Impact Factor
ISSN: 1837-9664
J Cancer 2024; 15(15):4969-4984. doi:10.7150/jca.98301 This issue Cite
Research Paper
Bidirectional Mendelian Randomization of Causal Relationship between Inflammatory Cytokines and Different Pathological Types of Lung Cancer
Department of Thoracic Surgery, Second Xiangya Hospital, Central South University, Changsha 410000, China.
Received 2024-5-10; Accepted 2024-7-7; Published 2024-7-16
Abstract
Prior research has proposed a potential association between lung cancer and inflammatory cytokines, yet the specific causal relationship remains unclear, especially across various lung cancer pathologies. This study utilized bidirectional Mendelian randomization (MR) to explore these causal connections, unveiling novel insights. Our research revealed distinctive inflammatory cytokine profiles for each subtype of lung cancer and identified potential biomarkers that could refine diagnostic and therapeutic approaches. We applied two-sample Mendelian randomization, leveraging genetic variance data from three extensive genome-wide association studies (GWAS) focusing on different lung cancer types (lung adenocarcinoma: 1590 cases and 314,193 controls of healthy individuals of European descent; lung squamous cell carcinoma: 1510 cases and 314,193 controls of European ancestry; small cell lung cancer: 717 cases and 314,193 controls of European ancestry). A separate GWAS summary on inflammatory cytokines from 8,293 healthy participants was also included. The inverse variance weighting method was utilized to examine causal relationships, with robustness confirmed through multiple sensitivity analyses, including MR-Egger, weighted median, and MR-PRESSO. Our analysis revealed that elevated levels of IL_1RA were associated with an increased risk of lung adenocarcinoma (OR: 1.29, 95% CI: 1.02-1.64, p = 0.031), while higher MCP_1_MCAF levels correlated with a decreased risk of lung squamous cell carcinoma (OR: 0.77, 95% CI: 0.61-0.98, p = 0.031). Furthermore, IL_10, IL_13, and TRAIL levels were positively associated with lung squamous cell carcinoma risk (IL_10: OR: 1.27, 95% CI: 1.06-1.53, p = 0.012; IL_13: OR: 1.15, 95% CI: 1.06-1.53, p = 0.036; TRAIL: OR: 1.15, 95% CI: 1.06-1.53, p = 0.043). No association was found between inflammatory cytokine levels and small cell lung cancer development, whereas SDF_1A and B-NGF were linked to an increased risk of this cancer type (SDF_1A: OR: 1.13, 95% CI: 1.05-1.21, p = 0.001; B-NGF: OR: 1.13, 95% CI: 1.01-1.27, p = 0.029). No significant relationship was observed between the 41 circulating inflammatory cytokines and lung adenocarcinoma or squamous cell carcinoma development. Our findings indicate distinct associations between specific inflammatory cytokines and different types of lung cancer. Elevated IL_1RA levels are a risk marker for lung adenocarcinoma, whereas higher MCP_1_MCAF levels appear protective against lung squamous cell carcinoma. Conversely, elevated levels of IL_10, IL_13, and TRAIL are linked with an increased risk of lung squamous cell carcinoma. The relationships of SDF_1A and B-NGF with small-cell lung cancer highlight the complexity of inflammatory markers in cancer development. This study provides a nuanced understanding of the role of inflammatory cytokines in lung cancer, underscoring their potential in refining diagnosis and treatment strategies.
Keywords: lung cancer, inflammatory cytokines, bidirectional Mendelian randomization
Introduction
Lung cancer remains the most prevalent cancer worldwide and is the primary cause of cancer-related deaths [1]. It is mainly classified into two types: non-small cell lung cancer (NSCLC), which constitutes approximately 85% of all cases, and small-cell lung cancer (SCLC) [2]. NSCLC is further categorized into subtypes such as lung squamous cell carcinoma, lung adenocarcinoma, and the less common large cell lung cancer [3]. Despite advances in medical research, the overall five-year survival rate for lung cancer patients is still distressingly low, under 21% [4, 5]. This low survival rate can be attributed largely to the lack of effective screening methods and the delayed diagnosis due to nonspecific symptoms, highlighting the critical need for improved diagnostic and therapeutic approaches [2].
Although smoking is a well-known risk factor, the underlying causes of lung cancer remain largely unknown. Recent advancements have shed light on the potential link between inflammation and lung cancer, with studies suggesting that inflammation within tumor tissues may play a significant role in cancer development, encompassing stages from initiation to metastasis [6]. Chronic inflammatory conditions are associated with a heightened risk of several cancers, including those of the lung, liver, and colon [7]. Inflammatory cytokines, crucial regulators of inflammation, are implicated in the progression of lung cancer. These cytokines serve dual functions: they activate immune responses to potentially curb tumor growth, but they can also facilitate the malignant transformation, proliferation, invasion, and spread of cancer cells [8]. P In particular, IL-6 and IL-8 have been identified as risk factors for lung cancer among smokers [9], with TNF-α, IL-10, and IL-17 also being considered in various contexts [6, 10]. The role of some cytokines, such as IL-17, in lung cancer remains contentious [11]. Additionally, the precise causal relationships between changes in levels of inflammatory cytokines and the development or progression of lung cancer, as well as their interaction with pharmaceutical interventions, continue to be subjects of ongoing research and debate. To date, several studies have examined the causal link between inflammatory cytokines and lung cancer. For instance, Zhao Yang et al. proposed a causal relationship between certain interleukin (IL)-1 family members and lung cancer [12], while Emmanouil Bouras et al. associated IL-18 with lung cancer [7]. However, our investigation reveals a gap in the literature concerning the exploration of causal relationships between different types of inflammatory cytokines and various pathological subtypes of lung cancer, as well as the absence of studies focusing on the potential variance in inflammatory cytokine profiles across distinct lung cancer pathologies. In our study, we employ a two-sample bidirectional Mendelian randomization (MR) approach to examine these causal associations and introduce new insights. Our objective is to delineate unique inflammatory cytokine profiles for each subtype of lung cancer and identify potential biomarkers to enhance noninvasive diagnostic and therapeutic strategies.
MR is a statistical approach that utilizes genetic variations from non-experimental data to infer causal relationships between exposures and outcomes, employing single nucleotide polymorphisms (SNPs) as genetic instrumental variables to represent those exposures [12-14]. Unlike traditional observational studies, MR leverages alleles randomly assigned during meiosis to overcome confounding factors and reverse causality issues, providing a more reliable basis for causal inference [15, 16]. In this study, we conducted a two-sample bidirectional MR analysis. We began by identifying effective genetic instrumental variables for 41 inflammatory markers using data from genome-wide association studies (GWAS). We then examined the relationship between these inflammatory markers and three different pathological types of lung cancer, further investigating the direction of causality through reverse MR analysis. This methodological approach helps lay the groundwork for the prevention of lung cancer via specific inflammatory cytokines, identifies potential therapeutic targets, and assists in differentiating between lung cancer types based on their inflammatory cytokine profiles.
Materials and Methods
Ethics and informed consent
The data for this study were obtained from four previously published GWAS. These studies were conducted in full compliance with ethical standards, having received the necessary approvals from relevant ethics committees. Additionally, all participants in these GWAS provided their informed consent in writing after being adequately informed about the research's objectives and procedures, ensuring the protection of the participants' rights, privacy, and well-being throughout the research process.
MR hypothesis
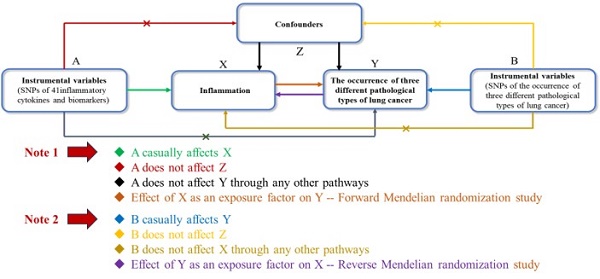
Figure 1 illustrates the structure and principles of our bidirectional two-sample MR study. The efficacy of MR analysis hinges on three fundamental assumptions:
1, Correlation: This principle requires that the genetic variants used as instruments must be significantly associated with the exposure of interest.
2, Independence: This criterion mandates that the genetic instruments must not be linked to potential confounders, ensuring their independence.
3, Exclusion restrictions: This assumption asserts that the genetic variants impact the outcome solely through their effect on the exposure, with no alternative pathways involved [18].
Bidirectional Two-Sample Mendelian Randomization Study Structure. This diagram illustrates the foundational principles necessary for the efficacy of Mendelian Randomization (MR) analysis, highlighting the critical assumptions: 1) Correlation, where genetic variants are significantly associated with the exposure; 2) Independence, ensuring genetic instruments are not linked to potential confounders; 3) Exclusion Restrictions, asserting that genetic variants influence the outcome exclusively through the exposure, without alternative pathways.
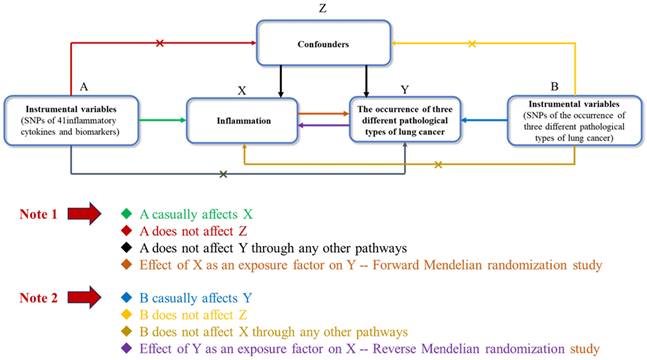
In our bidirectional study, we utilized data from four previously published GWAS to identify 41 significant SNPs associated with inflammatory cytokines and three types of lung cancer: SCLC, lung adenocarcinoma, and lung squamous cell carcinoma. Our methodological approach consisted of two primary phases. Initially, we identified genetic instrumental variables for each inflammatory cytokine to explore their potential causal relationships with SCLC, lung adenocarcinoma, and lung squamous cell carcinoma, respectively. Subsequently, we applied genetic instrumental variables related to SCLC, lung adenocarcinoma, and lung squamous cell carcinoma to examine the causal impact of these specific cancers on the levels of each circulating inflammatory cytokine. This stringent approach aligns with the principles of MR, offering a solid structure for evaluating the causal associations between inflammatory cytokines and various types of lung cancer.
Selection of genetic instrumental variables
The selection of genetic instrumental variables in this study commenced with a strict threshold set at p < 5 × 10-8, identifying SNPs strongly linked with lung cancer and inflammatory cytokines. When few SNPs were found for certain cytokines [19], a slightly relaxed threshold of p < 5 × 10-6 was applied [19]. Following this, we ensured that instrumental variables were independent by removing any with linkage disequilibrium (r² < 0.001 within 10,000 kb) [20]. Each SNP's contribution to explaining variance in the exposure was determined using the R² value, and the F-statistic was used to identify weak instrumental variable bias, with an F > 10 suggesting no significant bias [21]. Additionally, we utilized the PhenoScanner database, applying a genome-wide significance threshold (p < 5 × 10-8) to identify SNPs related to potential confounders such as smoking habits or specific outcomes. SNPs associated with such confounders were subsequently excluded from the MR analysis to ensure the integrity and reliability of the results.
Data sources
In this study, we utilized data from four publicly available GWAS summary datasets. The three GWAS summaries for lung cancer were sourced from the 10th generation Finnish database, categorizing lung cancer as LUAD, LUSC, and other types based on the international classification of diseases for oncology (ICD-10 or ICD-O-3). It is important to note that all lung cancer diagnoses underwent confirmation through histopathological analysis. The dataset for SCLC was extracted from the C3_SCLC_EXALLC section of the R10_manifest Finnish database (https://storage.googleapis.com/finngen-public-data r10/summary_stats/finngen_R10_C3_SCLC_EXALLC.gz), including 717 cases (584 male patients and 133 female patients) and 314,193 healthy controls of European descent. For lung adenocarcinoma, data were obtained from the C3_NSCLC_ADENO_EXALLC within the same database (https://storage.googleapis.com/finngen-public-data-r10/summary_stats/finngen_R10_C3_NSCLC_ADENO_EXALLC.gz), comprising 1,590 cases (1048 male patients and 542 female patients) and 314,193 controls of European origin. The data pertaining to lung squamous cell carcinoma were retrieved from the C3_NSCLC_SQUAM_EXALLC section (https://storage.googleapis.com/finngen-public-data-r10/summary_stats/finngen_R10_C3_NSCLC_SQUAM_EXALLC.gz), comprising 1,510 cases (1335 male patients and 175 female patients) and the same number of controls. Furthermore, the study integrated information on 41 circulating inflammatory cytokines from a comprehensive reanalysis of GWAS data related to serum cytokines [22] (https://doi.org/10.5523/bris.3g3i5smgghp0s2uvm1doflkx9x). This analysis amalgamated information from the Finnish Cardiovascular Risk in Young Adults study (n=1980; mean age: 37.4 years for men and 37.5 years for women) and the FINRISK survey (FINRISK1997: n=4608; average age: males -48.3 years; females -47.3 years / FINRISK2002: n=1705; mean age: men -60.4 years; women -60.1 years)., collectively involving 8,293 individuals of European ancestry, spanning from 1980 to 2011. Adjustments were made for age, sex, and the top ten genetic principal components to control for population structure, thereby minimizing potential confounding biases. The separation between exposure groups (inflammatory cytokines) and outcome groups (lung cancer types) in the population selection process was maintained to avoid overlap and ensure the integrity of the MR analysis.
Statistical analysis
In this study, we primarily applied the inverse variance weighting (IVW) method for MR analysis, selected for its efficiency and statistical strength [23]. It's important to note that IVW presumes all genetic variants to be valid instrumental variables, which might not always be the case in reality. To validate the causal inferences drawn, we employed additional analyses: the MR-Egger method was used to test for horizontal pleiotropy. Simple mode/Weighted mode methods helped assess the stability of our results against potential biases. Additionally, the MR-PRESSO test helped identify and remove SNPs potentially affected by confounding biases, thereby refining our dataset. A leave-one-out analysis evaluated the influence of each individual SNP on the overall results to ensure the robustness of our findings [24-27]. When significant results were obtained using the IVW method (p < 0.05), conclusions were deemed reliable if the consistency of the β-value was maintained across different methods, especially in scenarios free from horizontal pleiotropy, confounding biases, or heterogeneity. The MR-Egger approach was specifically applied in cases with suspected horizontal pleiotropy but without apparent heterogeneity, whereas the weighted median and random effects IVW methods were employed in instances of heterogeneity, provided there was no polymorphism [28, 29]. All data analyses were conducted using R version 4.3.2, utilizing the "TwosampleMR" and "MR-PRESSO" packages [29, 30], ensuring a comprehensive and methodical execution of MR analysis.
Results
Genetic predictors linked to 41 circulating inflammatory factors and diverse forms of lung cancer
To ensure a sufficient number of SNPs for the MR analysis, we established a significance threshold of p < 5 × 10-6 for SNPs linked to each circulating inflammatory factor and outcomes related to SCLC, lung adenocarcinoma, and lung squamous cell carcinoma. Following the exclusion of weak instrumental variables and SNPs associated with confounders related to outcomes, the analysis revealed 456 SNPs associated with the 41 inflammatory cytokines. Specifically, there were 10 SNPs associated with lung adenocarcinoma, 11 SNPs associated with lung squamous cell carcinoma, and 5 SNPs associated with SCLC. The subsequent screening of SNP analysis was employed in the ensuing MR process to effectively mitigate the impacts of confounding factors on the analysis. The F-statistics for the 41 inflammatory cytokine SNPs varied significantly, demonstrating a robust set of instrumental variables, with values ranging from 20.77 to 782.3. For SNPs associated with lung adenocarcinoma, F-statistics were observed to range from 21.04 to 55.18, while for lung squamous cell carcinoma, they ranged from 20.85 to 71.40. Additionally, F-statistics for SNPs associated with SCLC ranged from 20.93 to 26.34. Notably, all instrumental variables utilized in our study exhibited F-statistics greater than 10, confirming the absence of weak instrumental variable bias. Detailed information on the instrumental variables selected for this study and their corresponding F-statistics are illustrated in Supplementary Tables S1 and S2.
Effects of 41 inflammatory cytokines on the occurrence of three types of lung cancer
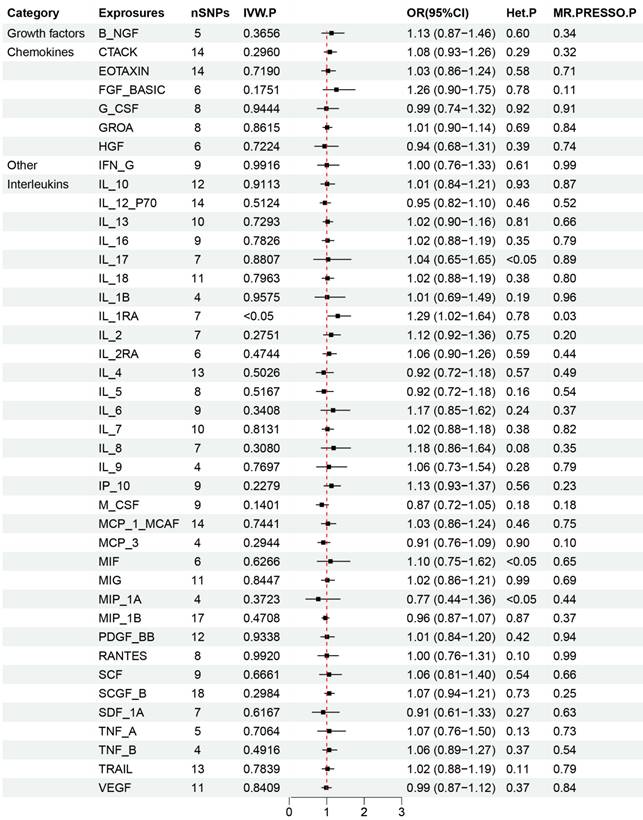
The critical outcomes of our MR analysis, illustrating the associations between 41 circulating inflammatory factors and lung adenocarcinoma, are visually detailed in Figure 2 and concisely summarized in Supplementary Table S3. In this analysis, we particularly focused on the role of inflammatory markers as predictive variables with lung adenocarcinoma as the outcome of interest. The primary findings from the Inverse Variance Weighting (IVW) method indicated a significant negative correlation between the increased genetic expression of IL_1RA and the likelihood of developing lung adenocarcinoma (OR: 1.29, 95% CI: 1.02-1.64, p=0.031). Additionally, MR-Egger analysis showed no indication of pleiotropy (p=0.85) or heterogeneity (p=0.67) concerning IL_1RA, reinforcing the reliability and sensitivity of these results. Moreover, the MR-PRESSO test confirmed no outliers among the IL_1RA SNPs, further solidifying the findings. To enhance the validity and sensitivity of these observations, a leave-one-out analysis was executed, yielding consistent β values compared to those derived from MR-Egger, Simple mode, Weighted median, and Weighted mode analyses, thus supporting the initial results from the IVW analysis. Figure 3 and Supplementary Figure S1 graphically delineates the causal linkage between IL_1RA levels and the incidence of lung adenocarcinoma, distinguishing this relationship from others between various inflammatory factors and the specified outcome. This set of analyses underpins the robustness and reliability of the discerned connection between IL_1RA and lung adenocarcinoma, as demonstrated by the collected data.
Results of Forward Mendelian randomization of inflammatory cytokines and lung adenocarcinoma.
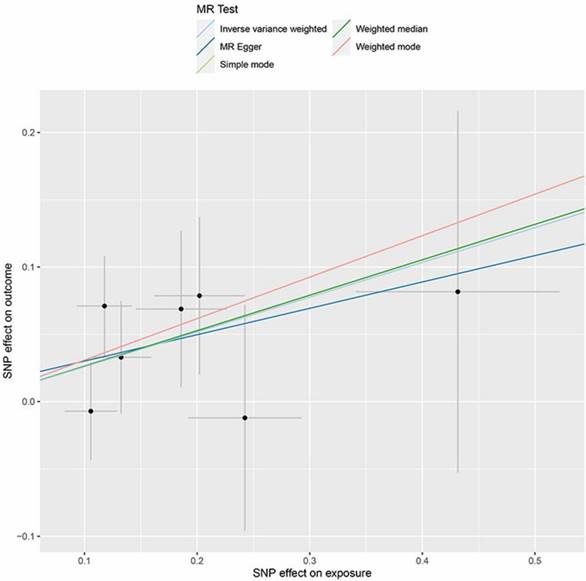
Scatter plot of the association between IL-1RA and lung adenocarcinoma. The five methods applied in the current manuscript were all depicted.
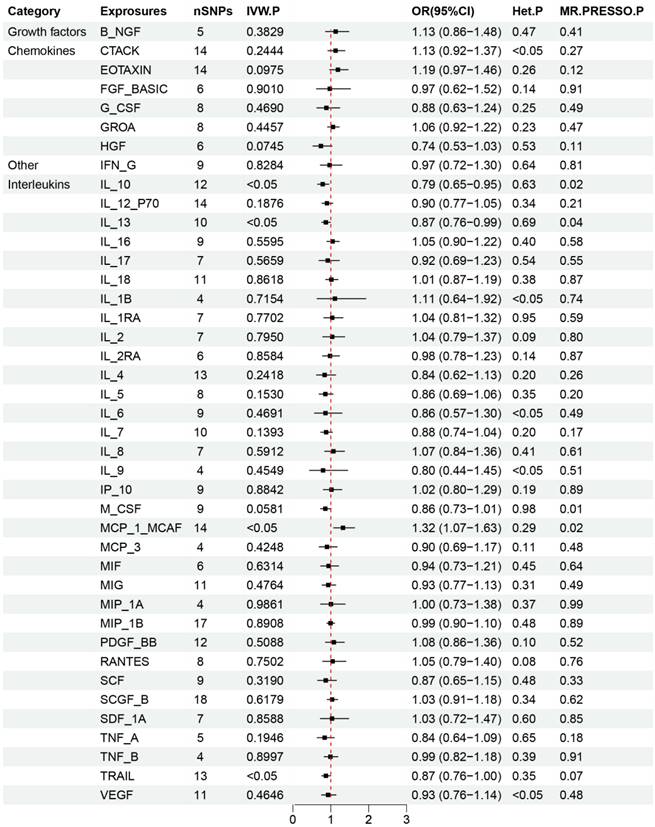
The primary results from the MR study are depicted in Figure 4 and detailed in Supplementary Table S4, where we analyzed the impact of 41 inflammatory markers on the risk of developing lung squamous cell carcinoma. The IVW analysis showed that higher levels of MCP_1_MCAF predicted by genetics were associated with a reduced risk of lung squamous cell carcinoma (OR: 0.77, 95% CI: 0.61-0.98, p=0.031). In contrast, elevated genetically predicted levels of IL_10, IL_13, and TRAIL were linked to an increased risk of this cancer type (IL_10: OR: 1.27, 95% CI: 1.06-1.53, p=0.012; IL_13: OR: 1.15, 95% CI: 1.06-1.53, p=0.036; TRAIL: OR: 1.15, 95% CI: 1.06-1.53, p=0.043). Further analyses, including MR-Egger, did not reveal any evidence of horizontal pleiotropy for these inflammatory factors (MCP_1_MCAF: p=0.56; IL_10: p=0.41; IL_13: p=0.47; TRAIL: p=0.18), nor did heterogeneity tests suggest discrepancies (MCP_1_MCAF: p=0.29; IL_10: p=0.63; IL_13: p=0.69; TRAIL: p=0.35). Additionally, the MR-PRESSO results demonstrated consistency with the identified associations of SNPs related to the investigated inflammatory factors, thus enhancing the overall robustness and sensitivity of this study. The validation of the robustness and sensitivity of our results was further confirmed through a leave-one-out analysis. Furthermore, the β values obtained from various alternative MR methods, such as MR-Egger, Simple mode, Weighted median, and Weighted mode analyses, consistently aligned with the results of the IVW analysis. These analytical layers strengthen the integrity of the conclusions. Figure 5 and Supplementary Figure S2-S4 elucidates the specific causal relationships identified between MCP_1_MCAF, IL_10, IL_13, TRAIL, and lung squamous cell carcinoma risk, distinguishing these particular cytokines from other studied factors in relation to lung cancer development.
The core findings from the MR study, as shown in Supplementary Figure S5 and detailed in Supplementary Table S5, explored the connection between 41 circulating inflammatory factors and the risk of SCLC. Our analysis revealed that there appeared to be no significant link between the evaluated circulating inflammatory cytokines and the development of SCLC. This conclusion is based on the compilation of comprehensive data, indicating that none of the studied inflammatory factors significantly contribute to the occurrence of SCLC.
Results of Forward Mendelian randomization of inflammatory cytokines and squamous cell lung carcinoma.
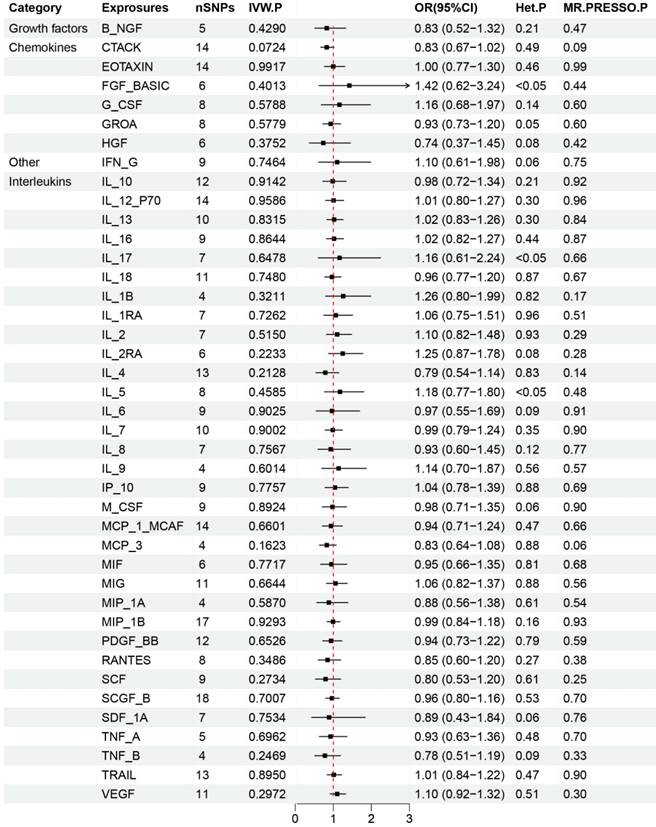
Effects of three different types of lung cancer on 41 inflammatory cytokines
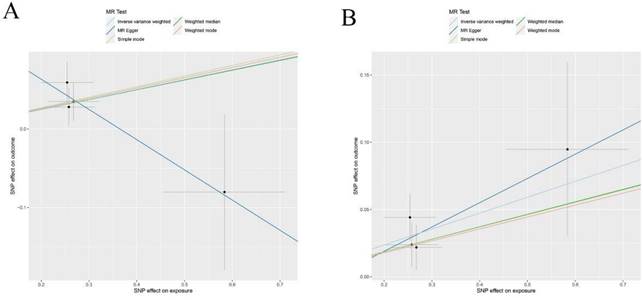
The main outcomes from our MR analysis, detailed in Supplementary Figure S5 and detailed in Supplementary Table S5, explored the relationship between SCLC incidence and 41 different circulating inflammatory markers. Due to the restricted number of SNPs, we could not establish causal links for IL17, TNFB, MIP-1B, and MCP3 concerning SCLC. Consequently, our discussion is limited to the association between SCLC and the remaining 37 inflammatory cytokines. Our IVW findings suggested a positive correlation between levels of SDF_1A and B-NGF and the occurrence of SCLC (SDF_1A: OR: 1.13, 95% CI: 1.05-1.21, p=0.001; B-NGF: OR: 1.13, 95% CI: 1.01-1.27, p=0.029). No horizontal pleiotropy or heterogeneity was detected in the MR-Egger analysis for these cytokines, ensuring the robustness and sensitivity of our results (SDF_1A: p=0.78, B_NGF: p=0.23 for pleiotropy; SDF_1A: p=0.72, B_NGF: p=0.31 for heterogeneity). The MR-PRESSO test, identifying no outliers, and a leave-one-out analysis, which yielded consistent β values, further validated these findings and enhanced the sensitivity of this study.
Scatter plots of Forward Mendelian randomization (MR) analyses for MCP-1, IL-10, IL-13 and TRAIL in squamous cell lung carcinoma, The five methods applied in the current manuscript were all depicted : (A) Scatter plot of the association between MCP-1and squamous cell lung carcinoma.; (B) Scatter plot of the association between IL-10 and squamous cell lung carcinoma; (C) Scatter plot of the association between IL-13 and squamous cell lung carcinoma.; (D) Scatter plot of the association between TRAIL and squamous cell lung carcinoma.
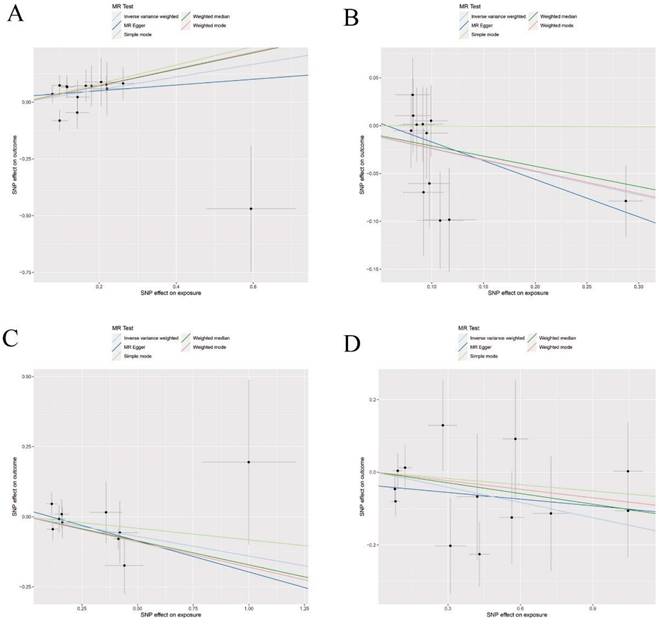
Figure 7 and Supplementary Figure S6-S8 provides a visual representation of the causal relationships between SCLC exposure and changes in levels of SDF_1A and B-NGF. Our analysis indicated no significant associations between SCLC and the other studied inflammatory cytokines. Additionally, as depicted in Supplementary Figure S9 and Supplementary Figure S10, alongside Supplementary Table S7 and Supplementary Table S8, no significant relationships were found between lung adenocarcinoma and lung squamous cell carcinoma with the levels of the 41 circulating inflammatory cytokines, underscoring the specificity of SCLC's link to SDF_1A and B-NGF.
Discussion
This study aimed to determine the causal relationships between inflammatory cytokines and three specific types of lung cancer using a two-sample two-way MR analysis. We investigated the connections between 41 inflammatory biomarkers, including chemokines, interleukins, and growth factors, and the onset of lung adenocarcinoma, squamous cell carcinoma, and SCLC. Key findings demonstrated that higher gene-predicted levels of IL_1RA were associated with an increased risk of lung adenocarcinoma. In contrast, elevated gene-predicted levels of IL_10, IL_13, and TRAIL were found to reduce the risk of squamous cell carcinoma. Additionally, higher levels of MCP_1_MCAF, as predicted by genetic analysis, were positively associated with a higher risk of lung squamous cell carcinoma. However, our analysis did not link any of the examined inflammatory cytokines with SCLC outcomes. When analyzing SCLC as an exposure factor, increased levels of SDF_1A and B_NGF were observed, indicating potential pathogenic pathways. Nonetheless, no association was found between the 41 inflammatory cytokines and the outcomes of lung adenocarcinoma or squamous cell carcinoma. In summary, our findings suggest that certain biomarkers may contribute to the pathogenesis of lung adenocarcinoma and squamous cell carcinoma, while others may act as downstream factors in the progression of SCLC. Our study uniquely identified IL-1RA as a potential risk factor for lung adenocarcinoma and revealed that MCP-1, IL-10, IL-13, and TRAIL were significant factors for lung squamous cell carcinoma. Additionally, a strong correlation was observed between SDF_1A and B_NGF with SCLC. These findings offer novel clinical insights into differentiating between various subtypes of lung cancer based on inflammatory profiles, potentially informing targeted therapeutic approaches and non-invasive diagnostic strategies.
Results of Forward Mendelian randomization of inflammatory cytokines and small cell lung cancer.
Causal relationship between 41 circulating inflammatory factors and lung adenocarcinoma
Our study highlighted IL_1RA as a potential risk factor for lung adenocarcinoma among 41 assessed circulating inflammatory cytokines. IL-1RA, known for its role in blocking IL-1-mediated cell signaling, typically functions as a tumor suppressor but can exhibit diverse effects depending on the cancer type and pathological context [31-34]. For instance, increased IL-1β levels in colon cancer have been linked to higher IL-1RA levels, impacting tumor behaviors such as invasion and apoptosis [35]. In esophageal cancer, IL-1RA contributes to the inhibition of cell growth by blocking IL-1α [36] and impacts lymphangiogenesis and metastasis through the regulation of VEGF-C and MMP9 [34]. Conversely, in cervical and oral squamous cell carcinoma, IL-1RA levels have been associated with patient prognosis and tumor progression through various pathways [37,38]. Alberto Mantovani et al. [33] suggested that IL-1RA could inhibit the "alarm" function of IL-1α, triggering protective immunity for skin cancer. While IL-1RA's role in lung cancer remains under debate, our findings suggested it may contribute to the development of lung adenocarcinoma. This might be due to the varying roles of IL-1RA in different disease stages, cell microenvironments, or its overall levels in circulation [39,40]. Although previous studies like Zhao Yang et al.'s MR analysis present differing data sources and outcomes [12], the general trend aligns with our findings, indicating IL-1RA's potential involvement in lung adenocarcinoma pathogenesis. Further research is necessary to solidify these associations and to better understand the differential roles of IL-1RA in cancer. Our focus on circulating IL-1RA levels and their implications on lung adenocarcinoma provides new directions for exploring inflammatory cytokines' influence on cancer development and progression.
Causal relationship between 41 circulating inflammatory factors and lung squamous cell carcinoma
Our results suggested that MCP_1_MCAF could act as a risk factor for lung squamous cell carcinoma. In contrast, IL_10, IL_13, and TRAIL may function as protective agents against this type of lung cancer. Interestingly, the occurrence of lung squamous cell carcinoma does not seem to significantly alter the levels of these inflammatory cytokines in the bloodstream. This distinction points towards the complex interplay between inflammation and cancer development, indicating specific cytokines might influence the pathogenesis of lung squamous cell carcinoma differently.
IL-13 is a multifunctional cytokine, mainly produced by T-helper type 2 (Th2) cells, with notable roles in inflammation and immune regulation [41,42]. It shares many features with IL-4, including receptor usage and regulatory elements, signaling predominantly through the type II IL4R (combining IL4Rα and IL13Rα1) or the IL13Rα2 receptor [43-45]. The impact of IL-13 on cancer is complex and not fully understood. While it is known to influence macrophage behavior, potentially supporting anti-inflammatory or tumor-friendly environments [46], its exact role can differ vastly between cancer types and stages. For instance, in certain contexts, IL-13 has been associated with promoting conditions favorable to cancer, such as through ROS-induced prooxidative environments linked to colorectal cancer progression [47-49]. Conversely, other studies highlight its potential in inhibiting cancer cell adhesion and promoting tissue homeostasis, thereby acting as a deterrent to the development of cancers like those found in the skin, lungs, and intestines [50-53]. In our study, IL-13 emerged as a potential protective factor against lung squamous cell carcinoma, suggesting a complex interplay in its function, highly dependent on the cancer's pathobiological context and disease stage [54].
Scatter plots of Reverse Mendelian randomization (MR) analyses for B-NGF and SDF-1A in squamous cell lung carcinoma, the five methods applied in the current manuscript were all depicted: (A) Scatter plot of the association between B-NGF and squamous cell lung carcinoma; (B) Scatter plot of the association between SDF-1A and squamous cell lung carcinoma.
The cytokine IL-10, produced by various immune cells and encoded by the IL-10 gene on chromosome 1q32, plays a critical role in modulating inflammatory responses [55,56]. The function of IL-10 in cancer development, however, is subject to debate due to its dual effects. Some researchers, such as Ling Chen et al. [57] and Yoon Ju Jung et al. [58] have reported that IL-10 may suppress the body's anti-tumor responses, particularly in gastric cancer. Conversely, Julius Malte Vahl et al. [59] and Y.W. Pan et al. [60] have found evidence suggesting IL-10 could support tumor tolerance and growth, particularly in lung cancers. Li Yang et al. [61] have also posited that IL-10 may encourage carcinogenesis, specifically in NSCLC. Yet, IL-10 is not universally detrimental in cancer contexts. For example, Daniel F. Zegarra Ruiz et al. found that IL-10 might mitigate colitis and inhibit colitis-associated colorectal cancer development [62], while Meng Qiao et al. identified IL-10's role in enhancing immunotherapy effectiveness in specific lung cancer types [63]. Narmeen Ahmad et al.'s study [64] associated IL-10 with a favorable prognosis in early invasive breast cancer. This dichotomy stems from IL-10's diverse roles in different tumor environments [65,66]. IL-10 may either support or inhibit tumor development, depending on factors such as the type of cancer, the stage of disease, and the local tumor microenvironment. In certain contexts, IL-10 can inhibit macrophage activity and promote regulatory T-cell function, potentially aiding tumor escape from immune surveillance [67,68]. Conversely, IL-10 can also bolster anti-tumor immune responses under certain conditions, for example, by stimulating cytotoxic CD8+ T cells and reducing inflammation [68-71]. Our study suggested that IL-10 could act as a protective factor against lung squamous cell carcinoma, adding to the complex narrative surrounding this cytokine. Despite the varying perspectives, it's clear IL-10's impact on cancer is multifaceted, emphasizing the need for context-specific evaluations in understanding its role in oncogenesis and treatment.
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL/Apo2L) is part of the tumor necrosis factor superfamily and operates as a type II transmembrane protein [72,73]. It is known for its unique ability to induce apoptosis selectively in cancer cells without harming normal cells by binding to death receptors DR4 and DR5 on the cancer cell surface [74,75]. TRAIL also plays a role in immune-mediated tumor surveillance and suppression, primarily through actions of T cells and natural killer cells [72,76]. This protein has been researched for its apoptotic effects in a variety of cancers, including those affecting the colon, lungs, bladder, breasts, kidneys, brain, prostate, and skin [77-87]. In our analysis, TRAIL was identified as a protective factor specifically against lung squamous cell carcinoma, highlighting its potential as a therapeutic agent. To enhance the delivery and effectiveness of TRAIL in chemotherapy, research is currently focusing on vehicles like mesenchymal stem cells and nanoparticles [88-91]. However, the challenge of TRAIL resistance in some cancer cells persists, prompting clinical trials [84] that explore overcoming this resistance through combination therapies with natural products or synthetic drugs such as disipramine, liensinine, and glipizide, aiming to sensitize these resistant tumor cells to TRAIL-induced apoptosis [87, 92-94].
Monocyte chemoattractant protein-1 (MCP-1 or CCL2) plays a critical role in immune responses and has been widely studied for its effects on monocyte attraction in vitro [95-98]. This chemokine has been linked to the progression and severity of various types of cancer, including breast, lung, and esophageal cancers, as it supports tumor growth, angiogenesis, and metastasis [98-109]. Our study also identified MCP-1 as a risk factor for lung squamous cell carcinoma. The CCL2/CCR2 pathway is particularly noted for its role in fostering environments conducive to cancer spread, as demonstrated by Yuqing Dong et al., who found its involvement in lymph node metastasis in tongue cancer through the activation of the RhoA and Rac1 pathways [103]. MCP-1 operates by binding to its specific receptor CCR2, initiating signaling pathways that promote cancer cell proliferation and suppress immune responses [110-112]. Current research efforts are focused on disrupting this pathway as a therapeutic strategy [113]. Clinical trials have explored the use of anti-MCP-1 antibodies or CCR2 antagonists, showing promising results in treating breast and prostate cancers [105,114,115]. These studies underscore the potential of targeting the MCP-1 signaling pathway in cancer therapy, offering new directions in the fight against tumor progression and metastasis.
Causal relationship between 41 circulating inflammatory factors and SCLC
Our analysis revealed no significant link between the 41 circulating inflammatory markers and the development of SCLC. However, an interesting observation emerged regarding SDF_1A and B_NGF levels: their elevation appeared to be associated with the occurrence of SCLC. This suggests that while these inflammatory cytokines may not initiate SCLC, they could play a role in its progression or be indicative of the disease's presence. This insight points to the potential utility of SDF_1A and B_NGF as biomarkers in the pathogenesis of SCLC, highlighting the need for further investigation into their roles and mechanisms within this specific cancer type.
Stromal-derived factor-1 (SDF-1), also recognized as CXCL12, is a chemokine found in multiple tissues such as the brain, heart, and lungs, among others. It significantly influences the immune system through its chemotactic effects on lymphocytes [116]. CXCL12 binds to its primary receptor, CXCR4 [117], and recent research has also identified CXCR7 as a second receptor, highlighting its importance in cellular signaling and function regulation [118]. Extracellular CXCL12 binds to either CXCR4 or CXCR7 on cell surfaces to regulate cellular functions by activating multiple signal transduction pathways [119]. The interaction between CXCL12 and its receptors has been implicated in the progression and metastasis of various cancers, including ovarian, lung, breast, cervical, colon, gastric, and esophageal cancers [117, 119-126]. Our research supported these findings by demonstrating an increase in circulating levels of SDF-1 in cases of SCLC, further emphasizing CXCL12's role in cancer biology. There is growing interest in targeting the CXCL12-CXCR4 and CXCL12-CXCR7 pathways for cancer therapy. Treatments developed so far, such as bicyclomycin (AMD3100), T22 peptide analogues, and dual inhibitors like GMI1359, focus on blocking these interactions [127-131]. Evidence suggests that inhibiting these pathways can significantly reduce cancer cell proliferation and metastasis, offering promising prospects for future therapeutic strategies against a range of cancers [68,121,132]. This approach could provide new avenues for cancer treatment, particularly for those cancers where SDF-1 levels are elevated.
B-NGF, an essential member of the nerve growth factor (NGF) family, shares similar biological activities with NGF. NGF is a critical neurotrophic factor that ensures the survival and differentiation of neuronal cells and is vital for the development and upkeep of the nervous system [133]. Research indicates that NGF, along with B-NGF, is involved in angiogenesis and can facilitate cancer progression [134-137]. In various types of cancers, including breast, prostate, liver, and oral cancers, B-NGF has been associated with promoting tumor initiation, progression, and metastasis [133, 136-142]. Our findings align with these studies, highlighting an increase in B-NGF levels in cases of SCLC, suggesting B-NGF might enhance tumor growth and the spread of SCLC. The involvement of B-NGF in the expansion and metastasis of SCLC suggests a potential positive feedback loop that could exacerbate the disease's progression. However, the specific mechanisms and potential therapeutic applications of B-NGF in cancer, especially SCLC, remain under-explored and merit further investigation to fully understand its role and impact.
Strengths and limitations
In a recent comprehensive study, we expanded upon previous single-item MR findings that linked CCL27/CTACK positively with lung cancer incidence in non-smokers, and IL-18 negatively with lung cancer and lung adenocarcinoma rates. For the first time, we conducted a two-way MR analysis focusing on 41 inflammatory cytokines across three distinct lung cancer types: lung adenocarcinoma, lung squamous cell carcinoma, and SCLC, utilizing data from the 10th-generation Finnish database. Our results demonstrated distinct associations between different inflammatory cytokines and the three types of lung cancer, with no overlapping cytokines among the different cancer types. This suggests that the specific inflammatory markers linked with each cancer type could serve as indirect indicators for classifying lung cancer pathologies. Consequently, this finding could potentially streamline the approach to lung cancer treatment, offering a method for guiding therapeutic decisions without necessitating invasive pathological biopsies.
Our study faces several limitations that should be considered: firstly, the data used were extracted from four large-scale GWAS, but specific demographic details and clinical records of the participants were unavailable. This absence restricts our ability to conduct subgroup analyses, which could provide more detailed insights. Secondly, we set the significance threshold for the GWAS data for exposure variables at p < 5 × 10-6, less stringent than the typical p < 5 × 10-8. This approach was necessary due to the limited number of SNPs meeting the more stringent threshold, which could otherwise hamper our ability to conduct comprehensive MR analyses. Thirdly, while results from alternative MR estimation methods such as MR-Egger, weighted mean, and simple models were not significant, the IVW method's higher statistical power and the consistency in the direction of β values allow us to still consider these findings relevant. Fourthly, the GWAS data were exclusively derived from European populations, limiting the generalizability of our findings across different racial and ethnic groups. It is essential to interpret these results with caution when applying them to non-European populations. Further research, incorporating diverse demographic groups and more detailed patient information, is needed to validate these findings and assess their applicability in clinical settings.
Conclusion
Gene analysis has shown that increased levels of IL_1RA are associated with a higher risk of developing lung adenocarcinoma. Conversely, genetic predictions of elevated IL_10, IL_13, and TRAIL levels appear to act as protective factors against lung squamous cell carcinoma. Furthermore, a rise in MCP_1_MCAF, as suggested by gene data, indicates a heightened risk for lung squamous cell carcinoma. Currently, there is no established link between circulating inflammatory cytokines and the prognosis of SCLC.
SCLC patients exhibit elevated levels of SDF_1A and B_NGF. Conversely, there is no indication that alterations in the levels of the 41 examined circulating inflammatory cytokines influence lung adenocarcinoma and lung squamous cell carcinoma. This differentiation implies that various types of lung cancer might possess unique inflammatory profiles, offering potential clinical insights into utilizing changes in inflammatory factor levels for future lung cancer diagnostics. This approach could be particularly valuable for distinguishing between different pathological types of lung cancer and tailoring non-invasive diagnostic and treatment strategies accordingly.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
Funding
The current work was supported by the Clinical Technology Innovation and Guidance Project of Hunan Province (2021SK53520), Hunan Provincial Key Area R&D Program (2021SK2020) and Natural Science Foundation of Hunan Province (2021JJ40871).
Ethics approval
The data for this study were obtained from four previously published GWAS. These studies were conducted in full compliance with ethical standards, having received the necessary approvals from relevant ethics committees. Additionally, all participants in these GWAS provided their informed consent in writing after being adequately informed about the research's objectives and procedures, ensuring the protection of the participants' rights, privacy, and well-being throughout the research process.
Author contributions
Conceptualization: Xinhang Hu, Muyun Peng, and Fenglei Yu; Writing-original draft preparation: Xinhang Hu, Xingchun Huang, Zhe Zhang; Writing-review and editing: Xinhang Hu, Shouzhi Xie, Yifan Ouyang and Li Wang; Visualization: Wangcheng Zhao and Xuyang Yi; Supervision: Zhi Yang, Muyun Peng and Fenglei Yu. All authors have read and agreed to the published version of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. HUANG G, YANG X, LI W. et al. A feasibility and safety study of computed tomography-guided percutaneous microwave ablation: a novel therapy for multiple synchronous ground-glass opacities of the lung. Int J Hyperthermia. 2020;37(1):414-22
2. DUTKOWSKA A, SZMYD B, KASZKOWIAK M. et al. Expression of inflammatory interleukins and selected miRNAs in non-small cell lung cancer. Sci Rep. 2021;11(1):5092
3. MOLINA J R, YANG P, CASSIVI S D. et al. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83(5):584-94
4. WU J, CHEN J, LV X. et al. Clinical value of serum and exhaled breath condensate inflammatory factor IL-11 levels in non-small cell lung cancer: Clinical value of IL-11 in non-small cell lung cancer. Int J Biol Markers. 2021;36(2):64-76
5. SIEGEL R L, MILLER K D, FUCHS H E. et al. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71(1):7-33
6. CHEN J, LI X, HUANG C. et al. Change of Serum Inflammatory Cytokines Levels in Patients With Chronic Obstructive Pulmonary Disease, Pneumonia and Lung Cancer. Technol Cancer Res Treat. 2020;19:1-7
7. BOURAS E, KARHUNEN V, GILL D. et al. Circulating inflammatory cytokines and risk of five cancers: a Mendelian randomization analysis. BMC Med. 2022;20(1):3
8. GAUR P, BHATTACHARYA S, KANT S. et al. Association of inflammatory biomarkers with lung cancer in North Indian population. Afr Health Sci. 2019;19(2):2147-55
9. BRENNER D R, FANIDI A, GRANKVIST K. et al. Inflammatory Cytokines and Lung Cancer Risk in 3 Prospective Studies. Am J Epidemiol. 2017;185(2):86-95
10. LI Q, HAN Y, FEI G. et al. IL-17 promoted metastasis of non-small-cell lung cancer cells. Immunol Lett. 2012;148(2):144-50
11. LIU L, LIU R, WEI C. et al. The role of IL-17 in lung cancer growth. Cytokine. 2023;169:156265
12. YANG Z, SCHOOLING C M, KWOK M K. Mendelian randomization study of interleukin (IL)-1 family and lung cancer. Sci Rep. 2021;11(1):17606
13. WOOTTON R E, RICHMOND R C, STUIJFZAND B G. et al. Evidence for causal effects of lifetime smoking on risk for depression and schizophrenia: a Mendelian randomisation study. Psychol Med. 2020;50(14):2435-43
14. HOLMES M V, ALA-KORPELA M, SMITH G D. Mendelian randomization in cardiometabolic disease: challenges in evaluating causality. Nat Rev Cardiol. 2017;14(10):577-90
15. LIU H, LIU Z, HUANG Y. et al. Exploring causal association between circulating inflammatory cytokines and functional outcomes following ischemic stroke: A bidirectional Mendelian randomization study. Eur J Neurol. 2024;31(2):e16123
16. DAVEY SMITH G, HEMANI G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23(R1):R89-98
17. XIANG M, WANG Y, GAO Z. et al. Exploring causal correlations between inflammatory cytokines and systemic lupus erythematosus: A Mendelian randomization. Front Immunol. 2022;13:985729
18. EMDIN C A, KHERA A V, KATHIRESAN S. Mendelian Randomization. Jama. 2017;318(19):1925-6
19. LI Y, LU J, WANG J. et al. Inflammatory Cytokines and Risk of Ischemic Stroke: A Mendelian Randomization Study. Front Pharmacol. 2021;12:779899
20. CHEN Y, SHEN J, WU Y. et al. Tea consumption and risk of lower respiratory tract infections: a two-sample mendelian randomization study. Eur J Nutr. 2023;62(1):385-93
21. BURGESS S, THOMPSON S G. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011;40(3):755-64
22. AHOLA-OLLI A V. et al. Genome-wide Association Study Identifies 27 Loci Influencing Concentrations of Circulating Cytokines and Growth Factors. Am J Hum Genet. 2017;100(1):40-50
23. HARTWIG F P, DAVEY SMITH G, BOWDEN J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017;46(6):1985-98
24. CHEN X, KONG J, PAN J. et al. Kidney damage causally affects the brain cortical structure: A Mendelian randomization study. EBioMedicine. 2021;72:103592
25. CHEN M, XIE C R, SHI Y Z. et al. Gut microbiota and major depressive disorder: A bidirectional Mendelian randomization. J Affect Disord. 2022;316:187-93
26. ZHANG F, XIAN D, FENG J. et al. Causal relationship between Alzheimer's disease and cardiovascular disease: a bidirectional Mendelian randomization analysis. Aging (Albany NY). 2023;15(17):9022-40
27. HEMANI G, BOWDEN J, DAVEY SMITH G. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum Mol Genet. 2018;27(R2):R195-r208
28. BIRNEY E. Mendelian Randomization. Cold Spring Harb Perspect Med. 2022;12(4):a041302
29. VERBANCK M, CHEN C Y, NEALE B. et al. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693-8
30. HEMANI G, ZHENG J, ELSWORTH B. et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:34408
31. DINARELLO C A, VAN DER MEER J W. Treating inflammation by blocking interleukin-1 in humans. Semin Immunol. 2013;25(6):469-84
32. ZHANG W, BORCHERDING N, KOLB R. IL-1 Signaling in Tumor Microenvironment. Adv Exp Med Biol. 2020;1240:1-23
33. MANTOVANI A, BARAJON I, GARLANDA C. IL-1 and IL-1 regulatory pathways in cancer progression and therapy. Immunol Rev. 2018;281(1):57-61
34. SHEN Z, ZHANG P, ZHANG W. et al. IL-1RA inhibits esophageal carcinogenesis and lymphangiogenesis via downregulating VEGF-C and MMP9. Funct Integr Genomics. 2023;23(2):164
35. CHEN Y, YANG Z, DENG B. et al. Interleukin 1α/1RA axis in colorectal cancer regulates tumor invasion, proliferation and apoptosis via autophagy. Oncol Rep. 2020;43(3):908-18
36. CHEN S, SHEN Z, LIU Z. et al. IL-1RA suppresses esophageal cancer cell growth by blocking IL-1α. J Clin Lab Anal. 2019;33(6):e22903
37. FUJIWAKI R, IIDA K, NAKAYAMA K. et al. Clinical significance of interleukin-1 receptor antagonist in patients with cervical carcinoma. Gynecol Oncol. 2003;89(1):77-83
38. YUAN S F, WANG Y M, CHAN L P. et al. IL-1RA promotes oral squamous cell carcinoma malignancy through mitochondrial metabolism-mediated EGFR/JNK/SOX2 pathway. J Transl Med. 2023;21(1):473
39. CAVALLI G, COLAFRANCESCO S, EMMI G. et al. Interleukin 1α: a comprehensive review on the role of IL-1α in the pathogenesis and treatment of autoimmune and inflammatory diseases. Autoimmun Rev. 2021;20(3):102763
40. RIDER P, CARMI Y, VORONOV E. et al. Interleukin-1α. Semin Immunol. 2013;25(6):430-8
41. SZYMANSKA B, SAWICKA E, JURKOWSKA K. et al. The relationship between interleukin-13 and angiogenin in patients with bladder cancer. J Physiol Pharmacol. 2021;72(4):10.26402
42. SHI J, SONG X, TRAUB B. et al. Involvement of IL-4, IL-13 and Their Receptors in Pancreatic Cancer. Int J Mol Sci. 2021;22(6):2998
43. SHI J, SHEN X, KANG Q. et al. Loss of Interleukin-13-Receptor-Alpha-1 Induces Apoptosis and Promotes EMT in Pancreatic Cancer. Int J Mol Sci. 2022;23(7):3659
44. MORAN A, PAVORD I D. Anti-IL-4/IL-13 for the treatment of asthma: the story so far. Expert Opin Biol Ther. 2020;20(3):283-94
45. HALLETT M A, VENMAR K T, FINGLETON B. Cytokine stimulation of epithelial cancer cells: the similar and divergent functions of IL-4 and IL-13. Cancer Res. 2012;72(24):6338-43
46. HU Q, WU G, WANG R. et al. Cutting edges and therapeutic opportunities on tumor-associated macrophages in lung cancer. Front Immunol. 2022;13:1007812
47. RAMASWAMI R, LURAIN K, MARSHALL V A. et al. Elevated IL-13 in effusions of patients with HIV and primary effusion lymphoma as compared with other Kaposi sarcoma herpesvirus-associated disorders. Aids. 2021;35(1):53-62
48. LIU H, ANTONY S, ROY K. et al. Interleukin-4 and interleukin-13 increase NADPH oxidase 1-related proliferation of human colon cancer cells. Oncotarget. 2017;8(24):38113-35
49. ZHOU R, QIAN S, GU X. et al. Interleukin-13 and its receptors in colorectal cancer (Review). Biomed Rep. 2013;1(5):687-90
50. DALESSANDRI T, CRAWFORD G, HAYES M. et al. IL-13 from intraepithelial lymphocytes regulates tissue homeostasis and protects against carcinogenesis in the skin. Nat Commun. 2016;7:12080
51. ALLAHVERDIAN S, HARADA N, SINGHERA G K. et al. Secretion of IL-13 by airway epithelial cells enhances epithelial repair via HB-EGF. Am J Respir Cell Mol Biol. 2008;38(2):153-60
52. DI NAPOLI A, GRECO D, SCAFETTA G. et al. IL-10, IL-13, Eotaxin and IL-10/IL-6 ratio distinguish breast implant-associated anaplastic large-cell lymphoma from all types of benign late seromas. Cancer Immunol Immunother. 2021;70(5):1379-92
53. SONG X, TRAUB B, SHI J. et al. Possible Roles of Interleukin-4 and -13 and Their Receptors in Gastric and Colon Cancer. Int J Mol Sci. 2021;22(2):727
54. FORMENTINI A, BRAUN P, FRICKE H. et al. Expression of interleukin-4 and interleukin-13 and their receptors in colorectal cancer. Int J Colorectal Dis. 2012;27(10):1369-76
55. KIM J M, BRANNAN C I, COPELAND N G. et al. Structure of the mouse IL-10 gene and chromosomal localization of the mouse and human genes. J Immunol. 1992;148(11):3618-23
56. CHEN D, HUANG L, ZHOU H. et al. Combining IL-10 and Oncolytic Adenovirus Demonstrates Enhanced Antitumor Efficacy Through CD8(+) T Cells. Front Immunol. 2021;12:615089
57. CHEN L, SHI Y, ZHU X. et al. IL-10 secreted by cancer-associated macrophages regulates proliferation and invasion in gastric cancer cells via c-Met/STAT3 signaling. Oncol Rep. 2019;42(2):595-604
58. JUNG Y J, WOO J S, HWANG S H. et al. Effect of IL-10-producing B cells in peripheral blood and tumor tissue on gastric cancer. Cell Commun Signal. 2023;21(1):320
59. VAHL J M, FRIEDRICH J, MITTLER S. et al. Interleukin-10-regulated tumour tolerance in non-small cell lung cancer. Br J Cancer. 2017;117(11):1644-55
60. PAN Y W, ZHOU Z G, WANG M. et al. Combination of IL-6, IL-10, and MCP-1 with traditional serum tumor markers in lung cancer diagnosis and prognosis. Genet Mol Res. 2016;15(4):10.4238
61. YANG L, DONG Y, LI Y. et al. IL-10 derived from M2 macrophage promotes cancer stemness via JAK1/STAT1/NF-κB/Notch1 pathway in non-small cell lung cancer. Int J Cancer. 2019;145(4):1099-110
62. ZEGARRA RUIZ D F, KIM D V, NORWOOD K. et al. Microbiota manipulation to increase macrophage IL-10 improves colitis and limits colitis-associated colorectal cancer. Gut Microbes. 2022;14(1):2119054
63. QIAO M, ZHOU F, LIU X. et al. Interleukin-10 induces expression of CD39 on CD8+T cells to potentiate anti-PD1 efficacy in EGFR-mutated non-small cell lung cancer. J Immunother Cancer. 2022;10(12):e005436
64. AHMAD N, AMMAR A, STORR S J. et al. IL-6 and IL-10 are associated with good prognosis in early stage invasive breast cancer patients. Cancer Immunol Immunother. 2018;67(4):537-49
65. MOORE K W, O'GARRA A, DE WAAL MALEFYT R. et al. Interleukin-10. Annu Rev Immunol. 1993;11:165-90
66. WANG X, WONG K, OUYANG W. et al. Targeting IL-10 Family Cytokines for the Treatment of Human Diseases. Cold Spring Harb Perspect Biol. 2019;11(2):a028548
67. GUO B. IL-10 Modulates Th17 Pathogenicity during Autoimmune Diseases. J Clin Cell Immunol. 2016;7(2):400
68. QIAO J, LIU Z, DONG C. et al. Targeting Tumors with IL-10 Prevents Dendritic Cell-Mediated CD8(+) T Cell Apoptosis. Cancer Cell. 2019;35(6):901-15.e4
69. STEWART C A, METHENY H, IIDA N. et al. Interferon-dependent IL-10 production by Tregs limits tumor Th17 inflammation. J Clin Invest. 2013;123(11):4859-74
70. EMMERICH J, MUMM J B, CHAN I H. et al. IL-10 directly activates and expands tumor-resident CD8(+) T cells without de novo infiltration from secondary lymphoid organs. Cancer Res. 2012;72(14):3570-81
71. KOHNO T, MIZUKAMI H, SUZUKI M. et al. Interleukin-10-mediated inhibition of angiogenesis and tumor growth in mice bearing VEGF-producing ovarian cancer. Cancer Res. 2003;63(16):5091-4
72. WU L S, WANG X W, HE W. et al. TRAIL inhibits platelet-induced colorectal cancer cell invasion. J Int Med Res. 2019;47(2):962-72
73. YI F, FRAZZETTE N, CRUZ A C. et al. Beyond Cell Death: New Functions for TNF Family Cytokines in Autoimmunity and Tumor Immunotherapy. Trends Mol Med. 2018;24(7):642-53
74. CARDOSO ALVES L, CORAZZA N, MICHEAU O. et al. The multifaceted role of TRAIL signaling in cancer and immunity. Febs j. 2021;288(19):5530-54
75. TWOMEY J D, KIM S R, ZHAO L. et al. Spatial dynamics of TRAIL death receptors in cancer cells. Drug Resist Updat. 2015;19:13-21
76. HUNG C M, LIU L C, HO C T. et al. Pterostilbene Enhances TRAIL-Induced Apoptosis through the Induction of Death Receptors and Downregulation of Cell Survival Proteins in TRAIL-Resistance Triple Negative Breast Cancer Cells. J Agric Food Chem. 2017;65(51):11179-91
77. OZAWA F, FRIESS H, KLEEFF J. et al. Effects and expression of TRAIL and its apoptosis-promoting receptors in human pancreatic cancer. Cancer Lett. 2001;163(1):71-81
78. DE LIMA FRAGELLI B D, CAMILLO L, DE ALMEIDA RODOLPHO J M. et al. Antitumor Effect of IL-2 and TRAIL Proteins Expressed by Recombinant Salmonella in Murine Bladder Cancer Cells. Cell Physiol Biochem. 2021;55(4):460-76
79. ABDEL SALAM N M, ABD-RABOU A A, SHARADA H M. et al. Combination Therapy of TRAIL and Thymoquinone Induce Breast Cancer Cell Cytotoxicity-Mediated Apoptosis and Cell Cycle Arrest. Asian Pac J Cancer Prev. 2021;22(5):1513-21
80. LI Y, JIN X, LI J. et al. Expression of TRAIL, DR4, and DR5 in bladder cancer: correlation with response to adjuvant therapy and implications of prognosis. Urology. 2012;79(4):968.e7-15
81. YOLDAS B, OZER C, OZEN O. et al. Clinical significance of TRAIL and TRAIL receptors in patients with head and neck cancer. Head Neck. 2011;33(9):1278-84
82. FRANK S, KöHLER U, SCHACKERT G. et al. Expression of TRAIL and its receptors in human brain tumors. Biochem Biophys Res Commun. 1999;257(2):454-9
83. HU J, WANG H, GU J. et al. Trail armed oncolytic poxvirus suppresses lung cancer cell by inducing apoptosis. Acta Biochim Biophys Sin (Shanghai). 2018;50(10):1018-27
84. ZINNAH K M A, NEWAZ MUNNA A, SEOL J W. et al. An Antidepressant Drug Increased TRAIL Receptor-2 Expression and Sensitized Lung Cancer Cells to TRAIL-induced Apoptosis. Anticancer Agents Med Chem. 2023;23(20):2225-36
85. LOTFOLLAHZADEH S, HOSSEINI E S, MAHMOUDI AZNAVEH H. et al. TRAIL/S-layer/graphene quantum dot nanohybrid enhanced stability and anticancer activity of TRAIL on colon cancer cells. Sci Rep. 2022;12(1):5851
86. NOGUEIRA D R, YAYLIM I, AAMIR Q. et al. TRAIL mediated signaling in pancreatic cancer. Asian Pac J Cancer Prev. 2014;15(15):5977-82
87. NAZIM U M, YIN H, PARK S Y. Neferine treatment enhances the TRAILinduced apoptosis of human prostate cancer cells via autophagic flux and the JNK pathway. Int J Oncol. 2020;56(5):1152-61
88. WANG Z, YU B, WANG B. et al. A novel capsid-modified oncolytic recombinant adenovirus type 5 for tumor-targeting gene therapy by intravenous route. Oncotarget. 2016;7(30):47287-301
89. ALIZADEH ZEINABAD H, SZEGEZDI E. TRAIL in the Treatment of Cancer: From Soluble Cytokine to Nanosystems. Cancers (Basel). 2022;14(20):5125
90. GUIMARãES P P G, GAGLIONE S, SEWASTIANIK T. et al. Nanoparticles for Immune Cytokine TRAIL-Based Cancer Therapy. ACS Nano. 2018;12(2):912-31
91. PARK S A, HAN H R, AHN S. et al. Combination treatment with VPA and MSCs-TRAIL could increase anti-tumor effects against intracranial glioma. Oncol Rep. 2021;45(3):869-78
92. NAZIM U M, MOON J H, LEE Y J. et al. Glipizide sensitizes lung cancer cells to TRAIL-induced apoptosis via Akt/mTOR/autophagy pathways. Oncotarget. 2017;8(59):100021-33
93. NAKAMURA H, TAGUCHI A, KAWANA K. et al. Therapeutic significance of targeting survivin in cervical cancer and possibility of combination therapy with TRAIL. Oncotarget. 2018;9(17):13451-61
94. LIU P C, LU G, DENG Y. et al. Inhibition of NF-κB Pathway and Modulation of MAPK Signaling Pathways in Glioblastoma and Implications for Lovastatin and Tumor Necrosis Factor-Related Apoptosis Inducing Ligand (TRAIL) Combination Therapy. PLoS One. 2017;12(1):e0171157
95. SINGH S, ANSHITA D, RAVICHANDIRAN V. MCP-1: Function, regulation, and involvement in disease. Int Immunopharmacol. 2021;101(Pt B):107598
96. DESHMANE S L, KREMLEV S, AMINI S. et al. Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res. 2009;29(6):313-26
97. HAO Q, VADGAMA J V, WANG P. CCL2/CCR2 signaling in cancer pathogenesis. Cell Commun Signal. 2020;18(1):82
98. DUTTA P, SARKISSYAN M, PAICO K. et al. MCP-1 is overexpressed in triple-negative breast cancers and drives cancer invasiveness and metastasis. Breast Cancer Res Treat. 2018;170(3):477-86
99. VALKOVIC T, FUCKAR D, STIFTER S. et al. Macrophage level is not affected by monocyte chemotactic protein-1 in invasive ductal breast carcinoma. J Cancer Res Clin Oncol. 2005;131(7):453-8
100. SHEN H, HE M, LIN R. et al. PLEK2 promotes gallbladder cancer invasion and metastasis through EGFR/CCL2 pathway. J Exp Clin Cancer Res. 2019;38(1):247
101. YOSHIMURA T, LI C, WANG Y. et al. The chemokine monocyte chemoattractant protein-1/CCL2 is a promoter of breast cancer metastasis. Cell Mol Immunol. 2023;20(7):714-38
102. MITTAL P, WANG L, AKIMOVA T. et al. The CCR2/MCP-1 Chemokine Pathway and Lung Adenocarcinoma. Cancers (Basel). 2020;12(12):3723
103. DONG Y, ZHANG S, ZHAO S. et al. CCL2 promotes lymphatic metastasis via activating RhoA and Rac1 pathway and predict prognosis to some extent in tongue cancer. Cancer Biol Ther. 2023;24(1):2205342
104. QIAN B Z, LI J, ZHANG H. et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475(7355):222-5
105. SUN C, LI X, GUO E. et al. MCP-1/CCR-2 axis in adipocytes and cancer cell respectively facilitates ovarian cancer peritoneal metastasis. Oncogene. 2020;39(8):1681-95
106. WANG W, SHEN F, WANG C. et al. MiR-1-3p inhibits the proliferation and invasion of bladder cancer cells by suppressing CCL2 expression. Tumour Biol. 2017;39(6):1010428317698383
107. OHTA M, KITADAI Y, TANAKA S. et al. Monocyte chemoattractant protein-1 expression correlates with macrophage infiltration and tumor vascularity in human esophageal squamous cell carcinomas. Int J Cancer. 2002;102(3):220-4
108. TANAKA K, KUREBAYASHI J, SOHDA M. et al. The expression of monocyte chemotactic protein-1 in papillary thyroid carcinoma is correlated with lymph node metastasis and tumor recurrence. Thyroid. 2009;19(1):21-5
109. OHTA M, KITADAI Y, TANAKA S. et al. Monocyte chemoattractant protein-1 expression correlates with macrophage infiltration and tumor vascularity in human gastric carcinomas. Int J Oncol. 2003;22(4):773-8
110. VAN COILLIE E, VAN DAMME J, OPDENAKKER G. The MCP/eotaxin subfamily of CC chemokines. Cytokine Growth Factor Rev. 1999;10(1):61-86
111. YOSHIMURA T. The chemokine MCP-1 (CCL2) in the host interaction with cancer: a foe or ally? Cell Mol Immunol. 2018;15(4):335-45
112. LIM S Y, YUZHALIN A E, GORDON-WEEKS A N. et al. Targeting the CCL2-CCR2 signaling axis in cancer metastasis. Oncotarget. 2016;7(19):28697-710
113. XU M, WANG Y, XIA R. et al. Role of the CCL2-CCR2 signalling axis in cancer: Mechanisms and therapeutic targeting. Cell Prolif. 2021;54(10):e13115
114. PIENTA K J, MACHIELS J P, SCHRIJVERS D. et al. Phase 2 study of carlumab (CNTO 888), a human monoclonal antibody against CC-chemokine ligand 2 (CCL2), in metastatic castration-resistant prostate cancer. Invest New Drugs. 2013;31(3):760-8
115. LI X, YAO W, YUAN Y. et al. Targeting of tumour-infiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinoma. Gut. 2017;66(1):157-67
116. SHEN B, ZHENG M Q, LU J W. et al. CXCL12-CXCR4 promotes proliferation and invasion of pancreatic cancer cells. Asian Pac J Cancer Prev. 2013;14(9):5403-8
117. MAO T L, FAN K F, LIU C L. Targeting the CXCR4/CXCL12 axis in treating epithelial ovarian cancer. Gene Ther. 2017;24(10):621-9
118. SáNCHEZ-MARTíN L S-M P, CABAñAS C. CXCR7 impact on CXCL12 biology and disease. Trends Mol Med. 2013;19(1):12-22
119. GUO J, TONG C Y, SHI J G. et al. C-X-C motif chemokine ligand 12 (CXCL12)/C-X-C motif chemokine receptor 7(CXCR7) regulates epithelial-mesenchymal transition process and promotes the metastasis of esophageal cancer by activating signal transducer and activator of transcription 3 (STAT3) pathway. Bioengineered. 2022;13(3):7425-38
120. WALD O, SHAPIRA O M, IZHAR U. CXCR4/CXCL12 axis in non small cell lung cancer (NSCLC) pathologic roles and therapeutic potential. Theranostics. 2013;3(1):26-33
121. IMAI H, SUNAGA N, SHIMIZU Y. et al. Clinicopathological and therapeutic significance of CXCL12 expression in lung cancer. Int J Immunopathol Pharmacol. 2010;23(1):153-64
122. GUO Q, GAO B L, ZHANG X J. et al. CXCL12-CXCR4 Axis Promotes Proliferation, Migration, Invasion, and Metastasis of Ovarian Cancer. Oncol Res. 2014;22(5-6):247-58
123. XU L, LI C, HUA F. et al. The CXCL12/CXCR7 signalling axis promotes proliferation and metastasis in cervical cancer. Med Oncol. 2021;38(5):58
124. QIN Y, WANG F, NI H. et al. Cancer-associated fibroblasts in gastric cancer affect malignant progression via the CXCL12-CXCR4 axis. J Cancer. 2021;12(10):3011-23
125. KHARE T, BISSONNETTE M, KHARE S. CXCL12-CXCR4/CXCR7 Axis in Colorectal Cancer: Therapeutic Target in Preclinical and Clinical Studies. Int J Mol Sci. 2021;22(14):7371
126. HAYASAKA H, YOSHIDA J, KURODA Y. et al. CXCL12 promotes CCR7 ligand-mediated breast cancer cell invasion and migration toward lymphatic vessels. Cancer Sci. 2022;113(4):1338-51
127. DONAHUE R E, JIN P, BONIFACINO A C. et al. Plerixafor (AMD3100) and granulocyte colony-stimulating factor (G-CSF) mobilize different CD34+ cell populations based on global gene and microRNA expression signatures. Blood. 2009;114(12):2530-41
128. LIANG Z, YOON Y, VOTAW J. et al. Silencing of CXCR4 blocks breast cancer metastasis. Cancer Res. 2005;65(3):967-71
129. PORVASNIK S, SAKAMOTO N, KUSMARTSEV S. et al. Effects of CXCR4 antagonist CTCE-9908 on prostate tumor growth. Prostate. 2009;69(13):1460-9
130. WONG D, KANDAGATLA P, KORZ W. et al. Targeting CXCR4 with CTCE-9908 inhibits prostate tumor metastasis. BMC Urol. 2014;14:12
131. FESTUCCIA C, MANCINI A, GRAVINA G L. et al. Dual CXCR4 and E-Selectin Inhibitor, GMI-1359, Shows Anti-Bone Metastatic Effects and Synergizes with Docetaxel in Prostate Cancer Cell Intraosseous Growth. Cells. 2019;9(1):32
132. LECAVALIER-BARSOUM M, CHAUDARY N, HAN K. et al. Targeting the CXCL12/CXCR4 pathway and myeloid cells to improve radiation treatment of locally advanced cervical cancer. Int J Cancer. 2018;143(5):1017-28
133. YUE X J, XU L B, ZHU M S. et al. Over-expression of nerve growth factor-β in human cholangiocarcinoma QBC939 cells promote tumor progression. PLoS One. 2013;8(4):e62024
134. EMANUELI C, SALIS M B, PINNA A. et al. Nerve growth factor promotes angiogenesis and arteriogenesis in ischemic hindlimbs. Circulation. 2002;106(17):2257-62
135. CANTARELLA G, LEMPEREUR L, PRESTA M. et al. Nerve growth factor-endothelial cell interaction leads to angiogenesis in vitro and in vivo. Faseb j. 2002;16(10):1307-9
136. ADRIAENSSENS E, VANHECKE E, SAULE P. et al. Nerve growth factor is a potential therapeutic target in breast cancer. Cancer Res. 2008;68(2):346-51
137. SIGALA S, BODEI S, MISSALE C. et al. Gene expression profile of prostate cancer cell lines: effect of nerve growth factor treatment. Mol Cell Endocrinol. 2008;284(1-2):11-20
138. YE Y, DANG D, ZHANG J. et al. Nerve growth factor links oral cancer progression, pain, and cachexia. Mol Cancer Ther. 2011;10(9):1667-76
139. OWUSU SEKYERE S, PORT K, DETERDING K. et al. Inflammatory patterns in plasma associate with hepatocellular carcinoma development in cured hepatitis C cirrhotic patients. United European Gastroenterol J. 2021;9(4):486-96
140. DAMM R, PECH M, CAVALLI P. et al. Correlation of chemokines and growth factors with radiation-induced liver injury after interstitial high dose rate (HDR) brachytherapy of liver metastases. J Cancer Res Clin Oncol. 2022;148(10):2815-26
141. LANKI M, MUSTONEN H, SALMI M. et al. Serum cytokine profiles in patients with pancreatic cancer and chronic pancreatitis. Pancreatology. 2023;23(6):657-62
142. CHENG Y, ZHENG S, PAN C T. et al. Analysis of aqueous humor concentrations of cytokines in retinoblastoma. PLoS One. 2017;12(5):e0177337
Author contact
![]() Corresponding authors: Muyun Peng & Fenglei Yu Department of Thoracic Surgery, Second Xiangya Hospital, Central South University, No. 139, Renmin Middle Road, Changsha 410000, Hunan Province, China. E-mail: pengmuyundeu.cn (MYP); yufengleiedu.cn (FLY); Tel.: +86-18273159365 (MYP); +86-13755182999 (FLY).
Corresponding authors: Muyun Peng & Fenglei Yu Department of Thoracic Surgery, Second Xiangya Hospital, Central South University, No. 139, Renmin Middle Road, Changsha 410000, Hunan Province, China. E-mail: pengmuyundeu.cn (MYP); yufengleiedu.cn (FLY); Tel.: +86-18273159365 (MYP); +86-13755182999 (FLY).

 Global reach, higher impact
Global reach, higher impact