 in the early and advanced stages of the tumor, inhibiting the expression of ALDH1 in the early stage and vice versa in the late stage.
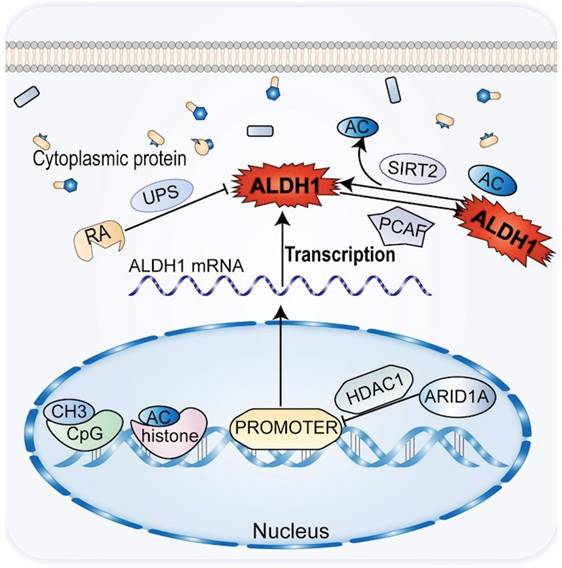
3.3 The epigenetics of ALDH1 is associated with CCA
Chemical modifications of epigenetic and post-translational proteins can impact the expression and activity of ALDH1 in CCA. Epigenetics plays an important role in the development and progression of CCA, affecting the tumor phenotype in the absence of changes in the DNA sequence [77]. Histone acetylation modifications and methylation of CpG islands regulate the transcriptional promoters of ALDH1A1 and ALDH1A3 to downregulate the corresponding ALDH1 [12]. Genes such as ALDH1A1, SPP1, and CD81 have been shown to be upregulated in ARID1A-deficient CCA cells, with the most significant changes in ALDH1A1 [78]. By reducing histone H3K27 acetylation, ARID1A introduces histone deacetylase into the promoter region of ALDH1A1, suppressing the expression of ALDH1A1 in CCA cells, thereby acting as a tumor suppressor of CCA (Figure 3). In CCA patients, deletion of ARID1A and upregulation of ALDH1A1 are independent prognostic factors [78].
Following post-translational chemical modification in CCA, ALDH1 can oxidize retinoids to form RA and its analogs (Figure 3). RA and its analogs, particularly tran-sretinoic acid, can reduce ALDH1 activity through the ubiquitin protease system without altering ALDH1 mRNA levels. However, RA may be altered by protein chemical modification of ALDH1 activity [79, 80]. Acetylation of the lysine 353 is another post-translational modification of ALDH1. ALDH1 activity decreases by acetylation of lysine 353, a process activated by acetyltransferase P300/CBP-associated factor (PCAF) [81, 82]. Notch signaling induces high ALDH1 activity in CCA. This process increases the expression of deacetylase sirtuin2 (SIRT2), which allows the elimination of ALDH1 acetylation [13, 81]. A reduction in ALDH1 activity leads to an increase in ROS and reactive aldehydes in CSC and promotes CSC apoptosis in CCA.
4. Relationship between ALDH1 and chemotherapy resistance in CCA
The GC and GEMOX regimens are standard first-line chemotherapy treatment for CCA [5]. Gemcitabine (GEM) plays a critical role in various first-line chemotherapy treatment. The occurrence of GEM chemotherapy resistance often predicts a poor prognosis in CCA. Elevated ALDH1 activity has been connected to the increased drug resistance and metastasis in a range of cancers [83-86]. Furthermore, in patients with advanced CCA, ALDH1 has been shown to play a crucial role in malignant behavior and resistance to GEM [87].
The common genetic epigenetic regulation of ALDH1 in CCA is that ALDH1 expression is down-regulated by methylation and acetylation before transcription, and ALDH1 protein activity is inhibited by ubiquitin and acetylation after transcriptional translation.
The interaction between ALDH1 and gemcitabine, in CCA, the expression of ALDH1 is often accompanied by gemcitabine resistance, and the up-regulation of ALDH1 is accompanied by an increase in the exclusion of gemcitabine and a decrease in gemcitabine entry. Related chemicals restore the sensitivity of gemcitabine to cholangiocarcinoma by inhibiting the expression of ALDH1.
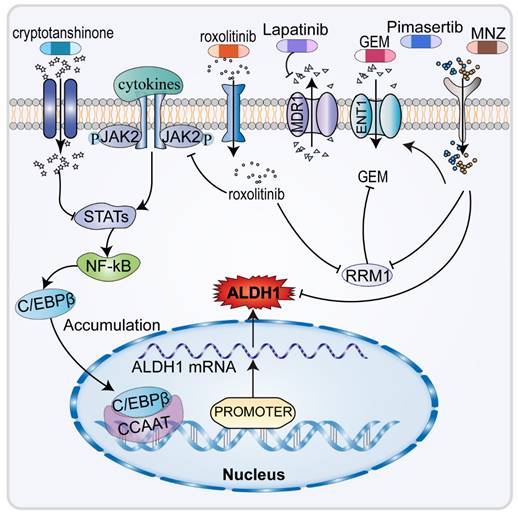
4.1 Mechanism of resistance to GEM through ALDH1 expression in CCA
Intrahepatic cholangiocarcinoma cell lines HuCCT1 and SNU1079 showed high expression of ALDH1, primarily by ALDH1A3 [87]. ALDH1 is a universal CSC marker, and most studies have reported that the long-term dormancy of CSC confers resistance to most drugs in fast-growing cancer cells. A high expression of ALDH1 is also closely related to EMT and promotes the acquisition of resistance to GEM. The occurrence of EMT is often associated with drug resistance in CCA cells [88]. ALDH1 upregulates and induces resistance mainly by increasing the population of CSC and promoting the appearance of EMT to most chemotherapeutic drugs administered for CCA [89].
In human CCA cell lines with high expression of ALDH1, CCA stem cells are resistant to GEM but not to cisplatin. The reason cannot be explained by the long-term dormancy of CSC as both drugs inhibit fast-growing cancer cells. GEM is a difluoro nucleoside antimetabolites anticancer drug that inhibits cell replication [90], by functioning as a water-soluble analogue of deoxycytidine and competitively inhibiting ribonucleotide reductase 1 (RRM1) [91]. It acts on pyrimidine deoxyribonucleic acid and requires conversion to gemcitabine triphosphate before acting with DNA to inhibit DNA synthesis and induce cell apoptosis [92]. The expression of RRM1, which is the molecular target of ALDH1A3, increased significantly in HuCCT1 and SNU1079 cells treated with high doses of GEM (Figure 4). RRM1 is a polymerase that converts ribonucleotides into deoxyribonucleosides and plays a critical role in DNA polymerization and repair [93]. RRM1 is also crucial in resistance to gemcitabine and can competitively inhibit the role of GEM [94]. This study demonstrated that RRM1 levels decreased, resulting in higher GEM response rates in patients with lower expression levels of ALDH1A3 who received GEM chemotherapy for CCA. The correlation between ALDH1A3 expression levels and chemotherapy responses was verified in 31 patients [87].
4.2 ALDH1 participates in the mechanisms underlying anti-GEM resistance
The therapeutic options for advanced chemotherapy refractory CCA are limited, highlighting the need to identify of new effective therapeutic agents (Figure 4). It has been found that the amatuximab and dasatinib can inhibit adhesion of cancer cells to the peritoneum and suppress the stemness and viability of cancer cells, reducing ALDH1 expression and improving tumor sensitivity to GEM [40, 95]. Infigratinib downregulates ALDH1 expression in CCA cells by inhibiting FGFR/AKT/MTOR and FGFR/STAT3, which also suppresses EMT, to increase the antitumor effect of GEM [96]. In patients with advanced CCA, ALDH1 plays an important role in malignant behavior and resistance to GEM [87]. As an inhibitor of Janus kinase 2 (JAK2), the targeted drug ruxolitinib inhibits downstream signal transduction and the expression of STAT1/3, downregulating the expression of ALDH1, thereby increasing tumor sensitivity to GEM [19]. Ruxolitinib has also been found to significantly inhibits the migratory capacity of CCA cells. Fraxetin and cryptotanshinone, the anti-dysentery drugs, can enhance the antitumor effect of GEM by inhibiting STAT3 which results in the downregulation of ALDH1 expression [97, 98].
Previous studies (Figure 4) have shown that ENT1 and RRM1 are key factors in the cytotoxicity of GEM [99-101]. ENT1 is a membrane transporter protein that promotes the efficient penetration of GEM into cells [93]. Upregulation of MDR1 within the ABC transporter protein family, responsible for drug efflux, lowers the concentration of many drugs, including GEM [102, 103]. In CCA cells, pimasertib [104] and metronidazole (MNZ), the MEK inhibitors, decrease ALDH1 activity, increase GEM sensitivity in CCA cells by increasing equilibrium nucleoside transporter protein 1 (ENT1), thus decreasing RRM1 [99]. Additionally, MNZ can decrease EMT by decreasing ALDH1 activity and decreasing tumor cell stemness, which ultimately lowers invasiveness and resistance to GEM [46, 105]. In CCA cells, hydroxyurea could be acted as a RRM inhibitor [106]. Moreover, lapatinib and emodin increased GEM sensitivity by inhibiting MDR, accompanied by decreased regulation of ALDH1 [107, 108]. ALDH1 is closely related to chemoresistance in CCA, thus opening new paths for the treatment of chemo-resistant CCA.
5. Conclusions
As one of the markers of tumor stem cells, ALDH1 is expressed in a variety of malignant tumors relating to the clinical prognosis of patients. Alteration of ALDH1 transcription or activity may be caused by common signal pathways and molecules like WNT, TGF-β, and STAT present in tumors. The enzymatic functions of ALDH1 can confer and preserve the characteristics of tumor CSC. Signaling pathways associated with ALDH1 can promote the completion of EMT as well as the acquisition of tumor dryness.
A comprehensive and current literature review revealed that ALDH1 plays an important role in the degree of malignancy, efficacy evaluation, and prognosis of CCA. On the other hand, the typical signaling pathways in CCA may impact the expression of ALDH1. In CCA, ALDH1 participates in the TGF-β/SMAD and WNT/β-catenin pathway, and its expression level is positively correlated with the tumor malignancy of CCA. Epigenetics regulates the expression of ALDH1 before and after translation and transcription, contributing to changes in the degree of malignancy of CCA.
ALDH1 is closely associated with chemotherapy resistance and EMT in CCA. GEM is the foundational drug for CCA chemotherapy, and the resistance to this drug is linked to the upregulation of ALDH1. Although new drugs have been demonstrated to inhibit the expression of ALDH1 and facilitate the recovery of chemosensitivity in CCA, the specific mechanisms underlying the expression of ALDH1 in CCA remain to be further discovered. Resistance to tumor radiation therapy, immunotherapy, and targeted therapy is not thought to be linked with the expression of ALDH1. A thorough understanding the function of ALDH1 is highly beneficial for the treatment and prognosis of CCA. Research on ALDH1 will also generate novel approaches for the treatment of CCA.
Acknowledgements
Funding
The present study was supported by grants from the following organizations: Education fund item of Hunan Province (20B380),The Hunan Provincial Natural Science Foundation of China (2020JJ5610), the Hunan Natural Science Fund for Excellent Young Scholars (2021JJ20003), the Natural Science Foundation of Changsha (kq2007023/kq2004115), Chen Xiao-Ping Foundation for Development of Science and Technology of Hubei Province (CXPJJH12000001-2020322), Project of Hunan Provincial Health Commission (202104010997) and Project of Changsha Biliary Diseases Clinical Medical Research Center(Grant No. kh2005011), Project of Changsha Biliary Diseases Clinical Medical Research Center(Grant No. kh2005011) and National Natural Science Foundation of China (82303511).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Author contributions
BD, YH and SL contributed to the analysis and manuscript preparation. YJ revised the manuscript. CP and SL contributed to the conception of the study. YJ and SL helped perform the analysis and participated in constructive discussions.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Nakeeb A, Pitt HA, Sohn TA, Coleman J, Abrams RA, Piantadosi S. et al. Cholangiocarcinoma. A spectrum of intrahepatic, perihilar, and distal tumors. Annals of surgery. 1996;224:463-73 discussion 73-5
2. DeOliveira ML, Cunningham SC, Cameron JL, Kamangar F, Winter JM, Lillemoe KD. et al. Cholangiocarcinoma: thirty-one-year experience with 564 patients at a single institution. Annals of surgery. 2007;245:755-62
3. Banales JM, Cardinale V, Carpino G, Marzioni M, Andersen JB, Invernizzi P. et al. Expert consensus document: Cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA). Nat Rev Gastroenterol Hepatol. 2016;13:261-80
4. Rizvi S, Khan SA, Hallemeier CL, Kelley RK, Gores GJ. Cholangiocarcinoma - evolving concepts and therapeutic strategies. Nat Rev Clin Oncol. 2018;15:95-111
5. Khan AS, Dageforde LA. Cholangiocarcinoma. Surg Clin North Am. 2019;99:315-35
6. Bertuccio P, Malvezzi M, Carioli G, Hashim D, Boffetta P, El-Serag HB. et al. Global trends in mortality from intrahepatic and extrahepatic cholangiocarcinoma. J Hepatol. 2019;71:104-14
7. Pavicevic S, Reichelt S, Uluk D, Lurje I, Engelmann C, Modest DP. et al. Prognostic and Predictive Molecular Markers in Cholangiocarcinoma. Cancers (Basel). 2022 14
8. Kim IG, Lee JH, Kim SY, Kim JY, Cho EW. Fibulin-3 negatively regulates ALDH1 via c-MET suppression and increases γ-radiation-induced sensitivity in some pancreatic cancer cell lines. Biochem Biophys Res Commun. 2014;454:369-75
9. Ghoneum A, Gonzalez D, Abdulfattah AY, Said N. Metabolic Plasticity in Ovarian Cancer Stem Cells. Cancers (Basel). 2020 12
10. Gui S, Xie X, O'Neill WQ, Chatfield-Reed K, Yu JG, Teknos TN. et al. p53 functional states are associated with distinct aldehyde dehydrogenase transcriptomic signatures. Sci Rep. 2020;10:1097
11. Fitzgerald TL, McCubrey JA. Pancreatic cancer stem cells: association with cell surface markers, prognosis, resistance, metastasis and treatment. Adv Biol Regul. 2014;56:45-50
12. Duan JJ, Cai J, Guo YF, Bian XW, Yu SC. ALDH1A3, a metabolic target for cancer diagnosis and therapy. Int J Cancer. 2016;139:965-75
13. Zhao D, Mo Y, Li MT, Zou SW, Cheng ZL, Sun YP. et al. NOTCH-induced aldehyde dehydrogenase 1A1 deacetylation promotes breast cancer stem cells. J Clin Invest. 2014;124:5453-65
14. Singh S, Brocker C, Koppaka V, Chen Y, Jackson BC, Matsumoto A. et al. Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radic Biol Med. 2013;56:89-101
15. Tomita H, Tanaka K, Tanaka T, Hara A. Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget. 2016;7:11018-32
16. Cojoc M, Peitzsch C, Kurth I, Trautmann F, Kunz-Schughart LA, Telegeev GD. et al. Aldehyde Dehydrogenase Is Regulated by β-Catenin/TCF and Promotes Radioresistance in Prostate Cancer Progenitor Cells. Cancer Res. 2015;75:1482-94
17. Sarabia-Sánchez M, Alvarado-Ortiz E, Toledo-Guzman ME, García-Carrancá A, Ortiz-Sánchez E. ALDH(HIGH) Population Is Regulated by the AKT/β-Catenin Pathway in a Cervical Cancer Model. Front Oncol. 2020;10:1039
18. Yamashita N, Kufe D. Addiction of Cancer Stem Cells to MUC1-C in Triple-Negative Breast Cancer Progression. Int J Mol Sci. 2022 23
19. Shao C, Sullivan JP, Girard L, Augustyn A, Yenerall P, Rodriguez-Canales J. et al. Essential role of aldehyde dehydrogenase 1A3 for the maintenance of non-small cell lung cancer stem cells is associated with the STAT3 pathway. Clin Cancer Res. 2014;20:4154-66
20. Canino C, Luo Y, Marcato P, Blandino G, Pass HI, Cioce M. A STAT3-NFkB/DDIT3/CEBPβ axis modulates ALDH1A3 expression in chemoresistant cell subpopulations. Oncotarget. 2015;6:12637-53
21. Ma M, He W, Zhao K, Xue L, Xia S, Zhang B. Targeting aldehyde dehydrogenase for prostate cancer therapies. Front Oncol. 2022;12:1006340
22. Casanova-Salas I, Masiá E, Armiñán A, Calatrava A, Mancarella C, Rubio-Briones J. et al. MiR-187 Targets the Androgen-Regulated Gene ALDH1A3 in Prostate Cancer. PLoS One. 2015;10:e0125576
23. Wu S, Lu H, Bai Y. Nrf2 in cancers: A double-edged sword. Cancer Med. 2019;8:2252-67
24. Kim D, Choi BH, Ryoo IG, Kwak MK. High NRF2 level mediates cancer stem cell-like properties of aldehyde dehydrogenase (ALDH)-high ovarian cancer cells: inhibitory role of all-trans retinoic acid in ALDH/NRF2 signaling. Cell Death Dis. 2018;9:896
25. Yu CC, Lo WL, Chen YW, Huang PI, Hsu HS, Tseng LM. et al. Bmi-1 Regulates Snail Expression and Promotes Metastasis Ability in Head and Neck Squamous Cancer-Derived ALDH1 Positive Cells. J Oncol. 2011. 2011
26. Jakob M, Sharaf K, Schirmer M, Leu M, Küffer S, Bertlich M. et al. Role of cancer stem cell markers ALDH1, BCL11B, BMI-1, and CD44 in the prognosis of advanced HNSCC. Strahlenther Onkol. 2021;197:231-45
27. Druesne-Pecollo N, Tehard B, Mallet Y, Gerber M, Norat T, Hercberg S. et al. Alcohol and genetic polymorphisms: effect on risk of alcohol-related cancer. Lancet Oncol. 2009;10:173-80
28. Rezaee M, Gheytanchi E, Madjd Z, Mehrazma M. Clinicopathological Significance of Tumor Stem Cell Markers ALDH1 and CD133 in Colorectal Carcinoma. Iran J Pathol. 2021;16:40-50
29. Gasparetto M, Pei S, Minhajuddin M, Khan N, Pollyea DA, Myers JR. et al. Targeted therapy for a subset of acute myeloid leukemias that lack expression of aldehyde dehydrogenase 1A1. Haematologica. 2017;102:1054-65
30. Petpiroon N, Bhummaphan N, Soonnarong R, Chantarawong W, Maluangnont T, Pongrakhananon V. et al. Ti(0.8)O(2) Nanosheets Inhibit Lung Cancer Stem Cells by Inducing Production of Superoxide Anion. Mol Pharmacol. 2019;95:418-32
31. Prasetyanti PR, Medema JP. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol Cancer. 2017;16:41
32. Vassalli G. Aldehyde Dehydrogenases: Not Just Markers, but Functional Regulators of Stem Cells. Stem Cells Int. 2019;2019:3904645
33. Marcato P, Dean CA, Giacomantonio CA, Lee PW. Aldehyde dehydrogenase: its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle. 2011;10:1378-84
34. Huang EH, Hynes MJ, Zhang T, Ginestier C, Dontu G, Appelman H. et al. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009;69:3382-9
35. Rodriguez-Torres M, Allan AL. Aldehyde dehydrogenase as a marker and functional mediator of metastasis in solid tumors. Clin Exp Metastasis. 2016;33:97-113
36. Melis M, Tang XH, Trasino SE, Gudas LJ. Retinoids in the Pathogenesis and Treatment of Liver Diseases. Nutrients. 2022 14
37. Toledo-Guzmán ME, Hernández MI, Gómez-Gallegos Á A, Ortiz-Sánchez E. ALDH as a Stem Cell Marker in Solid Tumors. Curr Stem Cell Res Ther. 2019;14:375-88
38. Zanoni M, Bravaccini S, Fabbri F, Arienti C. Emerging Roles of Aldehyde Dehydrogenase Isoforms in Anti-cancer Therapy Resistance. Front Med (Lausanne). 2022;9:795762
39. Wang N, Zou Q, Xu J, Zhang J, Liu J. Ligand binding and heterodimerization with retinoid X receptor α (RXRα) induce farnesoid X receptor (FXR) conformational changes affecting coactivator binding. J Biol Chem. 2018;293:18180-91
40. Matsuzawa F, Kamachi H, Mizukami T, Einama T, Kawamata F, Fujii Y. et al. Mesothelin blockage by Amatuximab suppresses cell invasiveness, enhances gemcitabine sensitivity and regulates cancer cell stemness in mesothelin-positive pancreatic cancer cells. BMC Cancer. 2021;21:200
41. Wakamatsu Y, Sakamoto N, Oo HZ, Naito Y, Uraoka N, Anami K. et al. Expression of cancer stem cell markers ALDH1, CD44 and CD133 in primary tumor and lymph node metastasis of gastric cancer. Pathol Int. 2012;62:112-9
42. Peng R, Zhang PF, Zhang C, Huang XY, Ding YB, Deng B. et al. Elevated TRIM44 promotes intrahepatic cholangiocarcinoma progression by inducing cell EMT via MAPK signaling. Cancer Med. 2018;7:796-808
43. Ryu HS, Chung JH, Lee K, Shin E, Jing J, Choe G. et al. Overexpression of epithelial-mesenchymal transition-related markers according to cell dedifferentiation: clinical implications as an independent predictor of poor prognosis in cholangiocarcinoma. Hum Pathol. 2012;43:2360-70
44. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871-90
45. Batlle E, Massagué J. Transforming Growth Factor-β Signaling in Immunity and Cancer. Immunity. 2019;50:924-40
46. Shuang ZY, Wu WC, Xu J, Lin G, Liu YC, Lao XM. et al. Transforming growth factor-β1-induced epithelial-mesenchymal transition generates ALDH-positive cells with stem cell properties in cholangiocarcinoma. Cancer Lett. 2014;354:320-8
47. Araki K, Shimura T, Suzuki H, Tsutsumi S, Wada W, Yajima T. et al. E/N-cadherin switch mediates cancer progression via TGF-β-induced epithelial-to-mesenchymal transition in extrahepatic cholangiocarcinoma. Br J Cancer. 2011;105:1885-93
48. Techasen A, Namwat N, Loilome W, Duangkumpha K, Puapairoj A, Saya H. et al. Tumor necrosis factor-α modulates epithelial mesenchymal transition mediators ZEB2 and S100A4 to promote cholangiocarcinoma progression. J Hepatobiliary Pancreat Sci. 2014;21:703-11
49. Wang M, Xiao J, Jiang J, Qin R. CD133 and ALDH may be the molecular markers of cholangiocarcinoma stem cells. Int J Cancer. 2011;128:1996-7
50. Minato T, Yamamoto Y, Seike J, Yoshida T, Yamai H, Takechi H. et al. Aldehyde dehydrogenase 1 expression is associated with poor prognosis in patients with esophageal squamous cell carcinoma. Ann Surg Oncol. 2013;20:209-17
51. Wu A, Luo W, Zhang Q, Yang Z, Zhang G, Li S. et al. Aldehyde dehydrogenase 1, a functional marker for identifying cancer stem cells in human nasopharyngeal carcinoma. Cancer Lett. 2013;330:181-9
52. Chen YB, Mu CY, Huang JA. Clinical significance of programmed death-1 ligand-1 expression in patients with non-small cell lung cancer: a 5-year-follow-up study. Tumori. 2012;98:751-5
53. Masugi Y, Nishihara R, Yang J, Mima K, da Silva A, Shi Y. et al. Tumour CD274 (PD-L1) expression and T cells in colorectal cancer. Gut. 2017;66:1463-73
54. Chen S, Crabill GA, Pritchard TS, McMiller TL, Wei P, Pardoll DM. et al. Mechanisms regulating PD-L1 expression on tumor and immune cells. J Immunother Cancer. 2019;7:305
55. Tamai K, Nakamura M, Mizuma M, Mochizuki M, Yokoyama M, Endo H. et al. Suppressive expression of CD274 increases tumorigenesis and cancer stem cell phenotypes in cholangiocarcinoma. Cancer Sci. 2014;105:667-74
56. Chien CS, Wang ML, Chu PY, Chang YL, Liu WH, Yu CC. et al. Lin28B/Let-7 Regulates Expression of Oct4 and Sox2 and Reprograms Oral Squamous Cell Carcinoma Cells to a Stem-like State. Cancer Res. 2015;75:2553-65
57. Puthdee N, Khramchantuk S, Nuwongsri P. LIN28B Enhanced STAT3 Signaling Regulates Inflammatory Response and Chemotherapeutic Resistance in Cholangiocytes. Asian Pac J Cancer Prev. 2021;22:3671-8
58. Degirmenci U, Wang M, Hu J. Targeting Aberrant RAS/RAF/MEK/ERK Signaling for Cancer Therapy. Cells. 2020 9
59. Kong B, Wu W, Cheng T, Schlitter AM, Qian C, Bruns P. et al. A subset of metastatic pancreatic ductal adenocarcinomas depends quantitatively on oncogenic Kras/Mek/Erk-induced hyperactive mTOR signalling. Gut. 2016;65:647-57
60. Sugawara H, Yasoshima M, Katayanagi K, Kono N, Watanabe Y, Harada K. et al. Relationship between interleukin-6 and proliferation and differentiation in cholangiocarcinoma. Histopathology. 1998;33:145-53
61. Komori J, Marusawa H, Machimoto T, Endo Y, Kinoshita K, Kou T. et al. Activation-induced cytidine deaminase links bile duct inflammation to human cholangiocarcinoma. Hepatology. 2008;47:888-96
62. Herberger B, Berger W, Puhalla H, Schmid K, Novak S, Brandstetter A. et al. Simultaneous blockade of the epidermal growth factor receptor/mammalian target of rapamycin pathway by epidermal growth factor receptor inhibitors and rapamycin results in reduced cell growth and survival in biliary tract cancer cells. Mol Cancer Ther. 2009;8:1547-56
63. Papoutsoglou P, Louis C, Coulouarn C. Transforming Growth Factor-Beta (TGFβ) Signaling Pathway in Cholangiocarcinoma. Cells. 2019 8
64. Hoshino Y, Nishida J, Katsuno Y, Koinuma D, Aoki T, Kokudo N. et al. Smad4 Decreases the Population of Pancreatic Cancer-Initiating Cells through Transcriptional Repression of ALDH1A1. Am J Pathol. 2015;185:1457-70
65. Kahlert C, Bergmann F, Beck J, Welsch T, Mogler C, Herpel E. et al. Low expression of aldehyde dehydrogenase 1A1 (ALDH1A1) is a prognostic marker for poor survival in pancreatic cancer. BMC Cancer. 2011;11:275
66. Derynck R, Turley SJ, Akhurst RJ. TGFβ biology in cancer progression and immunotherapy. Nat Rev Clin Oncol. 2021;18:9-34
67. You H, Ding W, Rountree CB. Epigenetic regulation of cancer stem cell marker CD133 by transforming growth factor-beta. Hepatology. 2010;51:1635-44
68. Buijs JT, van der Horst G, van den Hoogen C, Cheung H, de Rooij B, Kroon J. et al. The BMP2/7 heterodimer inhibits the human breast cancer stem cell subpopulation and bone metastases formation. Oncogene. 2012;31:2164-74
69. Sato Y, Harada K, Itatsu K, Ikeda H, Kakuda Y, Shimomura S. et al. Epithelial-mesenchymal transition induced by transforming growth factor-{beta}1/Snail activation aggravates invasive growth of cholangiocarcinoma. Am J Pathol. 2010;177:141-52
70. Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20:69-84
71. Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY. et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704-15
72. Boulter L, Guest RV, Kendall TJ, Wilson DH, Wojtacha D, Robson AJ. et al. WNT signaling drives cholangiocarcinoma growth and can be pharmacologically inhibited. J Clin Invest. 2015;125:1269-85
73. Monga SP. β-Catenin Signaling and Roles in Liver Homeostasis, Injury, and Tumorigenesis. Gastroenterology. 2015;148:1294-310
74. Fernández-Barrena MG, Perugorria MJ, Banales JM. Novel lncRNA T-UCR as a potential downstream driver of the Wnt/β-catenin pathway in hepatobiliary carcinogenesis. Gut. 2017;66:1177-8
75. Yoon JH, Canbay AE, Werneburg NW, Lee SP, Gores GJ. Oxysterols induce cyclooxygenase-2 expression in cholangiocytes: implications for biliary tract carcinogenesis. Hepatology. 2004;39:732-8
76. Komichi D, Tazuma S, Nishioka T, Hyogo H, Chayama K. Glycochenodeoxycholate plays a carcinogenic role in immortalized mouse cholangiocytes via oxidative DNA damage. Free Radic Biol Med. 2005;39:1418-27
77. Alpini G, Ueno Y, Glaser SS, Marzioni M, Phinizy JL, Francis H. et al. Bile acid feeding increased proliferative activity and apical bile acid transporter expression in both small and large rat cholangiocytes. Hepatology. 2001;34:868-76
78. Yoshino J, Akiyama Y, Shimada S, Ogura T, Ogawa K, Ono H. et al. Loss of ARID1A induces a stemness gene ALDH1A1 expression with histone acetylation in the malignant subtype of cholangiocarcinoma. Carcinogenesis. 2020;41:734-42
79. Moreb JS, Gabr A, Vartikar GR, Gowda S, Zucali JR, Mohuczy D. Retinoic acid down-regulates aldehyde dehydrogenase and increases cytotoxicity of 4-hydroperoxycyclophosphamide and acetaldehyde. J Pharmacol Exp Ther. 2005;312:339-45
80. Coyle KM, Maxwell S, Thomas ML, Marcato P. Profiling of the transcriptional response to all-trans retinoic acid in breast cancer cells reveals RARE-independent mechanisms of gene expression. Sci Rep. 2017;7:16684
81. Sullivan JP, Spinola M, Dodge M, Raso MG, Behrens C, Gao B. et al. Aldehyde dehydrogenase activity selects for lung adenocarcinoma stem cells dependent on notch signaling. Cancer Res. 2010;70:9937-48
82. Li Z, Xiang Y, Xiang L, Xiao Y, Li F, Hao P. ALDH maintains the stemness of lung adenoma stem cells by suppressing the Notch/CDK2/CCNE pathway. PLoS One. 2014;9:e92669
83. Sun QL, Sha HF, Yang XH, Bao GL, Lu J, Xie YY. Comparative proteomic analysis of paclitaxel sensitive A549 lung adenocarcinoma cell line and its resistant counterpart A549-Taxol. J Cancer Res Clin Oncol. 2011;137:521-32
84. Tanei T, Morimoto K, Shimazu K, Kim SJ, Tanji Y, Taguchi T. et al. Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential Paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin Cancer Res. 2009;15:4234-41
85. Croker AK, Allan AL. Inhibition of aldehyde dehydrogenase (ALDH) activity reduces chemotherapy and radiation resistance of stem-like ALDHhiCD44⁺ human breast cancer cells. Breast Cancer Res Treat. 2012;133:75-87
86. Attia YM, El-Kersh DM, Ammar RA, Adel A, Khalil A, Walid H. et al. Inhibition of aldehyde dehydrogenase-1 and p-glycoprotein-mediated multidrug resistance by curcumin and vitamin D3 increases sensitivity to paclitaxel in breast cancer. Chem Biol Interact. 2020;315:108865
87. Chen MH, Weng JJ, Cheng CT, Wu RC, Huang SC, Wu CE. et al. ALDH1A3, the Major Aldehyde Dehydrogenase Isoform in Human Cholangiocarcinoma Cells, Affects Prognosis and Gemcitabine Resistance in Cholangiocarcinoma Patients. Clin Cancer Res. 2016;22:4225-35
88. El Amrani M, Corfiotti F, Corvaisier M, Vasseur R, Fulbert M, Skrzypczyk C. et al. Gemcitabine-induced epithelial-mesenchymal transition-like changes sustain chemoresistance of pancreatic cancer cells of mesenchymal-like phenotype. Mol Carcinog. 2019;58:1985-97
89. Jin W. Role of JAK/STAT3 Signaling in the Regulation of Metastasis, the Transition of Cancer Stem Cells, and Chemoresistance of Cancer by Epithelial-Mesenchymal Transition. Cells. 2020 9
90. Liu K, Geng Y, Wang L, Xu H, Zou M, Li Y. et al. Systematic exploration of the underlying mechanism of gemcitabine resistance in pancreatic adenocarcinoma. Mol Oncol. 2022;16:3034-51
91. Han QL, Zhou YH, Lyu Y, Yan H, Dai GH. Effect of ribonucleotide reductase M1 expression on overall survival in patients with pancreatic cancer receiving gemcitabine chemotherapy: A literature-based meta-analysis. J Clin Pharm Ther. 2018;43:163-9
92. Huang P, Chubb S, Hertel LW, Grindey GB, Plunkett W. Action of 2',2'-difluorodeoxycytidine on DNA synthesis. Cancer Res. 1991;51:6110-7
93. Aughton K, Elander NO, Evans A, Jackson R, Campbell F, Costello E. et al. hENT1 Predicts Benefit from Gemcitabine in Pancreatic Cancer but Only with Low CDA mRNA. Cancers (Basel). 2021 13
94. Bergman AM, Pinedo HM, Peters GJ. Determinants of resistance to 2',2'-difluorodeoxycytidine (gemcitabine). Drug Resist Updat. 2002;5:19-33
95. Duong HQ, Yi YW, Kang HJ, Bae I, Jang YJ, Kwak SJ. et al. Combination of dasatinib and gemcitabine reduces the ALDH1A1 expression and the proliferation of gemcitabine-resistant pancreatic cancer MIA PaCa-2 cells. Int J Oncol. 2014;44:2132-8
96. Jaidee R, Kukongviriyapan V, Senggunprai L, Prawan A, Jusakul A, Laphanuwat P. et al. Inhibition of FGFR2 enhances chemosensitivity to gemcitabine in cholangiocarcinoma through the AKT/mTOR and EMT signaling pathways. Life Sci. 2022;296:120427
97. Chung SY, Hung YP, Pan YR, Chang YC, Wu CE, Hsu DS. et al. Ruxolitinib Combined with Gemcitabine against Cholangiocarcinoma Growth via the JAK2/STAT1/3/ALDH1A3 Pathway. Biomedicines. 2021 9
98. Guo Y, Xiao Y, Guo H, Zhu H, Chen D, Wang J. et al. The anti-dysenteric drug fraxetin enhances anti-tumor efficacy of gemcitabine and suppresses pancreatic cancer development by antagonizing STAT3 activation. Aging (Albany NY). 2021;13:18545-63
99. Kawamoto M, Umebayashi M, Tanaka H, Koya N, Nakagawa S, Kawabe K. et al. Combined Gemcitabine and Metronidazole Is a Promising Therapeutic Strategy for Cancer Stem-like Cholangiocarcinoma. Anticancer Res. 2018;38:2739-48
100. Maréchal R, Bachet JB, Mackey JR, Dalban C, Demetter P, Graham K. et al. Levels of gemcitabine transport and metabolism proteins predict survival times of patients treated with gemcitabine for pancreatic adenocarcinoma. Gastroenterology. 2012;143:664-74.e6
101. Sierzega M, Pach R, Kulig P, Legutko J, Kulig J. Prognostic Implications of Expression Profiling for Gemcitabine-Related Genes (hENT1, dCK, RRM1, RRM2) in Patients With Resectable Pancreatic Adenocarcinoma Receiving Adjuvant Chemotherapy. Pancreas. 2017;46:684-9
102. Martinez-Becerra P, Vaquero J, Romero MR, Lozano E, Anadon C, Macias RI. et al. No correlation between the expression of FXR and genes involved in multidrug resistance phenotype of primary liver tumors. Mol Pharm. 2012;9:1693-704
103. Wang C, Ye H, Zhang L, Cheng Y, Xu S, Zhang P. et al. Enhanced expression of ten-eleven translocation 1 reverses gemcitabine resistance in cholangiocarcinoma accompanied by a reduction in P-glycoprotein expression. Cancer Med. 2019;8:990-1003
104. Vena F, Jia R, Esfandiari A, Garcia-Gomez JJ, Rodriguez-Justo M, Ma J. et al. MEK inhibition leads to BRCA2 downregulation and sensitization to DNA damaging agents in pancreas and ovarian cancer models. Oncotarget. 2018;9:11592-603
105. Dembinski JL, Krauss S. Characterization and functional analysis of a slow cycling stem cell-like subpopulation in pancreas adenocarcinoma. Clin Exp Metastasis. 2009;26:611-23
106. Funamizu N, Kamata Y, Misawa T, Uwagawa T, Lacy CR, Yanaga K. et al. Hydroxyurea decreases gemcitabine resistance in pancreatic carcinoma cells with highly expressed ribonucleotide reductase. Pancreas. 2012;41:107-13
107. Bai Z, Guo Z, Liu J, Chen YA, Lu Q, Zhang P. et al. Lapatinib Suppresses HER2-Overexpressed Cholangiocarcinoma and Overcomes ABCB1- Mediated Gemcitabine Chemoresistance. Front Oncol. 2022;12:860339
108. Guo H, Liu F, Yang S, Xue T. Emodin alleviates gemcitabine resistance in pancreatic cancer by inhibiting MDR1/P-glycoprotein and MRPs expression. Oncol Lett. 2020;20:167
Author contact
![]() Corresponding authors: Professor Chuang Peng, Department of Hepatobiliary Surgery, Hunan Provincial People's Hospital (The First Affiliated Hospital of Hunan Normal University); 61 Jiefang Road, Changsha, Hunan 410005, P.R. China. Professor Yujing Zhang, Central Laboratory of Hunan Provincial People's Hospital (The First Affiliated Hospital of Hunan Normal University), Changsha, 410015, China.Key Laboratory of Molecular Epidemiology of Hunan Province, School of Medicine, Hunan Normal University, Changsha, China. E-mail: pengchuangcncom; E-mail: zhangyujingedu.cn.
Corresponding authors: Professor Chuang Peng, Department of Hepatobiliary Surgery, Hunan Provincial People's Hospital (The First Affiliated Hospital of Hunan Normal University); 61 Jiefang Road, Changsha, Hunan 410005, P.R. China. Professor Yujing Zhang, Central Laboratory of Hunan Provincial People's Hospital (The First Affiliated Hospital of Hunan Normal University), Changsha, 410015, China.Key Laboratory of Molecular Epidemiology of Hunan Province, School of Medicine, Hunan Normal University, Changsha, China. E-mail: pengchuangcncom; E-mail: zhangyujingedu.cn.

 Global reach, higher impact
Global reach, higher impact