Impact Factor
ISSN: 1837-9664
J Cancer 2024; 15(15):4902-4921. doi:10.7150/jca.94530 This issue Cite
Research Paper
Preliminary study on the active substances and cellular pathways of lactic acid bacteria for colorectal cancer treatment
1. Second Department of General Surgery, First People' s Hospital of Yunnan Province, Kunming 650032, Yunnan, China.
2. Second Department of General Surgery, Affiliated Hospital of Kunming University of Science and Technology, Kunming 650032, Yunnan, China.
3. Faculty of Life Science and Technology, Kunming University of Science and Technology, Kunming 650500, Yunnan, China.
4. Institute of Neuroscience, Kunming Medical University, Kunming 650500, Yunnan, China.
# Si-Hui Zhao, Shu-Ming Zhang, and Jin-Wei Yang contributed equally to this work.
* Yun Shang and Xiao-Ran Li contributed equally to this work.
Received 2024-1-21; Accepted 2024-6-30; Published 2024-7-16
Abstract
Colorectal cancer (CRC) is a common malignant tumor and is one of the three most common cancers worldwide. Traditional surgical treatment, supplemented by chemotherapy and radiotherapy, has obvious side effects on patients. Immunotherapy may lead to some unpredictable complications. Low introduction rate and high cost are some of the problems of gene therapy, so finding a safe, reliable and least toxic treatment method became the main research direction for this study. Lactic acid bacteria and their metabolites are widely used in functional foods or as adjuvant therapies for various diseases because they are safe to eat and have no adverse reactions. Research has shown that lactic acid bacteria and their metabolites play an auxiliary therapeutic role in colorectal cancer mainly by improving the intestinal flora composition, inhibiting the growth of pathogenic bacteria and inhibiting the proliferation of cancer cells. It is now widely believed that the substances that probiotics such as lactic acid bacteria exert anti-cancer effects are mainly secondary metabolites such as butyric acid. Lb. plantarum AY01 isolated from fermented food has good anti-cancer ability, and its main anti-cancer substance is 2'-deoxyinosine. Through flow cytometry detection, it was found that Lb. plantarum AY01 can block cell proliferation in the S phase. In addition, Lb. plantarum AY01 culture reduces the sensitivity of mice to colitis-associated CRC induced by azoxymethane (AOM)/dextran sulfate sodium salt (DSS) and exhibits the occurrence and promotion of tumors. According to transcriptome analysis, Lb. plantarum AY01 may induce apoptosis of colorectal cancer cells by activating the p38 MAPK pathway. This experiment provided possibilities for the treatment of CRC.
Keywords: CRC, Lb. plantarum AY01, 2'-Deoxyinosine
Introduction
Colorectal cancer (CRC) is one of the most common cancer types, with the third highest incidence rate and the fourth highest mortality rate of all cancer types worldwide [1]. Each year, 1.3 million new CRC cases occur, threatening 694,000 lives worldwide [2]. The occurrence and development of CRC are associated with inheritance, the environment, daily diets and other factors [3-5]. However, only 20-25% of cases are related to genetic factors [6]. Research has shown that improper eating habits, including the consumption of fat, alcohol and red meat, are the most important risk factors for CRC [7, 8]. An unhealthy intestinal state is usually accompanied by dysbacteriosis and disturbance of normal intestinal flora [9]. The intestinal microbial community composition of CRC patients changed significantly compared with that of healthy people [10-13]. Previous investigations have suggested that probiotics can improve the intestinal flora. Probiotics can alter the intestinal fora by reducing intestinal pH, competing for nutrition and secreting antibacterial compounds [14-16]. Clavijo et al. [17] reviewed the role of microbiota in the gastrointestinal tract of broilers and discussed the important role of probiotics in the control of pathogens. Le Leu et al. [18] examined whether the symbiotic combination of Bifidobacterium lactis (B. lactis) and resistant starch could change gut microbial composition. We found that yogurt fermented with different probiotics significantly changed the intestinal flora composition of 144 mice with slow transit constipation and improved defecation time and intestinal health [19].
At present, chemotherapy, radiotherapy and surgery are still the main methods for the treatment of CRC; however, their clinical utility remains hampered by their toxicity and side effects [20]. Surgery is the main treatment for CRC. However, the possibility of postsurgical trauma, decreased intestinal mucosal barrier function, intestinal flora disturbance, increased systemic inflammation, decreased immune function, and the risk of postoperative infection is also greater [21]. Therefore, enhancing the immune response and improving the gut microbiota are effective strategies to prevent CRC [22]. According to data from the World Health Organization and the Food and Agriculture Organization of the United Nations, a proper dose of probiotics has a beneficial effect on human health. Probiotics are retained in the body without any side effects, which has been recognized by the public [23]. Studies have shown that probiotics are closely connected with various health benefits, including the conditioning of the intestinal microflora, suppression of excess allergic responses and tumor suppressive effects [24, 25]. Dos Reis et al. [26] reviewed that treatment with probiotics for CRC could improve the composition of the intestinal microbial community, change the metabolic activity of intestinal microorganisms, break down carcinogens and reduce their carcinogenic activity, produce compounds with anticancer activity, enhance autoimmunity, strengthen the intestinal barrier, change host physiology, inhibit cancer cell proliferation and induce cancer cell apoptosis. Zhang et al. [27] used probiotics in postoperative CRC patients and found that the recurrence rate of the experimental group using probiotics was 10%, while that of the control group without probiotics was 33.3%. Mohania et al. [28] showed that the proliferation of cancer cells could be inhibited either by probiotics alone or in combination with cancer drugs in CRC mice. Many studies have confirmed that lactic acid bacteria (LAB) play an important role in preventing postoperative abdominal infection or intestinal complications [29]. Some researchers have attempted to find solutions from feces, but gut microbiota may pose potential risks, while probiotics from food sources are naturally safe. Ishikawa et al. [30] discovered that the occurrence rate of tumors with a grade of moderate atypia or higher was significantly lower in the group administered Lactiplantibacillus casei (Lb. casei). Konishi et al. [31] showed that the culture supernatant of Lb. casei ATCC334 has a strong tumor-suppressive effect on colon cancer cells. Sadeghi et al. [32] found that heat-inactivated Lactiplantibacillus plantarum (Lb. plantarum A7), Lactiplantibacillus rhamnosus GG (Lb. rhamnosus GG) and aseptic cell culture supernatant had good inhibitory effects on Caco-2 and HT-29 cells.
Studies have shown that LAB, especially Lb. plantarum, and their metabolites can inhibit CRC. Hu et al. [33] showed that oral administration of Lb. plantarum in BALB/c mice inhibited the growth of CT26 cells and prolonged the survival time of tumor-bearing mice by increasing the effector function of CD8+ and infiltration of natural killer (NK) cells into tumor tissues, upregulating IFN-γ and promoting Th1-type CD4+ differentiation. Faghfoori et al. [34] concluded that the culture supernatant of Lb. plantarum showed significant cytotoxic effects on Caco-2 cancer cells. Mushtaq et al. [35] observed that the water-soluble extract (WSE) of kalari cheese with added Lb. plantarum exhibited profound cell proliferation inhibition against MCF-7, HCT-116, IMR-32, and HEK-T cancer lines. These studies have shown that host microbial interactions bring health benefits to the host by regulating specific molecular mechanisms of symbiotic bacteria and probiotics. However, the antitumorigenic molecules produced by commensal bacteria and probiotics have not been identified. However, the antitumorigenic molecules involved remain unclear.
Therefore, this study screened ten strains of lactic acid bacteria isolated from fermented foods to test their resistance to colorectal cancer to find the most effective lactic acid bacteria for colorectal cancer treatment. Transcriptome analysis of colorectal cancer cells infected with freeze-dried powder of selected lactic acid bacteria was performed. At the same time, the lactic acid bacteria were screened for anti-colorectal cancer substances to obtain lactic acid bacteria with good anti-colorectal cancer ability and clarify the underlying mechanism.
Materials and Methods
Strains and LAB Culture
Ten strains of lactic acid bacteria were isolated from traditional fermented foods in Yunnan Province (Table 1).
Source of strain
| Source of strain | Source |
|---|---|
| Lb. plantarum QB3-1 | fermented soya bean |
| Lb. plantarum QB3-3 | fermented soya bean |
| Lb. plantarum YM4-3 | fermented soya bean |
| Lb. fermentum JSL8-2 | fermented soya bean |
| Lb. plantarum AY01 | fermented goat milk cheese |
| Lb. latics BZ06 | fermented goat milk cheese |
| Lb. casei AS02 | fermented goat milk cheese |
| Lb. brevis 3 M | fermented bean curd |
| Lb. curvatus 6 M | fermented bean curd |
| P. pentosus 2-4 L | sauerkraut |
Ten strains were activated in 5 mL Man-Rogosa-Sharpe (MRS). The strains were cultured at 37 ℃ for 24 hours. Ten strains were inoculated into 100 mL of MRS medium for expansion cultivation (inoculation concentration was 105 CFU/mL, cultivation time was 72 h). After 72 hours of culture, the LAB culture solution was centrifuged for 15 min at 10 000 g and 4 °C. After centrifugation, the supernatant (metabolite of LAB) was collected, and the precipitate was discarded. The culture supernatant of LAB was frozen to lyophilized powder using a laboratory vacuum freeze-drying machine SJIN-10 N (Ningbo Shuangjia Instrument Co., Ltd, China). The lyophilized powder was dissolved in RPMI 1640 (BI, Israel) medium without fetal bovine serum (FBS, BI, Israel) at a concentration of 0.5 g/mL. After dissolution, the solution was filtered through a 0.22 μm filter (Biosharp, China) and then stored in the refrigerator at -20 °C.
Cell Assay
Cell Culture
The human colon cancer cell lines HT-29, HCT-116, Caco-2, and SW-620 were all purchased from Kunming Institute of Zoology, Chinese Academy of Sciences. Cell lines were grown in high-glucose Roswell Park Memorial Institute (RPMI) 1640 (HT-29) or Dulbecco's Modified Eagle's Medium (DMEM, BI, Israel)) (HCT-116, Caco-2 and SW-620) supplemented with 10% (v*v-1) FBS, 2 mM L-glutamine, 50 IU/mL penicillin (BI, Israel) and 50 μg/mL streptomycin (BI, Israel) in a humidified atmosphere containing 5% CO2.
MTT Assay
Cells were plated in 96-well plates at a density of 105 cells/well. The cells were completely attached (approximately 12 h), and 100 μL of lyophilized powder solution of LAB culture supernatant at different concentrations was added (concentrations of 250 mg/mL, 125 mg/mL, 10 mg/mL, 5 mg/mL, and 1 mg/mL). Six biological replicates were set at each concentration. Serum-free RPMI 1640 medium of the same volume was added as a negative control. Cell growth was determined using the 3-(4,5-methylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, BBI, China) colorimetric growth assay. Every 12 hours, cell growth was determined by adding MTT solution (125 μg/well) and incubating for 4 h. Cellular MTT was resolved with dimethyl sulfoxide (DMSO, Sangon Biotech, China) and measured at 540 nm. Lb. plantarum AY01 and Lb. fermentum JSL8-2 were selected for further experiments. Lyophilized powder of Lb. plantarum AY01 and Lb. fermentum JSL8-2 culture supernatant were diluted to 1 mg/mL, 2 mg/mL, 4 mg/mL, 8 mg/mL, 16 mg/mL, 32 mg/mL, 64 mg/mL, and 128 mg/mL. In this study, 5-Fluorouracil (5-FU, Sangon Biotech, China) and RPMI 1640 medium (without serum) were used as positive and negative controls, respectively. Six biological replicates were set at each concentration. Then, 100 μL of different culture supernatant solutions, RPMI 1640 medium (without serum) and 5-FU (10 μg/mL) were used to treat HT-29, Caco-2, HCT-116 and SW-620 cells. As soon as the lyophilized powder solution was added, the HT-29 cells were cultured in a 5% CO2 incubator at 37 °C for 24 h, 36 h, 48 h, and 60 h, and the Caco-2, HCT-116 and SW-620 cells were cultured in a 5% CO2 incubator at 37 °C for 48 h. The MTT assay was used to determine the survival rate of all cancer cells.
Flow Cytometry Detection
HT-29 cells in 1 mL (106/mL) were added to 6-well plates and then cultured at 37 °C in a 5% CO2 incubator. After the cells were completely attached (approximately 12 h), 1 mL of lyophilized powder solution of Lb. plantarum AY01 and Lb. fermentum JSL8-2 culture supernatant were set at different concentrations. The final concentrations of lyophilized powder were 16 mg/mL, 32 mg/mL, and 64 mg/mL. As soon as the lyophilized powder solution was added, the HT-29 cells were cultured in a 5% CO2 incubator at 37 °C for 24 h and 48 h.
After cell culture, the cells were centrifuged at 10 000 g and 4 °C for 5 min. The supernatants were discarded, and the cells were collected. The cells were washed three times with precooled sterile PBS and centrifuged at 10 000 g and 4 °C for 5 min. Then, 1 mL 4',6-diamidino-2-phenylindole (DAPI, BBI, China) dye was added and mixed for detection by flow cytometry (Partec GmbH, Germany). The detection optical channel was FL-9. After detection, the data were analyzed by ModFit software.
Cell Apoptosis Detection
The cells were treated according to the above methods. The TransDetectTM Annexin V-FITC/PI Cell Apoptosis Detection Kit (TransGen Biotech, China) was used for detection: 1*Annexin V Binding Buffer was added into the cells to resuspend the cells, Annexin V-FITC and PI were added at 5 μL each and mixed gently, cell mixtures were placed at room temperature (20-25 °C) and reacted for 15 min without light, precooled 1*Annexin V Binding Buffer was added at 400 μL, mixed gently, and the samples were placed on ice without light. Flow cytometry was used to detect cell apoptosis within one hour. The detection channels were FL-1 and FL-3. In addition, three groups of control samples were set up: 1) negative control cells without any dye; 2) apoptotic-positive cells, single-stained Annexin V (without PI); and 3) apoptotic-positive cells, single-stained PI (without Annexin V).
Animal Modeling
Male Kunming mice were purchased from Kunming Medical University and were raised in the Animal Room of Kunming University of Science and Technology. All mice were randomly divided into cages, each containing five mice. The mice were placed in a temperature controlled room (25 ± 2 °C) with humidity (50-70%) and low noise and isolated from all other ongoing animal experiments. The mice were kept under a 12:12 h light:dark cycle. All mice were given food and water ad libitum. All animal experiments were approved by the Animal Ethics Committee of the Academy of Kunming University of Science and Technology.
In the mouse experiment, 40 mice were randomly divided into three groups: control + vehicle (n = 10), azoxymethane (AOM)/dextran sulfate sodium salt (DSS) + vehicle (n = 15), and AOM/DSS + Lb. plantarum AY01 culture supernatant (AOM/DSS + Lb. plantarum AY01) (n = 15). At 2 weeks of age (baseline week 0), mice received either an intraperitoneal injection of the carcinogen [36] AOM (10 mg/kg) (Sigma, St. Louis, MO, America) diluted in physiological saline (PS) (AOM/DSS) or PS alone (Control). Mice receiving the AOM injection were subjected to three cycles of DSS-supplemented water (36-50 kDa, MP Biomedical, America) at final concentrations of 2.5%, 2% and 2% at weeks 1, 4 and 7, respectively. Each DSS cycle lasted for a 1 weeek period. AOM was administered on the first day, and then 500 μL of physiological saline or Lb. plantarum AY01 cell supernatant (128 mg/mL) was administered via oral gavage for 10 weeks to mice daily.
Calculation of the symptom score was performed as previously described [37], taking into account stool consistency and rectal bleeding. Briefly, fresh colonic evacuates were smeared onto “Hemoccult” tape to assess the severity of diarrhea and were tested with a developer (Beckman Coulter, Brea, CA) to assess rectal bleeding. Bleeding was scored as follows: no positive detection of blood (0), detection of blood but not grossly visible (2), and gross visibility of blood (4). Diarrhea was scored as solid cylinder (0), soft cylinder and easily spreadable (2), and noncylindrical or runny (4).
Histological Analysis
Mice were euthanized by isoflurane (ChemeGen, America) at the end of 10 weeks. After sacrifice, the rectums and colons were harvested, flushed of feces and immediately fixed in 10% formalin (BBI, China) for 24 h for immunohistochemical and morphological analysis. Hematoxylin and eosin (BBI, China) staining of the rectum and colon was performed [38]. Paraffin sections of rectal and colon tissue samples were made, and then immunohistochemical tests based on Ki-67 and PCNA were performed on these samples. Immunohistochemical staining of Ki-67 and PCNA was performed as previously described [39].
Transcriptome Analysis of Lb. plantarum AY01 Anti-colorectal Cancer Cell (HT-29)
Cells with culture supernatant of L. plantarum AY01 treatment and control were collected by centrifugation. Total RNA was extracted using the TRIzol method according to the manufacturer's recommendations and assessed using an Agilent RNA 6000 Nano Kit on a Bioanalyzer 2100 (Agilent Technologies). Enriched mRNA using the Dynabeads® mRNA DIRECT™ Kit (Life Technologies) was used to construct mRNA-seq libraries using the KAPA Stranded mRNA-Seq Kit Illumina® platform according to the manufacturer's instructions.
The differentially expressed genes between the treatment and control were identified using the RUVSeq package (version 1.0.0, http://www.bioconductor.org). The DE genes had an average expression abundance of more than ten FKPM in either group with a false discovery rate less than 0.001 (FDR< 0.001) [40] and an absolute fold change (FC, treated/control) of more than three.
The hclust function of the stats package in R software (https://www.r-project.org/) was applied to perform hierarchical cluster analysis of DE genes between the treated cells and control cells. A heatmap for the DE genes was plotted using the heatmap.2 function in the gplots package.
The interaction network diagram of different genes was calculated by STRING [41], and the data with interaction coefficients greater than 0.7 were screened and imported into Cytoscape [42] to explore the network characteristics constructed by the whole transcriptome. The cell apoptosis process was obtained by GO enrichment using Clue GO [43] and Clue Pedia [44]. Cytohubba [45] was used to calculate the core genes in the interaction network (MCC algorithm). GEPIA2 (http://gepia2.cancer-pku.cn/) was used to identify core genes.
Crude Extraction and analysis of the Active Ingredients of Lb. plantarum AY01 in Anti-colorectal Cancer Cells
Lb. plantarum AY01 Supernatant Acquisition
The activated Lb. plantarum AY01 was inoculated into 10 L of fresh MRS liquid culture medium with a CFU of 106/mL and allowed to ferment at 37 ℃ for 96 hours. Subsequently, the samples were centrifuged with a large refrigerated centrifuge at 8000 rpm/min for 5 minutes, and the culture supernatant was collected.
Separation of the Lb. plantarum AY01 Crude Extraction Product
The supernatant of Lb. plantarum AY01 was separated by an MCI column and eluted with different proportions of methanol solution (the concentrations of the eluent were MeOH, MeOH:H2O=3:7, MeOH:H2O=6:4, and H2O). The separated solutions were subjected to thin layer chromatography, and elution segments with the same chromogenic point positions were merged. The combined eluent was subjected to water bath rotary evaporation (55 °C) to obtain the crude extraction product.
Liquid Chromatography‒mass Spectrometry (LC‒MS) Analysis of Crude Extraction Products from Lb. plantarum AY01
The crude extraction product of Lb. plantarum AY01 was dissolved in anhydrous ethanol and treated with 0.22 μm membrane filtration (Biosharp, China). The crude extraction products were separated using an Agilent ZORBAX SB-C18 column (4.6 × 150 mm, 5 μm) in LC‒MS, and the relevant mobile phase conditions are shown in Table 2. LC‒MS results are displayed in the Dictionary of Natural Product, NPEdia Compounds Search and The Natural Products Atlas to identify possible compounds by comparison. The toxicity of possible compounds can be calculated using the Cramer rules in Toxtree software.
High-performance Liquid Chromatography (HPLC) Analysis
The 2'-deoxyinosine standard and supernatant of Lb. plantarum AY01 was analyzed using HPLC (Agilent Technologies Inc., Santa Clara, CA). The supernatant of Lb. plantarum AY01 was filtered through a 0.45 μm filter (Biosharp, China). The supernatant was injected into an Agilent ZORBAX SB-C18 column (4.6 × 150 mm, 5 μm). The supernatant of Lb. plantarum AY01 were monitored at 220-320 nm [46]. The HPLC gradient is shown in the table 3.
Mobile phase conditions of LC‒MS
| Time(min) | C2H3N (%) | H2O(%) | Flow velocity(mL/min) |
|---|---|---|---|
| 0 | 20 | 80 | 1 |
| 15 | 22 | 78 | 1 |
| 30 | 45 | 55 | 1 |
| 35 | 55 | 45 | 1 |
| 40 | 55 | 45 | 1 |
| 50 | 100 | 0 | 1 |
| 60 | 100 | 0 | 1 |
HPLC gradient
| Time interval (min) | Gradient | Flow velocity(mL/min) |
|---|---|---|
| 0-34 | 1%(v/v) MeOH | 0.8 |
| 34-40 | 1-30%(v/v) MeOH | 0.8 |
| 40-48 | 30%(v/v) MeOH | 0.8 |
| 48-58 | 30%-1%(v/v) MeOH | 0.8 |
Statistical Analysis
Data from duplicate biological samples from the same time point were combined and expressed as the means and standard deviation (SD). The Mann-Whitney U test was used to compare groups when the variable did not show a normal distribution or did not exhibit equal variances. One-way ANOVA was used for statistical analysis for group comparisons. A p value of < 0.05 was considered significant. Analyses were performed using SigmaPlot 11.0 (Systat, Germany). One-way ANOVA was used for statistical analysis for group comparisons.
Nucleotide Sequence Accession Numbers
The transcriptomic data were deposited in the Bioproject database under accession number PRJNA643903.
Results
Screening for LAB with Tumor-Suppressive Effects
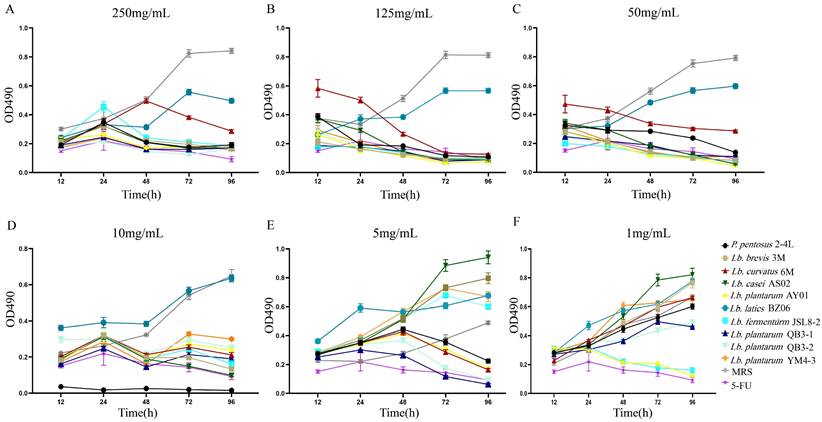
The lyophilized powder solutions of the culture supernatant of 10 strains of LAB showed different effects on HT-29 cells at different concentrations. Except for BZ06, the culture supernatants of the other nine strains of LAB showed good inhibitory effects on HT-29 cells at high concentrations (Figure 1A-D). With the decrease in the culture supernatant concentrations of nine strains of LAB, the inhibitory effect on HT-29 cells gradually decreased (Figure 1A-F). When the final concentrations were 1 mg/mL, only Lb. plantarum AY01 and Lb. fermentum JSL8-2 inhibited the proliferation of HT-29 cells (Figure 1F). Therefore, the preliminary screening results showed that Lb. plantarum AY01 and Lb. fermentum JSL8-2 had better inhibitory effects on HT-29 cells.
An MTT assay revealed that the numbers of HT-29 cells changed at different lyophilized powder concentrations, 250 mg/mL (A), 125 mg/mL (B), 50 mg/mL (C), 10 mg/mL (D), 5 mg/mL (E) and 1 mg/mL (F) of LAB culture supernatant. The error bars show the s.d. (n =6).
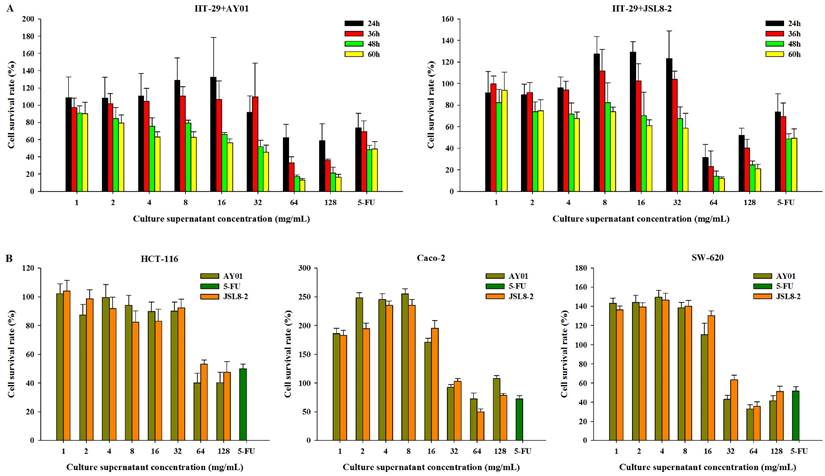
To determine the inhibitory effect of Lb. plantarum AY01 and Lb. fermentum JSL8-2 on colon cancer cells, HT-29 cells were treated with different lyophilized powder concentrations of Lb. plantarum AY01 and Lb. fermentum JSL8-2 for different times. The higher the lyophilized powder concentration of Lb. plantarum AY01 and Lb. fermentum JSL8-2 was, the lower the cell survival rate (Figure 2A), indicating a better inhibition effect of Lb. plantarum AY01 and Lb. fermentum JSL8-2 on HT-29 cells. At concentrations of 64 mg/mL and 128 mg/mL, Lb. plantarum AY01 and Lb. fermentum JSL8-2 had a good inhibitory effect (especially at 64 mg/mL). In addition, HT-29 cells treated with low concentrations of lyophilized powder solution grew faster than the blank control when the incubation time was less than 48 h, and the cell survival rate exceeded 100%. The treatment of HT-29 cells with two strains of bacteria showed the best effect at 48 h, so the final cultivation time of the subsequent experiment was set at 48 h. Lyophilized powders of Lb. plantarum AY01 and Lb. fermentum JSL8-2 also suppressed the growth of HCT-116, Caco-2 and SW-620 cells (Figure 2B). These data indicated that the secreted molecules of Lb. plantarum AY01 and Lb. fermentum JSL8-2 inhibited the growth of colon cancer cells.
The Cell Cycle and Apoptosis of HT-29 Cells
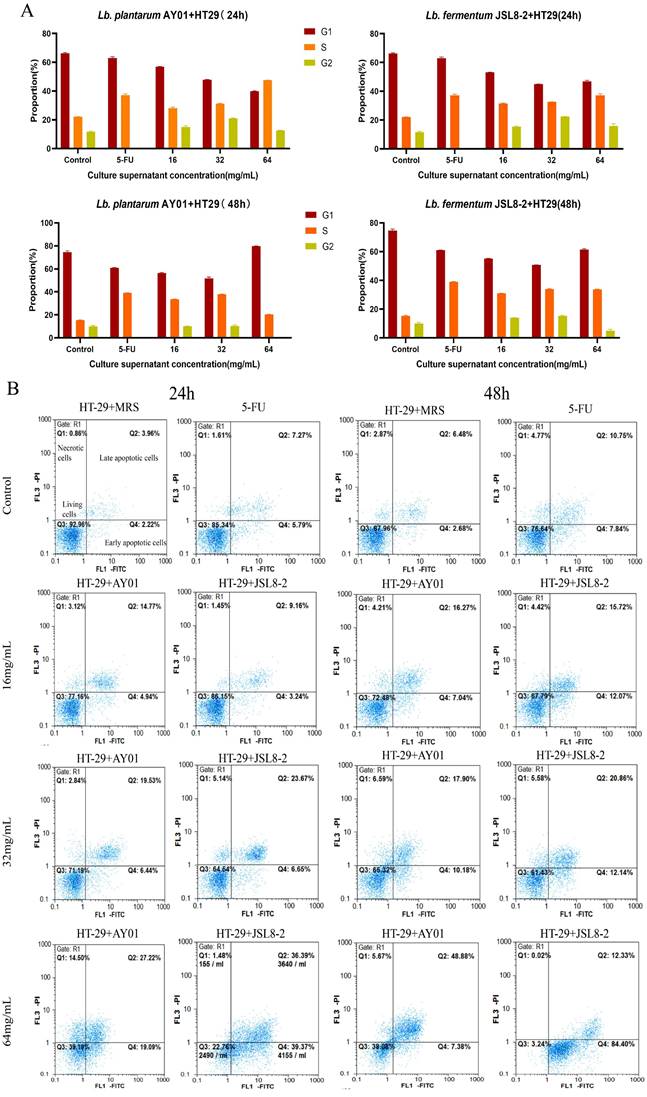
To investigate the inhibitory mechanism of Lb. plantarum AY01 and Lb. fermentum JSL8-2 on the growth of HT-29 cells, the cell cycle and apoptosis of HT-29 cells were detected by flow cytometry, MRS was used as a negative control (the cell cycle represented normal cells), and 10 μg/mL of 5-FU was used as a positive control. The cell cycle was divided into G1 phase, S phase, G2 phase and M phase. From the control group of MRS (24 h), two peaks were observed in the G1 phase and G2 phase, with proportions of 66.63% and 11.20%, respectively (Figure 3A). The 5-FU control group showed that the proportion of G2 phase had dropped to 0, and the S phase increased to 37.76%, indicating that 5-FU could block cells in S phase, which prevented cells from undergoing the complete cell cycle and the proliferation process was blocked (Figure 3A). Neither the concentration of Lb. plantarum AY01 nor Lb. fermentum JSL8-2 could cause HT-29 cell cycle arrest, and their cell cycle distribution was approximately the same as that of MRS, except that the ratios were slightly different (Figure 3A).
However, at a concentration of 64 mg/mL for 48 h, the cell cycle of HT-29 cells could block cells in S phase treated with Lb. plantarum AY01 and decreased the ratio of cells in G2 phase treated with Lb. fermentum JSL8-2 (Figure 3A), indicating that the culture supernatant of Lb. plantarum AY01 and Lb. fermentum JSL8-2 at 64 mg/mL could block the cell cycle of HT-29 cells. The proportion of S phase increased in the low concentration treatment group, and in the MTT experiment, the cell survival rate of the low concentration treatment group exceeded 100% when the incubation time was less than 48 hours. This indicates that short-term treatment with low concentrations of Lb. plantarum AY01 and Lb. fermentum JSL8-2 promotes the proliferation of HT-29, but long-term treatment still has an inhibitory effect on HT-29.
The results showed that Lb. plantarum AY01 and Lb. fermentum JSL8-2 induced apoptosis of HT-29 cells (Figure 3B). After treatment with MRS for 24 h, the proportion of living HT-29 cells was 92.96%, and the proportion of apoptotic cells was only 6.18%. The cells undergoing apoptosis were of normal growth, indicating that the cell status was good. When treated with 5-FU for 24 h, the percentage of HT-29 cells undergoing apoptosis increased from 6.18% to 13.06% compared with MRS, indicating that 5-FU can also induce apoptosis to some extent. After treatment of HT-29 cells with Lb. plantarum AY01 and Lb. fermentum JSL8-2 at concentrations of 16 mg/mL, 32 mg/mL and 64 mg/mL for 24 h, the apoptotic rates were 19.71% and 12.40%, 30.32% and 25.97%, 75.76% and 46.31%, respectively, and the inhibition rate was significantly higher than that of MRS. After treatment with MRS for 48 h, the apoptotic rate reached 9.16%, and after 48 h of treatment with 5-FU, the apoptotic rate was 18.59%, an increase of 5.53% compared with that at 24 h, indicating that the cells were naturally undergoing apoptosis under natural conditions with increasing culture time. After treatment with Lb. plantarum AY01 and Lb. fermentum JSL8-2 at concentrations of 16 mg/mL, 32 mg/mL and 64 mg/mL for 48 h, the apoptotic rate increased significantly to 27.79% and 23.31%, 33.00% and 28.08%, 96.73% and 56.26%, respectively. The induction effect was better as the concentration of the lyophilized powder solution increased. Obviously, as the treatment time increased, the effect of inducing apoptosis was also better, except for natural apoptosis. Notably, Lb. plantarum AY01 induced apoptosis of HT-29 cells significantly more than Lb. fermentum JSL8-2.
The results showed that the culture supernatant of Lb. plantarum AY01 and Lb. fermentum JSL8-2 inhibited the proliferation of HT-29 cells through cell cycle arrest and apoptosis. However, the culture supernatant of Lb. plantarum AY01 had a better inhibitory effect than Lb. fermentum JSL8-2. The best effect was achieved when HT-29 cells were treated with 64 mg mL-1 Lb. plantarum AY01 culture supernatant for 48 hours. The cell cycle of HT-29 cells was blocked in S phase, and the apoptotic rate reached 96.73%.
The survival rate of cancer cells detected by the MTT assay. (A) The survival rate of HT-29 cells treated with different culture supernatant concentrations of AY01 and JSL8-2 at different time. (B) The survival rate of HCT-116, Caco-2 and SW-620 cells treated with different culture supernatant concentrations of AY01 and JSL8-2 at 48 h. The error bars show the s.d. (n =6)
The cell cycle and apoptosis of HT-29 cells by flow cytometry. (A) The cell cycle of HT-29 cells treated with different concentrations of AY01, JSL8-2 culture supernatant and MRS (negative controls) and 5-FU (positive controls). (B) Apoptosis results of HT-29 cells treated with different concentrations of AY01, JSL8-2 culture supernatant and MRS (negative controls) and 5-FU (positive controls).
The Decrease in the Sensitivity of Mice to AOM/DSS-induced Colitis-associated CRC through Lb. plantarum AY01 Culture Supernatant
There was a main effect of Lb. plantarum AY01 culture supernatant on body weight loss, percent survival, rectal bleeding score and diarrhea score during DSS administration. In the second and third cycles, DSS caused noticeable weight loss and mortality in both AOM/DSS groups but that in the AOM/DSS + Lb. plantarum AY01 group was significantly lower than that in the AOM/DSS + Vehicle group (Figure 4B, C, P < 0.05), and by week 8, more than 50% of the mice in the AOM/DSS + Vehicle group had died. Consistent with this, a main effect of DSS on the severity of symptoms was seen during DSS administration, and the inflammation of AOM/DSS + Lb. plantarum AY01 group was alleviated. The AOM/DSS + Vehicle group exhibited greater symptom scores (including rectal bleeding score and diarrhea score) than the AOM/DSS + Lb. plantarum AY01 group (Figure 4D, E, P < 0.05). At the time of harvest after the third DSS cycle, H&E staining of colon tissue samples revealed highly differentiated adenoma in both the AOM/DSS + Vehicle and AOM/DSS + Lb. plantarum AY01 mice; remarkably, the mice in the AOM/DSS + Vehicle group had adenomas that infiltrated the muscle layer, while the mice in the AOM/DSS + Lb. plantarum AY01 group only had adenomas that infiltrated the submucosa (Figure 4F). We also counted Ki-67-stained cells and PCNA-stained cells and found a larger number of positive cells in AOM/DSS + Vehicle adenomas than in AOM/DSS + Lb. plantarum AY01 adenomas (Figure 5), suggesting that Lb. plantarum AY01 culture supernatant inhibits the proliferation of tumor cells.
These observations suggest that Lb. plantarum AY01 culture supernatant reduces the sensitivity of mice to AOM/DSS-induced colitis-associated CRC and then inhibits the occurrence and proliferation of tumors.
Transcriptomic Analysis of HT29 Cells Treated with Lb. plantarum AY01 Supernatant
To investigate the effect of the culture supernatant of Lb. plantarum AY01 on the gene expression of HT-29 cells, we examined the transcriptome analysis of HT-29 cells. Gene expression of HT-29 cells with and without Lb. plantarum AY01 culture supernatant were significantly different. The gene expression levels determined by the average RPKM values demonstrated that approximately 7000 of the total predicted genes were expressed, and there was a total of 1,215 significant differentially expressed genes. Among them, 628 genes were upregulated (HT-29 + Lb. plantarum AY01/HT-29), and 587 were downregulated. The results of GO enrichment analysis showed that the DEGs were mainly involved in the response to oxidative stress, multicellular organism growth and regulation of cell proliferation (Figure 6A-C). In response to oxidative stress, 14 genes, such as RCAN1, HMOX1, DUSP1, GCLM, and PPP1R15B, were upregulated, while 13 genes, such as TP53, NDUFS2, ERCC2, TXNIP, NAPRT, GSS, PON2, XPA, and MT-ND3, were downregulated (Figure 6A). Genes involved in multicellular organism growth were both up- and downregulated; upregulated genes included CDKN1C, SLC25A25, PLEKHA1, STIL, RARA, ZFP36L1, NCOA3, PLAG1, and KDM6A, and 10 genes were downregulated, including TP53, ERCC2, SPTBN2, TNS2, XPA, and SLITRK6 (Figure 6B). A total of 25 upregulated genes, including TNFSF9, Bcl-6, Klf4, HIF1A, NFkBIA, and JUN, and 11 downregulated genes, including STAT6, CDCA7 and MMP7, were involved in the regulation of cell proliferation (Figure 6C).
The KOBAS and DAVID databases were used for enrichment, and the list of main routes was obtained by sorting the P values from small to large (Table 4 and Table 5). It can be seen from the two tables that, at the transcription level, the anti-colorectal cancer effect of the Lb. plantarum AY01 strain mainly involves the FOXO pathway and p38 MAPK pathway. To explore the characteristics of the network constructed by the whole transcriptome, a network diagram of gene interactions and cell apoptosis was obtained by calculating and enriching genes. A gene interaction network diagram was calculated based on 1000 differentially expressed genes (Figure 7A). Two interaction networks composed of ten core genes, FBXO32, WDR34, DYNC2H1, DYNC2LI1, KLHL21, FBXO44, GAN, IFT88, IFT122 and WDR35, were generated using the cytoHubba calculation (MCC algorithm) (Figure 7B).
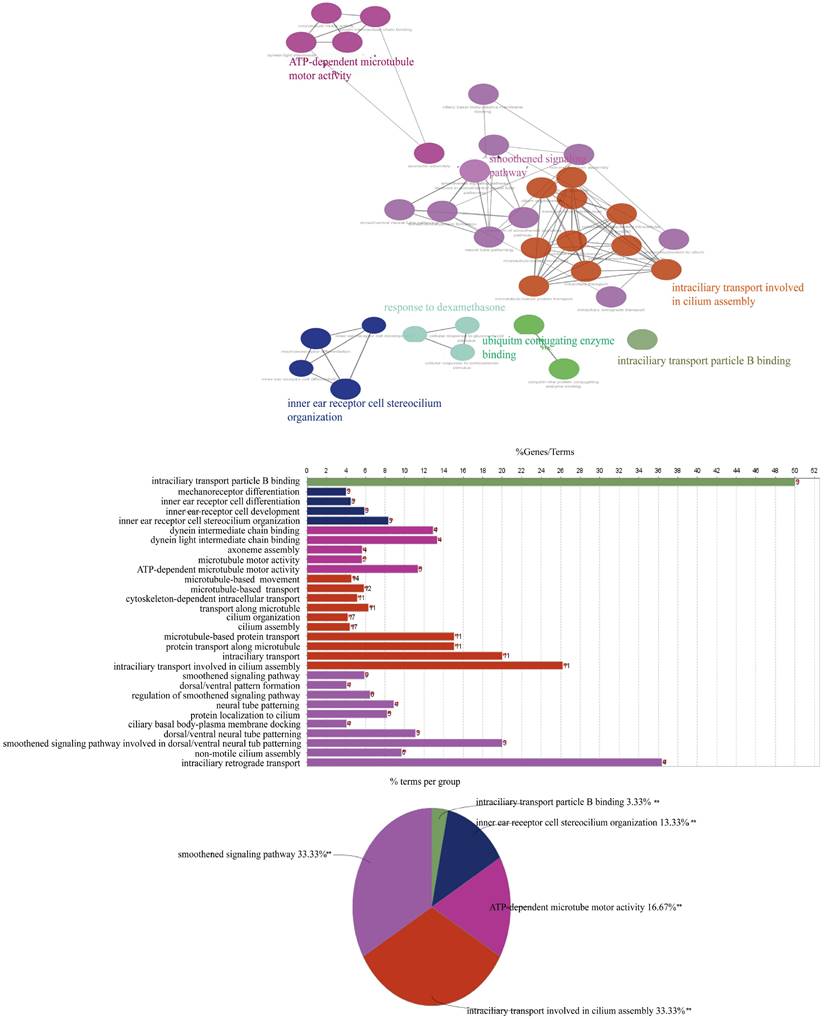
According to the annotation of GO molecular function, biological process and immune process on the basis of these two subnets, 35 points and 112 edges were obtained, which belong to five groups, among which the most important are cilia assembly and ubiquitination process (Figure 8). According to previous studies, the above two subnetworks connect downstream of the p38 MAPK pathway through the FOXO pathway, and the p38 MAPK pathway is involved in the process of apoptosis. According to the analysis of the above transcriptome data, AYO1 can activate p38 MAPK to dredge collaterals and promote the apoptosis of cancer cells.
Lb. plantarum AY01 culture supernatant reduces sensitivity to AOM/DSS-induced colitis-associated CRC. (A) Animal experiment design. Mice were treated with AOM/DSS as Material and methods described. (B) Percent body weight change. The mean changes in body weight of the three groups mice were measured at the indicated time until week 10. (C) Survival curve. Their survival was monitored until week 10 after treatment with AOM. Rectal bleeding score (D) and diarrhea score (E). Bleeding and diarrhea score were scored as Material and methods described. # Indicates statistical significance (p < 0.05) for Control + Vehicle group vs. all groups, * indicates statistical signficiance (p < 0.05) for AOM/DSS + Vehicle vs. AOM/DSS + AY01, n=7-15/group. (F) H&E staining of colon tissue samples.
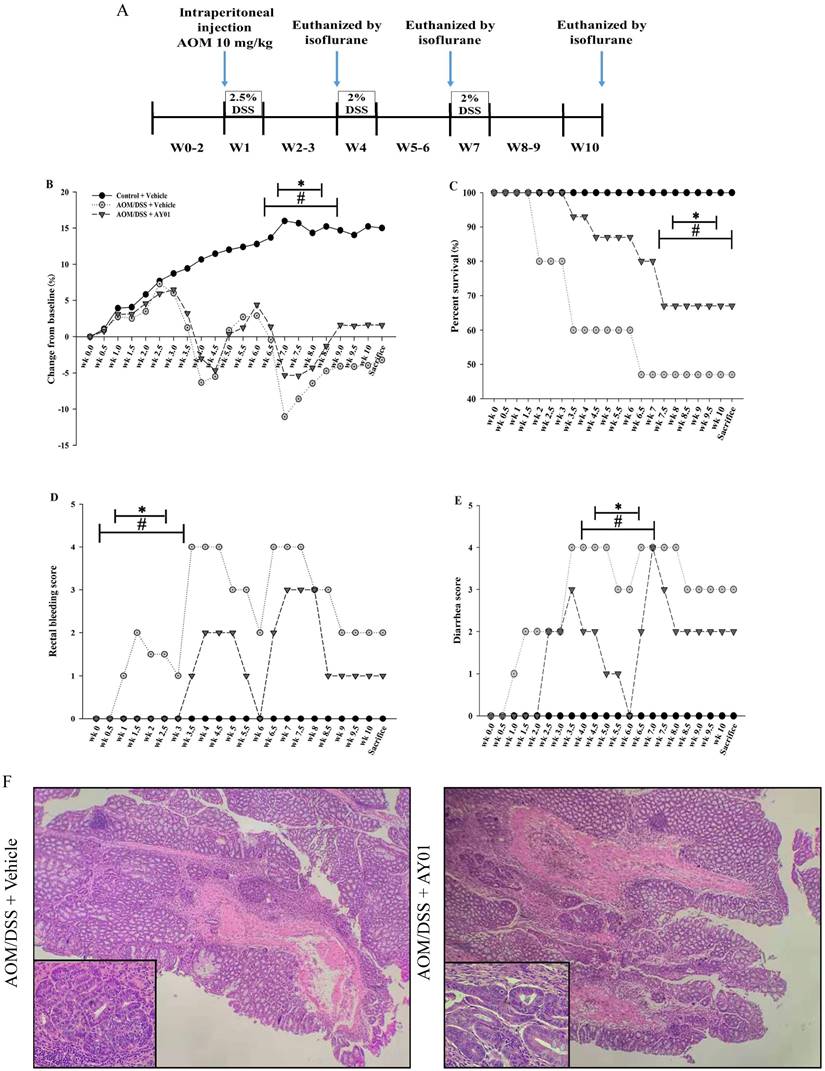
Ki-67 and PCNA immunohistochemistry staining (left panel) and percentage of Ki-67 and PCNA positive cells (right panel). * indicates statistical signficiance (p < 0.05) for AOM/DSS + Vehicle vs. AOM/DSS + AY01.
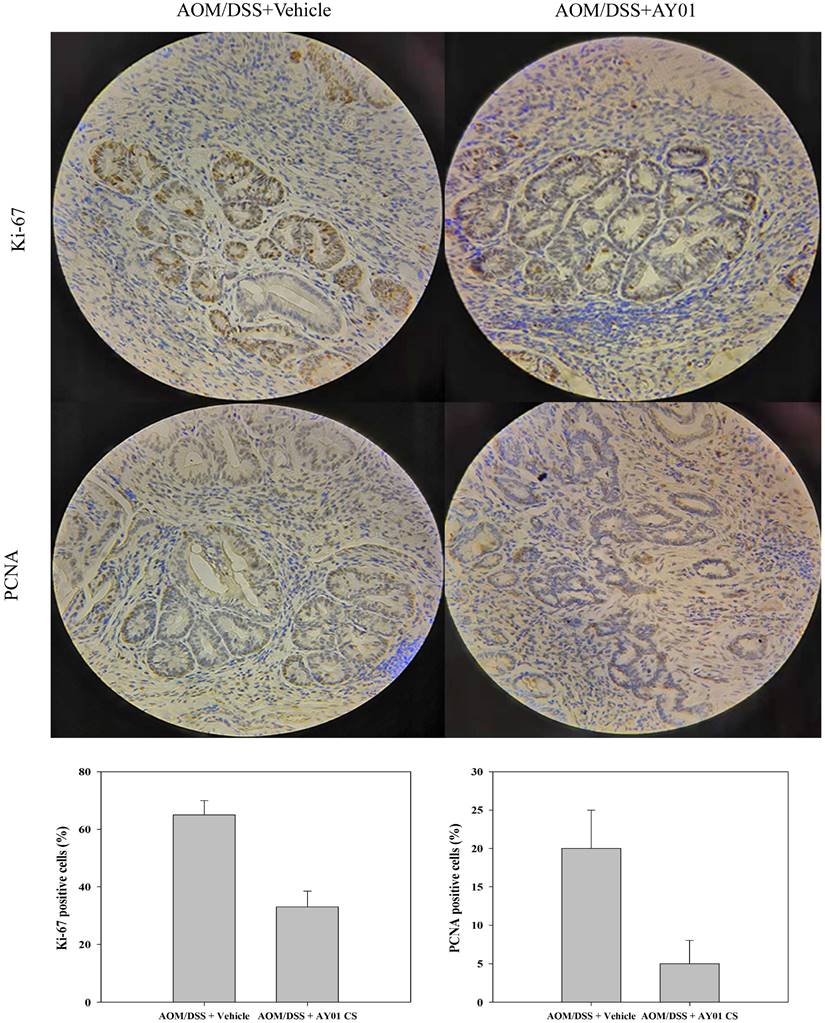
The difference of the gene expression levels in HT-29 cells with and without Lb. plantarum AY01 culture supernatant. The DEGs involved in response to oxidative stress (A), multicellular organism growth (B) and regulation of cell proliferation (C) by GO enrichment analyses. The DEGs involved in FoxO signaling pathway (D), Ferroptosis (E) and NOD-like receptor signaling pathway (F) by KEGG enrichment analyses.
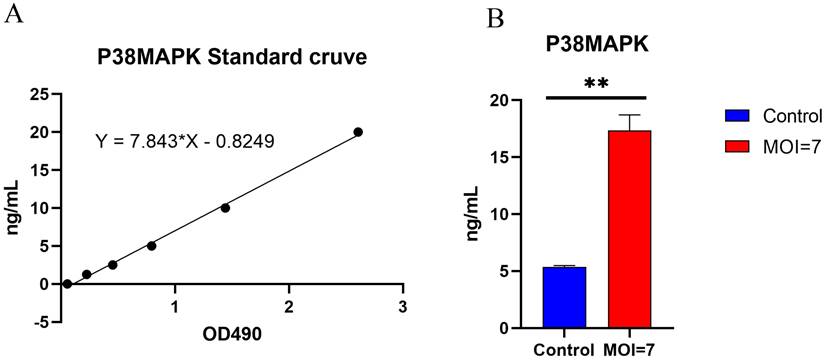
The p38 protein content in the co-cultured supernatant of AY01 and HCT116 was detected by Elisa human p38 MAPK assay kit (Figure 9). The p38 protein in the co-cultured supernatant of AY01 and HCT116 was significantly higher than that in the control group (HCT116 supernatant).
2'-Deoxyinosine is an Anti-colorectal Substance in Lb. plantarum AY01
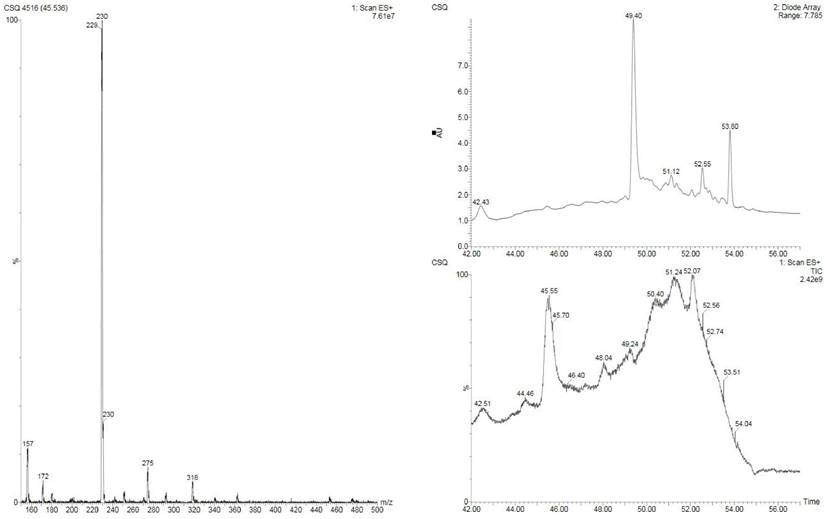
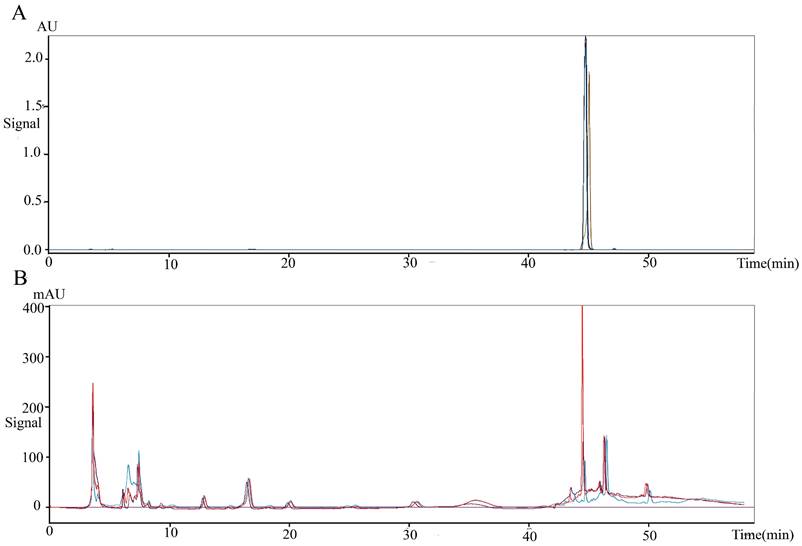
To obtain the anti-colorectal cancer active substance, the fermentation products of Lb. plantarum AY01 were preliminarily isolated. The four fractions obtained for the first time were screened by the MTT method, and all of them had certain biological activity (pH was neutral), among which the activity of the fraction eluted with 100% methanol was the best (inhibition rate was 86.8%). Subsequently, the elution section of CH2Cl2: MeOH = 20:1 was found to have the best activity by further separation of this section. The substance segment with the highest concentration was subjected to subsequent separation. According to the molecular weight displayed by LC‒MS, the final separated product was compared with compounds with similar molecular weights (Figure 10). 2'-Deoxyinosine, found after comparison, can inhibit the growth of human cancer cell lines and is related to purine nucleoside phosphorylase (PNP) deficiency. The fermentation product of Lb. plantarum AY01 was detected by HPLC, and the peak time was consistent with that of the standard sample (Figure 11). Therefore, it is preliminarily considered that the active substance of Lb. plantarum AY01 against colorectal cancer is 2'-deoxyinosine.
Top 10 KEGG pathways enriched by DAVID
| KEGG pathway | Gene number | The proportion of enriched genes in total genes (%) | P value |
|---|---|---|---|
| FoxO signaling pathway | 24 | 0.015249423 | P < 0.01 |
| Chagas disease (American trypanosomiasis) | 16 | 0.010166282 | P < 0.01 |
| MAPK signaling pathway | 28 | 0.017790994 | P < 0.01 |
| p53 signaling pathway | 12 | 0.007624712 | P < 0.01 |
| Insulin signaling pathway | 17 | 0.010801675 | P < 0.01 |
| Type II diabetes mellitus | 9 | 0.005718534 | P < 0.01 |
| TNF signaling pathway | 14 | 0.008895497 | P < 0.01 |
| Proximal tubule bicarbonate reclamation | 6 | 0.003812356 | P < 0.01 |
| Renal cell carcinoma | 10 | 0.006353926 | P < 0.01 |
| NOD-like receptor signaling pathway | 9 | 0.005718534 | P < 0.01 |
Top 10 pathways of KEGG enriched by KOBAS
| KEGG pathway | Gene number | Number of background genes | P value |
|---|---|---|---|
| FoxO signaling pathway | 24 | 134 | P < 0.01 |
| Metabolic pathways | 77 | 1243 | P < 0.01 |
| MAPK signaling pathway | 28 | 255 | P < 0.01 |
| Pathways in cancer | 32 | 397 | P < 0.01 |
| Chagas disease (American trypanosomiasis) | 16 | 104 | P < 0.01 |
| p53 signaling pathway | 13 | 69 | P < 0.01 |
| Insulin signaling pathway | 17 | 139 | P < 0.01 |
| AGE-RAGE signaling pathway in diabetic complications | 14 | 101 | P < 0.01 |
| AMPK signaling pathway | 15 | 125 | P < 0.01 |
| cAMP signaling pathway | 19 | 199 | P < 0.01 |
Discussion
In this study, Lb. plantarum AY01 and Lb. fermentum JSL8-2 were screened against human CRC cells by using the culture supernatant of LAB to test the inhibitory effect on CRC cell (HT-29, HCT-116, Caco-2 and SW-620) proliferation. Flow cytometry showed that Lb. plantarum AY01 and Lb. fermentum JSL8-2 could inhibit the proliferation of HT-29 cells by inducing apoptosis and blocking the cell cycle. However, Lb. plantarum AY01 had a more significant inhibitory effect than Lb. fermentum JSL8-2. Lb. plantarum AY01 significantly reduced the susceptibility of mice to AOM/DSS-induced colorectal cancer. Through transcriptome analysis, it was preliminarily judged that the anti-colorectal cancer effect of Lb. plantarum AY01 mainly involved the FOXO pathway and p38 MAPK pathway. The interaction network formed by ten core differentially expressed genes connects downstream of the p38 MAPK pathway through the FOXO pathway, which is involved in cell apoptosis. Lb. plantarum AY01 plays an important role in regulating cell cilia assembly and protein ubiquitination during the process of inducing apoptosis in colorectal cancer cells. The separation and extraction of anticancer substances from the supernatant of Lb. plantarum AY01 revealed that 2'-deoxyinosine was its anticancer substance. Moreover, 2'-deoxyinosine can block the growth of cancer cells in the S phase. Meanwhile, flow cytometry detected that the supernatant of Lb. plantarum AY01 also blocked the growth of colorectal cancer cells in the S phase.
Potentially related compounds based on their molecular weight
| Name | Toxicity | Role |
|---|---|---|
| (R)-Pantetheine | high | The synthetic precursor of CoA plays a crucial role in metabolism |
| (-2-Hydroxy-3-(3-indol yl) propanoic acid | high | / |
| (3R,5Z) 3-Hydroxy-5-dod ecenoic acid | low | / |
| (1R,2S)-2-Hexylcyclopro panedecanoic acid | middle | Lipid composition of Lactobacillus |
| 2'-Deoxyinosine | high | Inhibits the growth of human cancer cell lines and is related to the lack of Purine nucleoside phosphorylase (PNP) |
| Pyroglutaminylphenylala nine | / | / |
| Pyroglutaminylisoleucine | / | / |
| Pyroglutaminylleucine | / | / |
Both Lb. plantarum and Lb. fermentum have been widely used in yogurt fermentation and kimchi production, and their safety and health promotion effects have been recognized. There have been studies using different concentrations of Lb. fermentum (180 mg/kg, 360 mg/kg and 720 mg/kg) to treat nude mice transplanted with the human colorectal cancer cell line HT-29 and it was found that the subcutaneous tumor inhibition rates in nude mice reached 63%, 79%, and 83%, respectively. This study found that the inhibition rate of Lb. plantarum AY01 culture supernatant on HT-29 cells was better, reaching 96.73% after 48 h. In another in vitro experiment, Lb. fermentum RM28, which was found from fermented milk, could inhibit the proliferation of Caco-2 cells with an inhibition rate of 23% [47]. In this study, the inhibition rates of Caco-2 were 35% and 53% when treated with Lb. plantarum AY01 and Lb. fermentum JSL8-2 culture supernatants, respectively.
(A) Interaction network diagram of 1000 significantly different genes (500 significantly upregulated genes, 500 significantly downregulated genes, red indicating significant upregulation, green dots indicating significant downregulation, and blue dots indicating interaction related genes supplemented by STRING). (B) Ten core genes and their formation modules in differential gene interaction networks.
Molecular functions, biological processes, and immune annotations of the two networks in Figureure 6B. A total of 35 points and 112 edges were obtained, belonging to 5 groups, with the most important being ciliary assembly and ubiquitination processes.
Expression of human p38 MAPK protein from HCT116 after co-cultured with AY01. (A) p38 MAPK ELISA standard curve. (B) the expression of p38 MAPK protein.
LC-MS Mass Spectrometry Results of AY01 Crude Extract.
Studies have shown that probiotics use the glycolysis, NF-κB or p38 MAPK pathways to exert anticancer effects [48-50]. Through the analysis of transcriptome data, we found that Lb. plantarum AY01 may inhibit the growth of colorectal cancer cells by activating the p38 MAPK pathway. The p38 MAPK family plays an important role in complex cellular processes such as proliferation, differentiation, development, transformation, and apoptosis [51]. In the protein‒protein interaction network constructed by introducing significantly different genes into STRING, many genes that were not enriched were filled between significantly different genes, forming an interaction network. Based on the criteria of incorporating interaction coefficients greater than 0.7 into subsequent analysis, we simplified the network graph generated by STRING and imported it into Cytoscape to construct a PPI with closer and more accurate interactions. In the topology analysis of the network, it is found that the network does not conform to the power distribution; that is, we cannot find a key gene connecting the whole network. After using cytoHubba for network analysis and key node mining, two core submodules could be extracted from the entire network. The differential genes in these two modules showed that Lb. plantarum AY01 upregulated the ubiquitination process and downregulated the cell cilia assembly process. Ubiquitination regulates various complex cellular processes, including protein degradation, protein‒protein interactions, endocytosis, cell cycle progression, and activation or deactivation of substrates [52]. Therefore, any functional mutation or abnormal expression of the ubiquitination system may lead to a variety of diseases, including cancer, degenerative disease, and adaptive and innate immune-related diseases [53]. In cancer, ubiquitination may lead to activation or inhibition of tumor-related pathways. In the extraction of modules involved in the ubiquitination process, most genes showed an upregulation trend, indicating that the ubiquitination process was enhanced at the transcriptional level in colorectal cancer cells during this process [54].
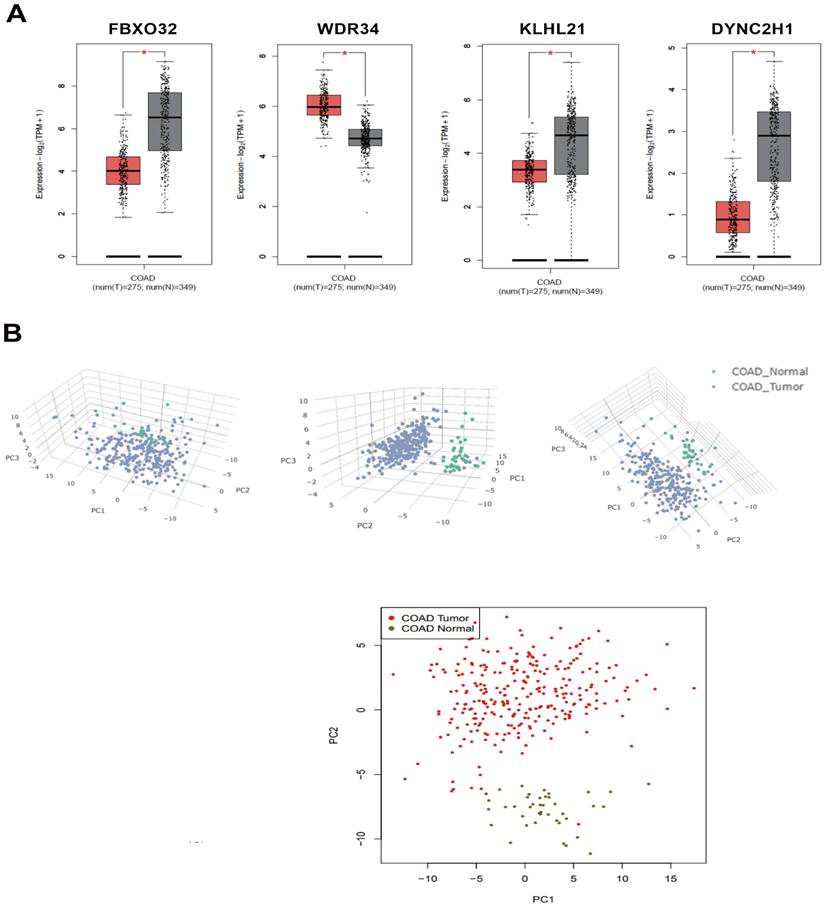
These two modules are connected by the FOXO pathway, and the p38 protein in the p38 MAPK pathway can activate the FOXO pathway. The key genes of these two modules were mined in the GEPIA2 database, and four of them were found to have differences in expression between colorectal cancer patients and normal individuals. FBXO32, KLHL21, and DYNC2H1 were significantly downregulated and WDR34 was significantly upregulated in colorectal cancer patients. In the transcriptome, DYNC2H1 and WDR34 were significantly downregulated, and FBXO32 and KLHL21 were significantly upregulated. This showed that the treatment of colorectal cancer cells with Lb. plantarum AY01 can correct the expression of FBXO32, KLHL21, and WDR34 (Figure 12). However, the p38 MAPK pathway of Lb. plantarum AY01 anti-colorectal cancer cells was only speculated by bioinformatics analysis, and further experimental confirmation is needed. At the same time, the three genes mentioned above may be the main target genes activated by Lb. plantarum AY01 in the Figureht against colorectal cancer, and further gene suppression or overexpression is needed to confirm this speculation.
(A) 2'-Deoxyinosine standard peak appears between 40-48 minutes (repeat three times). (B) The peak appears in the supernatant of AY01 between 40-48 minutes (repeat three times).
(A) There is a significant difference in the expression levels of FBXO32, WDR34, KLHL21, and DYNC2H1 between colon cancer tissue and normal colon tissue (black represents normal colon tissue, red represents colon cancer tissue). (B) PCA dimensionality reduction analysis of normal colon tissue and colon cancer tissue.
Anti-colorectal cancer substances from Lb. plantarum AY01 were isolated and purified in this study. According to the LC‒MS results, there are still many compounds present in the final separated components. When comparing compounds in the database using their molecular weights, we discovered the substance 2'-deoxyinosine. Further confirmed by HPLC, it was preliminarily determined that 2'-deoxyinosine is the main anti-colorectal cancer substance of Lb. plantarum AY01. Previous studies have shown that 2'-deoxyinosine is produced by macrophages in the body and may play an important role in inflammation-driven carcinogenesis [55]. After modifying the four-stranded oligonucleotide with 2'-deoxyinosine as an additional group, the obtained compound enhanced the inhibitory effect of the four-stranded oligonucleotide on the growth of tumor cells and induced S-phase arrest in tumor cells [56]. But no research has shown that 2'-deoxyinosine can induce apoptosis in colorectal cancer cells. When used in combination with 5-fluorouracil, 2'-deoxyinosine can enhance the sensitivity of colorectal cancer cells to 5-fluorouracil [57]. This study found that Lb. plantarum AY01 supernatant can inhibit colorectal cancer cells. There may be other substances in the supernatant of Lb. plantarum AY01 that combine with 2'-deoxyinosine to inhibit colorectal cancer cells.
This study identified the main substances and cellular pathways of Lb. plantarum AY01 in the treatment of colorectal cancer. New possibilities have been proposed in this study for the treatment and medication of colorectal cancer patients in the future.
Conclusions
Overall, most of the ten strains of lactic acid bacteria in this study had certain anti-colorectal cancer capabilities, with Lb. plantarum AY01 having the best anti-colorectal cancer effect. The optimal conditions for Lb. plantarum AY01 to inhibit colorectal cancer cells were at a concentration of 64 and time of 48 h. The anti-colorectal cancer effects of Lb. plantarum AY01 may involve the p38 MAPK pathway. Currently, it is widely believed that the anticancer substances that probiotics such as lactic acid bacteria exert are mainly secondary metabolites such as butyric acid. The main anti-colorectal cancer substance of Lb. plantarum AY01 is 2'-deoxyinosine. These studies provide a favorable basis for further research on the anti-colorectal cancer effect of lactic acid bacteria.
Acknowledgements
Funding
Yunnan First People's Hospital Clinical Medicine Center Open Project (2022LCZXKF-XH05); The Yunnan Digestive Endoscopy Clinical Medical Center Foundation for Health Commission of Yunnan Province (2022LCZXKF-XH05 of ZX2019-01-02).
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding authors.
Author Contributions
Si-Hui Zhao: Writing-original draft. Shu-Ming Zhang: Data curation, Writing-original draft, HPLC analysis. Jin-Wei Yang: Data curation. Chen-Jian Liu: Inhibition of HT-29 cell proliferation by Lb. plantarum AY01. Xue-Qin Zeng: Mice to AOM/DSS-induced colitis-associated CRC. Yuan-Lian Zhang: Transcriptome analysis. Si-Qian Chen: Lb. plantarum AY01 extraction of the main anti-colorectal cancer substances. Zhi-Min Zhao: Screening for LAB with tumor-suppressive effects. Yun-xin Xia: Inhibition of HT-29 cell proliferation by Lb. fermentum JSL8-2. Xiao-Ran Li: Writing - review & editing, Project administration. Yun Shang: Funding acquisition.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490-502
2. Chen J, Domingue JC, Sears CL. Microbiota dysbiosis in select human cancers: Evidence of association and causality. Seminars in Immunology. 2017;32:25-34
3. Barbara G, Feinle-Bisset C, Ghoshal UC, Santos J, Vanner SJ, Vergnolle N. et al. The Intestinal Microenvironment and Functional Gastrointestinal Disorders. Gastroenterology. 2016;150:1305-1318
4. Pickard JM, Zeng MY, Caruso R, Núñez G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunological Reviews. 2017;279:70-89
5. Fearon ER. Molecular Genetics of Colorectal Cancer. Annual Review of Pathology: Mechanisms of Disease. 2011;6:479-507
6. Burt RW. Colon cancer screening. Gastroenterology. 2000;119:837-53
7. Platz EA, Willett WC, Colditz GA, Rimm EB, Spiegelman D, Giovannucci E. Proportion of colon cancer risk that might be preventable in a cohort of middle-aged US men. Cancer Causes & Control. 2000;11:579-88
8. Watson AJM, Collins PD. Colon Cancer: A Civilization Disorder. Digestive Diseases. 2011;29:222-8
9. Luo P, Yang ZW, Chen B, Zhong XM. The multifaceted role of CARD9 in inflammatory bowel disease. Journal of Cellular and Molecular Medicine. 2020;24:34-9
10. Gao ZG, Guo BM, Gao RY, Zhu QC, Qin HL. Microbiota disbiosis is associated with colorectal cancer. Frontiers in Microbiology. 2015;6:20-29
11. Sobhani I, Amiot A, Le Baleur Y, Levy M, Auriault M-L, Van Nhieu JT. et al. Microbial dysbiosis and colon carcinogenesis: could colon cancer be considered a bacteria-related disease? Therapeutic advances in gastroenterology. 2013;6:215-29
12. Kostic AD, Chun EY, Robertson L, Glickman JN, Gallini CA, Michaud M. et al. Fusobacterium nucleatum Potentiates Intestinal Tumorigenesis and Modulates the Tumor-Immune Microenvironment. Cell Host & Microbe. 2013;14:207-15
13. Liu CJ, Zhang YL, Shang Y, Wu B, Yang E, Luo YY. et al. Intestinal bacteria detected in cancer and adjacent tissue from patients with colorectal cancer. Oncology Letters. 2019;17:1115-27
14. Gerritsen J, Smidt H, Rijkers GT, de Vos WM. Intestinal microbiota in human health and disease: the impact of probiotics. Genes and Nutrition. 2011;6:209-40
15. Fooks LJ, Gibson GR. Probiotics as modulators of the gut flora. British Journal of Nutrition. 2002;88:S39-S49
16. Ng SC, Hart AL, Kamm MA, Stagg AJ, Knight SC. Mechanisms of Action of Probiotics: Recent Advances. Inflammatory Bowel Diseases. 2009;15:300-10
17. Clavijo V, Flórez MJV. Non-Invited Review The gastrointestinal microbiome and its association with the control of pathogens in broiler chicken production: A review. Poultry Science. 2018;97:1006-21
18. Le Leu RK, Hu Y, Brown IL, Woodman RJ, Young GP. Synbiotic intervention of Bifidobacterium lactis and resistant starch protects against colorectal cancer development in rats. Carcinogenesis. 2010;31:246-51
19. Liu CJ, Tang XD, Yu J, Zhang HY, Li XR. Gut microbiota alterations from different Lactobacillus probiotic-fermented yoghurt treatments in slow-transit constipation. Journal of Functional Foods. 2017;38:110-8
20. Österlund P, Ruotsalainen T, Korpela R, Saxelin M, Ollus A, Valta P. et al. Lactobacillus supplementation for diarrhoea related to chemotherapy of colorectal cancer:: a randomised study. British Journal of Cancer. 2007;97:1028-34
21. McMillan DC, Canna K, McArdle CS. Systemic inflammatory response predicts survival following curative resection of colorectal cancer. British Journal of Surgery. 2003;90:215-9
22. Geier MS, Butler RN, Howarth GS. Probiotics, prebiotics and synbiotics - A role in chemoprevention for colorectal cancer? Cancer Biology & Therapy. 2006;5:1265-9
23. Bezkorovainy A. Probiotics: determinants of survival and growth in the gut. American Journal of Clinical Nutrition. 2001;73:399S-405S
24. Rowland IR, Rumney CJ, Coutts JT, Lievense LC. Effect of Bifidobacterium longum and inulin on gut bacterial metabolism and carcinogen-induced aberrant crypt foci in rats. Carcinogenesis. 1998;19:281-5
25. Kalliomäki M, Salminen S, Arvilommi H, Kero P, Koskinen P, Isolauri E. Probiotics in primary prevention of atopic disease:: a randomised placebo-controlled trial. Lancet. 2001;357:1076-9
26. dos Reis SA, da Conceiçao LL, Siqueira NP, Rosa DD, da Silva LL, Peluzio MDG. Review of the mechanisms of probiotic actions in the prevention of colorectal cancer. Nutrition Research. 2017;37:1-19
27. Zhang JW, Du P, Gao J, Yang BR, Fang WJ, Ying CM. Preoperative Probiotics Decrease Postoperative Infectious Complications of Colorectal Cancer. American Journal of the Medical Sciences. 2012;343:199-205
28. Mohania D, Kansal VK, Kruzliak P, Kumari A. Probiotic Dahi Containing Lactobacillus acidophilus and Bifidobacterium bifidum Modulates the Formation of Aberrant Crypt Foci, Mucin-Depleted Foci, and Cell Proliferation on 1,2-Dimethylhydrazine-Induced Colorectal Carcinogenesis in Wistar Rats. Rejuvenation Research. 2014;17:325-33
29. Sugawara G, Nagino M, Nishio H, Ebata T, Takagi K, Asahara T. et al. Perioperative synbiotic treatment to prevent postoperative infectious complications in biliary cancer surgery - A randomized controlled trial. Annals of Surgery. 2006;244:706-14
30. Ishikawa H, Akedo I, Otani T, Suzuki T, Nakamura T, Takeyama I. et al. Randomized trial of dietary fiber and Lactobacillus casei administration for prevention of colorectal tumors. International Journal of Cancer. 2005;116:762-7
31. Konishi H, Fujiya M, Tanaka H, Ueno N, Moriichi K, Sasajima J. et al. Probiotic-derived ferrichrome inhibits colon cancer progression via JNK-mediated apoptosis. Nature Communications. 2016;7:12365-77
32. Sadeghi-Aliabadi H, Mohammadi F, Fazeli H, Mirlohi M. Effects of Lactobacillus plantarum A7 with probiotic potential on colon cancer and normal cells proliferation in comparison with a commercial strain. Iranian Journal of Basic Medical Sciences. 2014;17:815-9
33. Hu JT, Wang CF, Ye LP, Yang WT, Huang HB, Meng F. et al. Anti-tumour immune effect of oral administration of Lactobacillus plantarum to CT26 tumour-bearing mice. Journal of Biosciences. 2015;40:269-79
34. Faghfoori Z, Gargari BP, Saber A, Seyyedi M, Fazelian S, Khosroushahi AY. Prophylactic effects of secretion metabolites of dairy lactobacilli through downregulation of ErbB-2 and ErbB-3 genes on colon cancer cells. European Journal of Cancer Prevention. 2020;29:201-9
35. Mushtaq M, Gani A, Masoodi FA. Himalayan cheese (Kalari/Kradi) fermented with different probiotic strains: In vitro investigation of nutraceutical properties. Lwt-Food Science and Technology. 2019;104:53-60
36. Enos RT, Velázquez KT, McClellan JL, Cranford TL, Nagarkatti M, Nagarkatti PS. et al. High-fat diets rich in saturated fat protect against azoxymethane/dextran sulfate sodium-induced colon cancer. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2016;310:G906-G19
37. Kaur K, Saxena A, Debnath I, O'Brien JL, Ajami NJ, Auchtung TA. et al. Antibiotic-mediated bacteriome depletion in Apc(Min)(/+) mice is associated with reduction in mucus-producing goblet cells and increased colorectal cancer progression. Cancer Medicine. 2018;7:2003-12
38. Velazquez KT, Enos RT, Carson MS, Cranford TL, Bader JE, Sougiannis AT. et al. miR155 deficiency aggravates high-fat diet-induced adipose tissue fibrosis in male mice. Physiological reports. 2017;5:13412-23
39. Liu C, Liu ED, Meng YX, Dong XM, Bi YL, Wu HW. et al. Keratin 8 reduces colonic permeability and maintains gut microbiota homeostasis, protecting against colitis and colitis-associated tumorigenesis. Oncotarget. 2017;8:96774-90
40. Benjamini Y, Hochberg Y. Controlling the false discovery rate a practical and powerful approach to multiple testing. Journal of the Royal statistical society: series B (Methodological). 1995;57(1):289-300
41. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J. et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research. 2019;47:D607-D13
42. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D. et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13:2498-504
43. Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A. et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091-3
44. Bindea G, Galon J, Mlecnik B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29:661-3
45. Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. Bmc Systems Biology. 2014;8(Suppl 4):S11-S18
46. Lim KS, Jenner A, Halliwell B. Quantitative gas chromatography mass spectrometric analysis of 2'-deoxyinosine in tissue DNA. Nat Protoc. 2006;1:1995-2002
47. Thirabunyanon M, Boonprasom P, Niamsup P. Probiotic potential of lactic acid bacteria isolated from fermented dairy milks on antiproliferation of colon cancer cells. Biotechnology Letters. 2009;31:571-6
48. Khedr OMS, El-Sonbaty SM, Moawed FSM, Kandil EI, Abdel-Maksoud BE. Lactobacillus acidophilus ATCC 4356 Exopolysaccharides Suppresses Mediators of Inflammation through the Inhibition of TLR2/STAT-3/P38-MAPK Pathway in DEN-Induced Hepatocarcinogenesis in Rats. Nutrition and Cancer-an International Journal. 2022;74:1037-47
49. Lee HJ, Lim SM, Kim DH. Lactobacillus johnsonii CJLJ103 Attenuates Scopolamine-Induced Memory Impairment in Mice by Increasing BDNF Expression and Inhibiting NF-kappa B Activation. Journal of Microbiology and Biotechnology. 2018;28:1443-6
50. Lu X, Qiao SP, Peng C, Yan WY, Xu Z, Qu JX. et al. Bornlisy Attenuates Colitis-Associated Colorectal Cancer via Inhibiting GPR43-Mediated Glycolysis. Frontiers in Nutrition. 2021;8:706382
51. Lin FH, Chang JSB, Brigman BE. Role of Mitogen-Activated Protein Kinase in Osteoblast Differentiation. Journal of Orthopaedic Research. 2011;29:204-10
52. Pickart C. M, Eddins, M. J. Ubiquitin: structures, functions, mechanisms. BIOCHIMICA ET BIOPHYSICA ACTA. 2004;1695:55-72
53. Popovic D, Vucic D, Dikic I. Ubiquitination in disease pathogenesis and treatment. Nature Medicine. 2014;20:1242-53
54. Wang Y, Lu JH, Wu QN, Jin Y, Wang DS, Chen YX. et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Molecular Cancer. 2019;18:174-192
55. Yasui M, Suenaga E, Koyama N, Masutani C, Hanaoka F, Gruz P. et al. Miscoding properties of 2'-deoxyinosine, a nitric oxide-derived DNA adduct, during translesion synthesis catalyzed by human DNA polymerases. Journal of Molecular Biology. 2008;377:1015-23
56. Fan XM, Sun LD, Li KF, Yang XT, Cai BB, Zhang YF. et al. The Bioactivity of D-/L-Isonucleoside- and 2 '-Deoxyinosine-Incorporated Aptamer AS1411s Including DNA Replication/MicroRNA Expression. Molecular Therapy-Nucleic Acids. 2017;9:218-29
57. Ciccolini J, Peillard L, Evrard A, Cuq P, Aubert C, Pelegrin A. et al. Enhanced antitumor activity of 5-fluorouracil in combination with 2′-deoxyinosine in human colorectal cell lines and human colon tumor xenografts. Clinical Cancer Research. 2000;6:1529-35
Author contact
![]() Corresponding authors: Yun Shang, E-mail: doc_shangcom; Tel.: 13888357770; Xiao-Ran Li, E-mail: starkeyrancom; Tel.: 13700602921.
Corresponding authors: Yun Shang, E-mail: doc_shangcom; Tel.: 13888357770; Xiao-Ran Li, E-mail: starkeyrancom; Tel.: 13700602921.

 Global reach, higher impact
Global reach, higher impact