Impact Factor
ISSN: 1837-9664
J Cancer 2024; 15(18):6103-6109. doi:10.7150/jca.99928 This issue Cite
Research Paper
YTHDF3 gene polymorphisms increase Wilms tumor risk in Chinese girls
1. Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, Guangzhou 510623, Guangdong, China.
2. Department of Hematology, The Key Laboratory of Pediatric Hematology and Oncology Diseases of Wenzhou, The Second Affiliated Hospital and Yuying Children's Hospital of Wenzhou Medical University, Wenzhou 325027, Zhejiang, China.
3. Department of Pediatric Surgery, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, Henan, China.
4. Department of Pediatric Surgery, The Second Affiliated Hospital of Xi'an Jiaotong University, Xi'an 710004, Shaanxi, China.
5. Department of Pathology, Children Hospital and Women Health Center of Shanxi, Taiyuan 030013, Shannxi, China.
#These authors contributed equally to this work.
Received 2024-6-21; Accepted 2024-9-28; Published 2024-10-7
Abstract
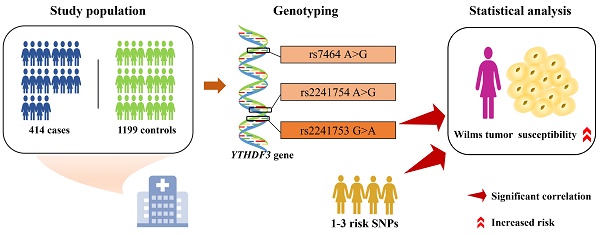
Wilms tumor is a prevalent pediatric tumor influenced by various genetic factors. m6A modification is a common nucleotide modification that plays a role in a variety of cancers. As a “reader”, YTHDF3 is essential for recognizing m6A modifications. However, the association between YTHDF3 gene polymorphisms and Wilms tumor susceptibility has not been previously reported. A five-center case‒control study including 414 patients and 1199 controls was conducted to explore the relationship between YTHDF3 gene polymorphisms and Wilms tumor susceptibility. The samples were genotyped via TaqMan real-time quantitative polymerase chain reaction. Odds ratios (ORs) and 95% confidence intervals (CIs) were utilized as indicators to assess their correlation. The YTHDF3 rs2241753 AA genotype was significantly associated with an increased risk of Wilms tumor in females (adjusted OR=1.74, 95% CI=1.05-2.88, P=0.033). The risk of Wilms tumor was also notably elevated in female children with 1-3 risk genotypes (adjusted OR=1.47, 95% CI=1.04-2.07, P=0.028). The YTHDF3 rs2241753 AA genotype and the presence of 1-3 risk genotypes were significantly associated with increased Wilms tumor risk in female children.
Keywords: YTHDF3, Wilms tumor, m6A, polymorphism, susceptibility
Introduction
Wilms tumors, also known as nephroblastoma, are common childhood renal malignancies, accounting for 6% of pediatric oncologic diseases and 95% of renal tumors in children [1]. The majority of cases occur in children under 5 years of age, with the highest incidence in children 2-3 years of age [1, 2]. In Western countries, the incidence of Wilms tumor is approximately 7-10 cases per million people, whereas in China, it is approximately 3.3 cases per million people [3, 4]. Wilms tumor manifests as an abnormality in the developmental process of the renal embryo, resulting in the coexistence of different stages of renal development, including persistent blastema, tubular epithelial and mesenchymal components [5, 6]. It exhibits diverse morphological structures, and its histopathological features are believed to be correlated with its prognosis [6]. Advancements in modern medicine have led to considerable success in treating Wilms tumor, which has an overall survival rate exceeding 90% [7, 8]. However, notably, patients with adverse histologic features still have a poor prognosis, and the survival rate for patients with recurrence remains relatively low [8]. In addition, approximately 25% of survivors have serious chronic diseases [9]. Therefore, further refinement in the treatment of Wilms tumor is necessary to reduce the incidence of complications and sequelae and to develop therapeutic approaches for high-risk Wilms tumor types.
Genetic factors are widely recognized to play crucial roles in the development of Wilms tumor. The inactivation of WT1, the first identified tumor suppressor gene associated with Wilms tumor, is an important factor in Wilms tumor development [10]. Subsequent studies have revealed that mutations in WTX, TP53, CTNNB1, IGF2, and MYCN affect the development of Wilms tumor [11-13]. There is increasing strong evidence supporting the contribution of genetic variation to Wilms tumor. Single nucleotide polymorphisms (SNPs) are common genetic variants that affect cancer susceptibility. A genome-wide association study (GWAS) revealed the presence of genes associated with Wilms tumor susceptibility in the 2p24 and 11q14 regions. Moreover, 22q12, Xp22 and 5q14 were also predicted to be potential risk regions [14]. Studies on candidate genes have shown that polymorphisms in base excision repair genes and nucleotide excision repair genes are significantly associated with Wilms tumor susceptibility [5, 15]. There is evidence demonstrating an association between Wilms tumor susceptibility and polymorphisms in various tumor-associated genes, such as BARD1 [16], METTL3 [17], FTO [18], and ALKBH5 [19]. However, further exploration into the genetic etiology of Wilms tumor is warranted.
N6-methyladenosine (m6A) is a common, widely distributed posttranscriptional modification of mRNAs in eukaryotes [20]. Mounting evidence suggests that m6A modification plays a pivotal role in cancer development and regulation [21-23]. This modification is regulated by "writers", "erasers", and "readers" [24, 25]. RNA methyltransferases, known as "writers", primarily include METTL3/METTL14/WTAP, which are responsible for adding methyl groups to the adenine residues of mRNAs [26, 27]. Demethylases such as FTO and ALKBH5 act as "erasers" by removing methylations. YTHDF1/2/3 and IGF2BP1 are m6A-binding proteins referred to as “readers”, which specifically recognize and bind to m6A modification sites on mRNAs and play essential roles in regulating mRNA stability, translation, splicing and export [28-31]. Consequently, dysregulation of m6A modification is often closely associated with various diseases, especially cancer [32, 33].
YTHDF3, a member of the YTH domain family, is believed to regulate the translation and degradation of methylated mRNAs in concert with YTHDF1 and YTHDF2 [29]. Currently, the relationship between YTHDF3 gene variants and cancers such as Wilms tumor remains unclear. The associations between SNPs of YTHDF2 and YTHDC1 (other members of the YTH domain family) and Wilms tumor susceptibility have been elucidated [34, 35]. However, further analysis is needed to understand the relationship between the YTHDF3 gene SNPs and the risk of Wilms tumor. The objective of this study was to investigate the association between YTHDF3 polymorphisms and Wilms tumor susceptibility in Chinese children while providing a theoretical basis for subsequent studies.
Materials and methods
Study population
The participation criteria and clinical characteristics of the selected subjects have been described in previous studies [19]. Patients with Wilms tumor who were included were required to meet the following criteria: (1) Han Chinese ancestry; (2) new diagnosis confirmed by pathology; (3) no family history of disease or cancer; and (4) age 14 years or younger. Patients who had received medical interventions or did not provide signed informed consent were excluded. Ultimately, we recruited 414 patients who were diagnosed with Wilms tumor from five cities in China and 1199 healthy controls who were matched for age and sex (Table S1). All the subjects were from the Chinese Han population. The parents or guardians of the subjects signed an informed consent form. The study was approved by the Ethics Committee of Guangzhou Women and Children's Medical Center (Ethical Approval No: 202016601).
Polymorphism selection and genotyping
SNPs with potential functions in YTHDF3 were screened via the dbSNP database (https://www.ncbi.nlm.nih.gov/snp), SNPinfo software (http://snpinfo.niehs.nih.gov/), and LDlink (https://ldlink.nci.nih.gov/), and the screening criteria were described in detail in previous studies [36]. Briefly, the following criteria were used: (1) SNPs located at both ends of the YTHDF3 gene, i.e., the 5' nearest gene, the 3' nearest gene, and the 3' and 5' untranslated regions (UTRs); (2) SNPs with a minor allele frequency (MAF) ≥ 5% in Chinese Han subjects; (3) SNPs affecting the activity of transcription factor-binding sites (TFBSs) or miRNA binding sites; and (4) SNPs with a low linkage disequilibrium (LD) with other selected SNPs (R2<0.8). Ultimately, YTHDF3 rs2241753 G>A, rs2241754 A>G, and rs7464 A>G were selected for further study; rs2241753 G>A and rs2241754 A>G affect TFBS activity and rs7464 A>G affect miRNA binding site activity (Table S2). There was no significant LD among these selected SNPs (R2<0.8). R2=0.372 between rs2241753 G>A and rs2241754 A>G; R2=0.178 between rs2241753 G>A and rs7464 A>G; R2=0.149 between rs2241754 G>A and rs7464 A>G (Figure S1).
Genomic DNA was extracted from peripheral blood via the TIANamp Blood DNA Kit (TianGen Biotech, Beijing, China). DNA samples were genotyped for SNPs via TaqMan real-time PCR (Applied Biosystems, Foster City, CA) [18, 37]. To ensure accuracy, negative controls without DNA templates were included on each plate, and laboratory technicians were blinded to the sample information. Additionally, a random selection of 10% of the samples was regenerated to assess the error rate, with the results showing 100% concordance.
Statistical analysis
Depending on the type of variable, either the chi-square test or t test was used to compare the clinical differences between the patients and controls. The degree of deviation of each SNP genotype from Hardy‒Weinberg equilibrium (HWE) in the controls was assessed via a goodness-of-fit chi-square test. The homozygotes of the common allele were designated as the reference group, whereas the remaining genotypes were classified as variants [38]. An unconditional logistic regression analysis was conducted to examine the association between YTHDF3 SNPs and Wilms tumor risk, with adjustments made for age and sex to minimize their potential interference with the correlation analysis. Odds ratios (ORs) and 95% confidence intervals (CIs) were employed as indicators to evaluate this correlation. The effects of the YTHDF3 SNPs on Wilms tumor susceptibility were subsequently analyzed in patients stratified by age, sex, and clinical stage. eQTL analysis was performed via the GTEx portal (https://www.gtexportal.org/home/) to determine the effects of SNPs on nearby gene expression levels. All the statistical significance tests were performed with a two-sided P<0.05 as the threshold. All data analysis was performed via SAS 9.1 (SAS Institute Inc.).
Results
Association between YTHDF3 polymorphisms and Wilms tumor risk
The YTHDF3 SNPs were successfully genotyped in 391 of 414 cases and 1198 of 1199 controls. The correlation between YTHDF3 gene polymorphisms and Wilms tumor susceptibility is shown in Table 1. The genotype frequencies of all three SNPs in the control group were consistent with HWE (HWE>0.05). However, none of the three SNPs were significantly correlated with Wilms tumor risk. Subsequently, rs2241753 AA, rs2241754 AA, and rs7464 AG/GG were identified as risk genotypes. However, combined analysis revealed no significant difference in Wilms tumor susceptibility between carriers of 1-3 risk genotypes and noncarriers.
Association of YTHDF3 gene polymorphisms with Wilms tumor susceptibility
| Genotype | Cases (N=391) | Controls (N=1198) | P a | Crude OR (95% CI) | P | Adjusted OR (95% CI) b | P b |
|---|---|---|---|---|---|---|---|
| rs2241753 G>A (HWE=0.273) | |||||||
| GG | 182 (46.55) | 538 (44.91) | 1.00 | 1.00 | |||
| GA | 163 (41.69) | 542 (45.24) | 0.89 (0.70-1.13) | 0.342 | 0.89 (0.70-1.13) | 0.341 | |
| AA | 46 (11.76) | 118 (9.85) | 1.15 (0.79-1.69) | 0.464 | 1.15 (0.78-1.68) | 0.482 | |
| Additive | 0.943 | 1.01 (0.85-1.20) | 0.943 | 1.00 (0.85-1.20) | 0.960 | ||
| Dominant | 209 (53.45) | 660 (55.09) | 0.572 | 0.94 (0.74-1.18) | 0.572 | 0.94 (0.74-1.18) | 0.565 |
| GG/GA | 345 (88.24) | 1080 (90.15) | 1.00 | 1.00 | |||
| AA | 46 (11.76) | 118 (9.85) | 0.280 | 1.22 (0.85-1.75) | 0.280 | 1.21 (0.85-1.74) | 0.294 |
| rs2241754 A>G (HWE=0.672) | |||||||
| AA | 125 (31.97) | 380 (31.72) | 1.00 | 1.00 | |||
| AG | 190 (48.59) | 583 (48.66) | 0.99 (0.76-1.29) | 0.944 | 1.00 (0.77-1.30) | 0.996 | |
| GG | 76 (19.44) | 235 (19.62) | 0.98 (0.71-1.37) | 0.919 | 0.98 (0.71-1.36) | 0.914 | |
| Additive | 0.917 | 0.99 (0.84-1.17) | 0.917 | 0.99 (0.84-1.17) | 0.924 | ||
| Dominant | 266 (68.03) | 818 (68.28) | 0.927 | 0.99 (0.77-1.26) | 0.927 | 1.00 (0.78-1.27) | 0.970 |
| AA/AG | 315 (80.56) | 963 (80.38) | 1.00 | 1.00 | |||
| GG | 76 (19.44) | 235 (19.62) | 0.938 | 0.99 (0.74-1.32) | 0.939 | 0.98 (0.74-1.31) | 0.899 |
| rs7464 A>G (HWE=0.728) | |||||||
| AA | 207 (52.94) | 662 (55.26) | 1.00 | 1.00 | |||
| AG | 152 (38.87) | 454 (37.90) | 1.07 (0.84-1.36) | 0.579 | 1.08 (0.85-1.37) | 0.554 | |
| GG | 32 (8.18) | 82 (6.84) | 1.25 (0.81-1.93) | 0.321 | 1.26 (0.81-1.95) | 0.301 | |
| Additive | 0.317 | 1.10 (0.92-1.31) | 0.317 | 1.10 (0.92-1.32) | 0.294 | ||
| Dominant | 184 (47.06) | 536 (44.74) | 0.424 | 1.10 (0.87-1.38) | 0.424 | 1.10 (0.88-1.39) | 0.400 |
| AA/AG | 359 (91.82) | 1116 (93.16) | 1.00 | 1.00 | |||
| GG | 32 (8.18) | 82 (6.84) | 0.373 | 1.21 (0.79-1.86) | 0.373 | 1.22 (0.80-1.87) | 0.356 |
| Combined effect of risk genotypes c | |||||||
| 0 | 163 (41.69) | 541 (45.16) | 1.00 | 1.00 | |||
| 1-3 | 228 (58.31) | 657 (54.84) | 0.231 | 1.15 (0.91-1.45) | 0.231 | 1.16 (0.92-1.46) | 0.221 |
OR, odds ratio; CI, confidence interval, HWE, Hardy‒Weinberg equilibrium.
a c2 test for genotype distributions between Wilms tumor patients and cancer-free controls.
b Adjusted for age and gender.
c Risk genotypes were carriers with rs2241753 AA, rs2241754 AA and rs7464 AG/GG genotypes.
Stratification analysis for association between YTHDF3 genotypes and Wilms tumor susceptibility
| Variables | rs2241753 (case/control) | AOR (95% CI)a | Pa | rs7464 (case/control) | AOR (95% CI)a | Pa | Risk genotypes (case/control) | AOR (95% CI)a | Pa | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG/GA | AA | AA/AG | GG | 0 | 1-3 | |||||||
| Age, months | ||||||||||||
| ≤18 | 119/416 | 18/49 | 1.32 (0.74-2.35) | 0.352 | 125/436 | 12/29 | 1.37 (0.68-2.78) | 0.382 | 61/210 | 76/255 | 1.02 (0.70-1.50) | 0.907 |
| >18 | 226/664 | 28/69 | 1.22 (0.76-1.94) | 0.409 | 234/680 | 20/53 | 1.09 (0.63-1.86) | 0.766 | 102/331 | 152/402 | 1.25 (0.94-1.67) | 0.132 |
| Sex | ||||||||||||
| Female | 157/474 | 27/47 | 1.74 (1.05-2.88) | 0.033 | 167/494 | 17/27 | 1.86 (0.99-3.51) | 0.053 | 72/253 | 112/268 | 1.47 (1.04-2.07) | 0.028 |
| Male | 188/606 | 19/71 | 0.86 (0.50-1.46) | 0.574 | 192/622 | 15/55 | 0.88 (0.48-1.59) | 0.661 | 91/288 | 116/389 | 0.94 (0.69-1.29) | 0.712 |
| Clinical stage | ||||||||||||
| I | 109/1080 | 19/118 | 1.60 (0.94-2.70) | 0.082 | 122/1116 | 6/82 | 0.69 (0.29-1.62) | 0.392 | 49/541 | 79/657 | 1.33 (0.91-1.94) | 0.136 |
| II | 97/1080 | 15/118 | 1.41 (0.79-2.50) | 0.246 | 102/1116 | 10/82 | 1.36 (0.68-2.71) | 0.383 | 54/541 | 58/657 | 0.89 (0.60-1.31) | 0.557 |
| III | 81/1080 | 8/118 | 0.93 (0.44-1.96) | 0.840 | 83/1116 | 6/82 | 1.00 (0.42-2.36) | 0.993 | 35/541 | 54/657 | 1.27 (0.82-1.98) | 0.284 |
| IV | 43/1080 | 2/118 | 0.43 (0.10-1.81) | 0.250 | 39/1116 | 6/82 | 2.14 (0.88-5.23) | 0.094 | 17/541 | 28/657 | 1.37 (0.74-2.54) | 0.313 |
| I+II | 206/1080 | 34/118 | 1.50 (1.00-2.26) | 0.053 | 224/1116 | 16/82 | 0.99 (0.57-1.73) | 0.976 | 103/541 | 137/657 | 1.10 (0.83-1.45) | 0.516 |
| III+IV | 124/1080 | 10/118 | 0.75 (0.38-1.48) | 0.410 | 122/1116 | 12/82 | 1.37 (0.73-2.59) | 0.332 | 52/541 | 82/657 | 1.31 (0.91-1.89) | 0.152 |
AOR, adjusted odds ratio; CI, confidence interval.
a Adjusted for age and sex without the stratify factor.
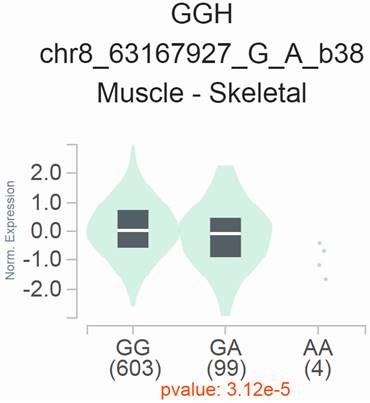
eQTL analysis of YTHDF3 rs2241753 G>A in the GTEx database. The rs2241753 G genotype was significantly associated with increased GGH mRNA levels in skeletal muscle tissue (P=3.12×10-5).
Subgroup analysis
Subgroup analysis was conducted by stratifying the subjects on the basis of age, sex, and clinical stage. The results are presented in Table 2. The rs2241753 AA genotype was significantly associated with an increased risk of Wilms tumor in the female subgroup (adjusted OR=1.74, 95% CI=1.05-2.88, P=0.033). Furthermore, the presence of 1-3 risk genotypes was also significantly associated with an increased risk of Wilms tumor in this subgroup (adjusted OR=1.47, 95% CI=1.04-2.07, P=0.028).
Effect of rs2241753 G>A on the mRNA expression of nearby genes
We conducted expression quantitative trait locus (eQTL) analyses to explore the potential impact of rs2241753 G>A on the mRNA expression of its neighboring genes, utilizing data from GTEx. The results revealed a significant association between the presence of the rs2241753 G allele and increased GGH mRNA expression in skeletal muscle (Figure 1).
Discussion
The m6A modification gene plays a crucial role in cancer regulation, and previous studies have partially elucidated the correlation between m6A gene SNPs and Wilms tumor susceptibility [17-19, 38]. Here, we sought to investigate the potential role of YTHDF3 gene SNPs in the risk of Wilms tumor. Our data indicate that rs2241753 in YTHDF3 is significantly associated with an increased risk of Wilms tumor in female children. Furthermore, there is an increased risk of Wilms tumor in female children with 1-3 risk genotypes.
YTH domain family proteins, including YTHDF1, YTHDF2, YTHDF3, and YTHDC1, are essential for mediating m6A functions. YTHDC1 is responsible for regulating mRNA splicing and export in the nucleus [39]. Unlike YTHDC1, YTHDF 1/2/3 are cytoplasmic m6A readers [40]. YTHDF2 binds to m6A-modified mRNAs in the cytoplasm and localizes them to mRNA decay sites such as processing bodies, thereby promoting mRNA degradation [30]. YTHDF1 interacts with initiation factors to promote ribosome production and ultimately enhances the translational efficiency of its target mRNAs [41]. In addition, YTHDF3 potentially affects the specificity of YTHDF1 and YTHDF2 binding to mRNAs. YTHDF3 can act synergistically with YTHDF1 and YTHDF2 to affect the translation and degradation of m6A-modified mRNAs [29, 31]. Therefore, YTHDF3 affects normal biological functions by regulating the translation and degradation of m6A-modified transcripts [29]. Dysregulation of its expression may contribute to biological abnormalities and ultimately to diseases, including cancer.
YTHDF3 can synergize with poly(A)-binding protein cytoplasmic 1 (PABPC1) and eukaryotic translation initiation factor 4gamma2 (eIF4G2), but not YTHDF1, to bind to the m6A motif in the 5'UTR of CCND1 mRNA and promote CCND1 translation. CCND1 can further increase the renewal capacity of hematopoietic stem cells [42]. In addition, YTHDF3 has been found to induce cellular autophagy in response to nutritional deficiencies. YTHDF3 expression is significantly increased in malnourished mouse embryonic fibroblasts (MEFs), and it promotes FOXO3 translation by recognizing the m6A sites of FOXO3 mRNA, which further induces cellular autophagy [43, 44]. Furthermore, YTHDF3 is capable of negatively regulating antiviral immunity by upregulating FOXO3 to inhibit the expression of interferon-stimulated genes (ISGs) [45]. Notably, during the induction of autophagy, YTHDF3 is believed to bind to m6A around the termination codon of the FOXO3 transcript, promoting the translation of FOXO3 by recruiting eukaryotic translation initiation factor 3 subunit A (eIF3a) and eukaryotic initiation factor 4B (eIF4B) at the 5' end [43]. Conversely, in regulating antiviral immunity, YTHDF3 is believed to bind directly to the start region of the FOXO3 mRNA in a non-m6A-dependent manner, promoting the translation of FOXO3 by synergizing with PABP1 and eIF4G2 [45]. According to previous reports, the overexpression of YTHDF3 induces brain metastasis in breast cancer patients, which leads to a poor patient prognosis. YTHDF3 promotes the translation of m6A-modified mRNAs of breast cancer brain metastasis-associated genes (e.g., ST6GALNAC5, GJA1, and EGFR) by recruiting eIF3a and increasing the expression of the corresponding proteins, which ultimately affects the brain metastasis of breast cancer [46]. YTHDF3 also enhances its own translation efficiency by binding to the m6A residue of the 5'UTR of its mRNA. In addition, the copy number of the YTHDF3 gene was found to be significantly increased in breast cancer, which led to the transcriptional upregulation of YTHDF3 [46, 47]. Patients harboring genetic alterations in YTHDF3 have a relatively poor survival rate [47]. Moreover, miR-106b-5p was found to downregulate YTHDF3 expression in breast cancer [48]. Both the mRNA and protein levels of YTHDF3 increased under the condition of glucose deficiency. YTHDF3 overexpression was found to increase the expression of glycolytic genes and promotes the degradation of the antisense RNA DICER1 (DICER1-AS1). DICER1-AS1 restricts glycolysis, proliferation and metastasis in PC cells and acts as a PC inhibitory factor. The effect of YTHDF3 on DICER1-AS1 was found to be significantly enhanced in glucose-deficient pancreatic cancer cells. In addition, miR-5586-5p counteracts the inhibitory effect of YTHDF3 on DICER1-AS1 [49].
On the basis of the aforementioned findings, YTHDF3 overexpression promotes cancer. Differential analysis utilizing the TCGA database revealed significant differences in m6A modification regulatory genes between Wilms tumor tissues and normal tissues, with upregulation of YTHDF3 expression in Wilms tumor tissues [50]. The relationship between YTHDF3 gene polymorphisms and Wilms tumor remains incompletely understood. Moreover, the effect of genetic variation in YTHDF3 on its protein expression has not been adequately investigated, and whether YTHDF3 SNPs affect its mRNA stability or protein expression levels needs to be further verified. Our study revealed that the YTHDF3 rs2241753 AA genotype has an enhancing effect on Wilms tumor susceptibility in female children. The mechanism underlying this effect requires additional investigation, possibly because the variant of YTHDF3 rs2241753 affects the expression of its downstream Wilms tumor-associated genes. YTHDF3 rs2241753 is located in the TFBS, and single nucleotide variants here may affect the binding of mRNA to transcription factors, which in turn causes changes in gene expression levels. Furthermore, female carriers of 1-3 risk genotypes also exhibited increased Wilms tumor susceptibility, suggesting that a potential interaction among these variants impacts the risk of Wilms tumor. However, no significant association was observed between YTHDF3 SNPs and Wilms tumor susceptibility in the overall sample included our study. This may be because the effect of YTHDF3 SNPs on Wilms tumor is relatively subtle or population specific.
Our study presents a multicenter analysis with a large sample size, demonstrating for the first time the relationship between YTHDF3 SNPs and Wilms tumor susceptibility. However, there are some limitations that should be acknowledged. First, the study was restricted to one race, as all participants were from the Chinese Han population. Second, the effects of lifestyle and environmental factors on Wilms tumor incidence were not considered. Third, the number of SNPs in the study was relatively small, indicating the need to include more YTHDF3 SNPs and analyze their combined effects on Wilms tumor risk. Finally, additional protein-level studies are needed to determine the potential mechanisms by which YTHDF3 SNPs influence Wilms tumor susceptibility.
In conclusion, our findings suggest that the presence of the YTHDF3 rs2241753 AA genotype or 1-3 YTHDF3 risk genotypes may increase the risk of Wilms tumor occurrence in female children. Our findings need to be validated in different populations after controlling for multiple confounders.
Abbreviations
SNP: single nucleotide polymorphism; GWAS: genome-wide association study; m6A: N6-methyladenosine; UTR: untranslated region; TFBS: transcription factor-binding site; LD: linkage disequilibrium; HWE: Hardy‒Weinberg equilibrium; OR: odds ratio; CI: confidence interval; eQTL: expression quantitative trait loci; PABPC1: poly (A) binding protein cytoplasmic 1; eIF4G2: eukaryotic translation initiation factor 4 gamma 2; MEF: mouse embryonic fibroblast; ISG: interferon-stimulated gene; eIF3a: eukaryotic translation initiation factor 3 subunit A; eIF4B: eukaryotic initiation factor 4B; DICER1-AS1: antisense RNA1 of DICER1.
Supplementary Material
Supplementary figure and tables.
Acknowledgements
This study was funded by grants from the Youth Medical Innovation and Practice Research Program of Guangzhou (No: 2023QNYXYB010), the National Natural Science Foundation of China (No: 82003523), the Major Science and Technology Special Project of Wenzhou (No: ZY2020021), and the Natural Science Foundation of Zhejiang Province (No: LGF21H260012).
Author contributions
All the authors contributed significantly to this work. WF, RXH, GL, and JH designed the research study; HZ, JZ, JC, SL, JR, GL, JH, and WF performed the research study and collected the samples and clinical data; CD and RXH analyzed the data; CD, YH, RXH, and WF wrote the paper; and JH prepared all the tables. All the authors have read and approved the final version of the manuscript.
Data availability statement
All the data are available upon request.
ORCID
Rui-Xi Hua, https://orcid.org/0000-0001-5319-656X.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Davidoff AM. Wilms' tumor. Curr Opin Pediatr. 2009;21:357-64
2. Pastore G, Znaor A, Spreafico F, Graf N, Pritchard-Jones K, Steliarova-Foucher E. Malignant renal tumours incidence and survival in European children (1978-1997): report from the Automated Childhood Cancer Information System project. Eur J Cancer. 2006;42:2103-14
3. Bao PP, Li K, Wu CX, Huang ZZ, Wang CF, Xiang YM. et al. [Recent incidences and trends of childhood malignant solid tumors in Shanghai, 2002-2010]. Zhonghua Er Ke Za Zhi. 2013;51:288-94
4. Breslow N, Olshan A, Beckwith JB, Green DM. Epidemiology of Wilms tumor. Med Pediatr Oncol. 1993;21:172-81
5. Zhu J, Fu W, Jia W, Xia H, Liu GC, He J. Association between NER Pathway Gene Polymorphisms and Wilms Tumor Risk. Mol Ther Nucleic Acids. 2018;12:854-60
6. Coppes MJ. Wilms tumor: to cure and understanding. Crit Rev Oncol Hematol. 1995;18:179-96
7. Dome JS, Graf N, Geller JI, Fernandez CV, Mullen EA, Spreafico F. et al. Advances in Wilms Tumor Treatment and Biology: Progress Through International Collaboration. J Clin Oncol. 2015;33:2999-3007
8. Sonn G, Shortliffe LM. Management of Wilms tumor: current standard of care. Nat Clin Pract Urol. 2008;5:551-60
9. Gibson TM, Mostoufi-Moab S, Stratton KL, Leisenring WM, Barnea D, Chow EJ. et al. Temporal patterns in the risk of chronic health conditions in survivors of childhood cancer diagnosed 1970-99: a report from the Childhood Cancer Survivor Study cohort. Lancet Oncol. 2018;19:1590-601
10. Haber DA, Buckler AJ, Glaser T, Call KM, Pelletier J, Sohn RL. et al. An internal deletion within an 11p13 zinc finger gene contributes to the development of Wilms' tumor. Cell. 1990;61:1257-69
11. Rivera MN, Kim WJ, Wells J, Driscoll DR, Brannigan BW, Han M. et al. An X chromosome gene, WTX, is commonly inactivated in Wilms tumor. Science. 2007;315:642-5
12. Treger TD, Chowdhury T, Pritchard-Jones K, Behjati S. The genetic changes of Wilms tumour. Nat Rev Nephrol. 2019;15:240-51
13. Andrade RC, Cardoso LC, Ferman SE, Faria PS, Seuánez HN, Achatz MI. et al. Association of TP53 polymorphisms on the risk of Wilms tumor. Pediatr Blood Cancer. 2014;61:436-41
14. Turnbull C, Perdeaux ER, Pernet D, Naranjo A, Renwick A, Seal S. et al. A genome-wide association study identifies susceptibility loci for Wilms tumor. Nat Genet. 2012;44:681-4
15. Zhu J, Jia W, Wu C, Fu W, Xia H, Liu G. et al. Base Excision Repair Gene Polymorphisms and Wilms Tumor Susceptibility. EBioMedicine. 2018;33:88-93
16. Fu W, Zhu J, Xiong SW, Jia W, Zhao Z, Zhu SB. et al. BARD1 Gene Polymorphisms Confer Nephroblastoma Susceptibility. EBioMedicine. 2017;16:101-5
17. Lin A, Zhou M, Hua RX, Zhang J, Zhou H, Li S. et al. METTL3 polymorphisms and Wilms tumor susceptibility in Chinese children: A five-center case-control study. J Gene Med. 2020;22:e3255
18. Hua RX, Fu W, Lin A, Zhou H, Cheng J, Zhang J. et al. Role of FTO gene polymorphisms in Wilms tumor predisposition: A five-center case-control study. J Gene Med. 2021;23:e3348
19. Hua RX, Liu J, Fu W, Zhu J, Zhang J, Cheng J. et al. ALKBH5 gene polymorphisms and Wilms tumor risk in Chinese children: A five-center case-control study. J Clin Lab Anal. 2020;34:e23251
20. Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA Modifications in Gene Expression Regulation. Cell. 2017;169:1187-200
21. Chang LL, Xu XQ, Liu XL, Guo QQ, Fan YN, He BX. et al. Emerging role of m6A methylation modification in ovarian cancer. Cancer Cell Int. 2021;21:663
22. Zhang W, Xiao P, Tang J, Wang R, Wang X, Wang F. et al. m6A Regulator-Mediated Tumour Infiltration and Methylation Modification in Cervical Cancer Microenvironment. Front Immunol. 2022;13:888650
23. Liu Z, He J, Han J, Yang J, Liao W, Chen N. m6A Regulators Mediated Methylation Modification Patterns and Tumor Microenvironment Infiltration Characterization In Nasopharyngeal Carcinoma. Front Immunol. 2021;12:762243
24. Meyer KD, Jaffrey SR. Rethinking m(6)A Readers, Writers, and Erasers. Annu Rev Cell Dev Biol. 2017;33:319-42
25. Guan Q, Lin H, Hua W, Lin L, Liu J, Deng L. et al. Variant rs8400 enhances ALKBH5 expression through disrupting miR-186 binding and promotes neuroblastoma progression. Chin J Cancer Res. 2023;35:140-62
26. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93-5
27. Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ. et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177-89
28. Patil DP, Pickering BF, Jaffrey SR. Reading m(6)A in the Transcriptome: m(6)A-Binding Proteins. Trends Cell Biol. 2018;28:113-27
29. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ. et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27:315-28
30. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117-20
31. Li A, Chen YS, Ping XL, Yang X, Xiao W, Yang Y. et al. Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 2017;27:444-7
32. Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC. et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18:110
33. Li Z, Weng H, Su R, Weng X, Zuo Z, Li C. et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N(6)-Methyladenosine RNA Demethylase. Cancer Cell. 2017;31:127-41
34. Wang Z, Zhuo Z, Li L, Hua RX, Li L, Zhang J. et al. The contribution of YTHDF2 gene rs3738067 A>G to the Wilms tumor susceptibility. J Cancer. 2021;12:6165-9
35. Lin A, Hua RX, Zhou M, Fu W, Zhang J, Zhou H. et al. YTHDC1 gene polymorphisms and Wilms tumor susceptibility in Chinese children: A five-center case-control study. Gene. 2021;783:145571
36. He J, Wang F, Zhu J, Zhang R, Yang T, Zou Y. et al. Association of potentially functional variants in the XPG gene with neuroblastoma risk in a Chinese population. J Cell Mol Med. 2016;20:1481-90
37. Chen YP, Liao YX, Zhuo ZJ, Yuan L, Lin HR, Miao L. et al. Association between genetic polymorphisms of base excision repair pathway and glioma susceptibility in Chinese children. World J Pediatr. 2022;18:632-5
38. Zhuo Z, Lu H, Zhu J, Hua RX, Li Y, Yang Z. et al. METTL14 Gene Polymorphisms Confer Neuroblastoma Susceptibility: An Eight-Center Case-Control Study. Mol Ther Nucleic Acids. 2020;22:17-26
39. Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomed Pharmacother. 2019;112:108613
40. Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z. et al. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 2021;6:74
41. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H. et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388-99
42. Zhang X, Cong T, Wei L, Zhong B, Wang X, Sun J. et al. YTHDF3 modulates hematopoietic stem cells by recognizing RNA m(6)A modification on Ccnd1. Haematologica. 2022;107:2381-94
43. Hao W, Dian M, Zhou Y, Zhong Q, Pang W, Li Z. et al. Autophagy induction promoted by m(6)A reader YTHDF3 through translation upregulation of FOXO3 mRNA. Nat Commun. 2022;13:5845
44. Hao W, Dian M, Wang J, Sun Y, Xiao D. Epitranscriptomic turbo for autophagy boost: m(6)A reader YTHDF3. Autophagy. 2023;19:1882-4
45. Zhang Y, Wang X, Zhang X, Wang J, Ma Y, Zhang L. et al. RNA-binding protein YTHDF3 suppresses interferon-dependent antiviral responses by promoting FOXO3 translation. Proc Natl Acad Sci U S A. 2019;116:976-81
46. Chang G, Shi L, Ye Y, Shi H, Zeng L, Tiwary S. et al. YTHDF3 Induces the Translation of m(6)A-Enriched Gene Transcripts to Promote Breast Cancer Brain Metastasis. Cancer Cell. 2020;38:857-71.e7
47. Anita R, Paramasivam A, Priyadharsini JV, Chitra S. The m6A readers YTHDF1 and YTHDF3 aberrations associated with metastasis and predict poor prognosis in breast cancer patients. Am J Cancer Res. 2020;10:2546-54
48. Liu M, Zhou S, Wang J, Zhang Q, Yang S, Feng J. et al. Identification of genes associated with survival of breast cancer patients. Breast Cancer. 2019;26:317-25
49. Hu Y, Tang J, Xu F, Chen J, Zeng Z, Han S. et al. A reciprocal feedback between N6-methyladenosine reader YTHDF3 and lncRNA DICER1-AS1 promotes glycolysis of pancreatic cancer through inhibiting maturation of miR-5586-5p. J Exp Clin Cancer Res. 2022;41:69
50. Jia C, Gao H, Ma W, Liu X, Chang M, Sun F. Identification of the expression patterns and potential prognostic role of m6A-RNA methylation regulators in Wilms Tumor. BMC Med Genomics. 2023;16:222
Author contact
![]() Corresponding authors: Wen Fu or Rui-Xi Hua, Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, 9 Jinsui Road, Guangzhou 510623, Guangdong, China, Email: lydia_fwcom (Wen Fu) or huaruixsysu.edu.cn (Rui-Xi Hua).
Corresponding authors: Wen Fu or Rui-Xi Hua, Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, 9 Jinsui Road, Guangzhou 510623, Guangdong, China, Email: lydia_fwcom (Wen Fu) or huaruixsysu.edu.cn (Rui-Xi Hua).

 Global reach, higher impact
Global reach, higher impact