Impact Factor
ISSN: 1837-9664
J Cancer 2025; 16(1):351-367. doi:10.7150/jca.99278 This issue Cite
Research Paper
SIGLEC1 as a novel prognostic factor is regulated by Yiqi Huayu Jiedu Decoction in colorectal cancer
1. School of Medicine, Nanjing University of Chinese Medicine, Nanjing, 210023, China.
2. Affiliated Hospital of Nanjing University of Chinese Medicine, Jiangsu Province Hospital of Chinese Medicine, Nanjing, 210029, China.
3. School of Elderly Care Services and Management, Nanjing University of Chinese Medicine, Nanjing, 210023, China.
Received 2024-6-4; Accepted 2024-10-19; Published 2025-1-1
Abstract
We studied the prognostic value of SIGLEC1 in colorectal cancer (CRC) using bioinformatics. SIGLEC1 exhibited differential expression between the tumor and control samples, and improved survival was observed in patients with increased SIGLEC1 expression. The univariate and multivariate analyses confirmed SIGLEC1 as an independent prognostic factor for CRC, based on which a nomogram was constructed for predicting survival in patients with CRC. Additionally, higher SIGLEC1 expression was correlated with increased immunological/stromal/ESTIMATE scores as well as immune cell infiltration and was strongly and positively associated with T helper cells and macrophages. Furthermore, significant positive correlations were observed between SIGLEC1 expression and inhibitory/coinhibitory immunological genes. Additionally, the TIDE results of patients with CRC showed that increased SIGLEC1 expression was related to poorer immunotherapeutic responses. In clinical samples from patients with CRC, a decrease in SIGLEC1 expression was noted compared to para-cancerous tissues and samples from patients who received Yiqi Huayu Jiedu Decoction (YHJD) treatment. The in vivo results indicated the superior efficacy of YHJD against CRC in inhibiting tumor metastasis. Our study demonstrates that SIGLEC1 serves as a prognostic factor for CRC, strongly linked to immune response, and can be modulated by YHJD, suggesting novel avenues for CRC treatment.
Keywords: SIGLEC1, Colorectal Cancer, Bioinformatics, Yiqi Huayu Jiedu Decoction, Traditional Chinese Medicine
Introduction
Colon cancer, also known as colorectal cancer (CRC), is a serious form of cancer typically arising at the junction of the rectum and sigmoid colon. This cancer frequently evolves from precancerous polyps in the colon [1]. CRC ranks as the third most prevalent and the second most lethal cancer globally, with approximately 1.9 million people being diagnosed each year [2] Colon cancer development is attributed to complex related factors, among which the genetic factors should be attach importance to [3]. Individuals suffering from colon cancer commonly endure gastrointestinal symptoms, including alterations in bowel habits, the presence of an abdominal mass, and abdominal pain, significantly impacting their everyday lives [4].
The accurate diagnosis of colon cancer and devising effective treatment options against it is a global concern among healthcare professionals. Current treatment strategies for CRC are chemotherapy, radiotherapy, and surgical resection, among which surgery is the most used approach. However, even after surgery, CRC exhibits a recurrence rate of 30% [5]. Moreover, surgical resection may cause postoperative complications such as constipation or diarrhea owing to the removal of a part of the healthy colon [6]. In summary, there is a lack of a comprehensive strategy for the early diagnosis and treatment of colon cancer patients, resulting in their prognosis remaining less than optimal. Therefore, future studies should be performed to elucidate the molecular mechanisms of colon cancer, screen new markers and therapeutic targets, and provide innovative methods for managing patients with colon cancer.
Sialic acid-binding immunoglobulin-like lectin 1 (SIGLEC1/CD169) is a receptor found on the surfaces of macrophages and dendritic cells, and its expression can be induced by interferons. It is a critical candidate gene for disease surveillance [7, 8]. The SIGLEC family comprises transmembrane proteins that interact with sialic acid and contain C2-set immunoglobulin domains, thereby regulating the function and immune response of innate and adaptive immune cells [9, 10]. Previous studies suggest that SIGLEC1 contributes to the inflammatory immune response following a pathogen infection because it is elevated by inflammatory macrophages and affects T-cell activation and functions [11, 12]. SIGLEC1 is markedly related to inflammatory disorders, including asthma, pulmonary active tuberculosis, COVID-19, and chronic obstructive pulmonary disease [13, 14]. Furthermore, SIGLEC1 has been linked to conditions such as type 1 diabetes and systemic lupus erythematosus [15, 16]. CD169, which is encoded by the SIGLEC1 gene, is a protein found in a specific subset of macrophages located in secondary lymphoid organs. CD169+ macrophages elicit anti-tumor immune responses by capturing and presenting antigens derived from tumors, thereby inhibiting tumor growth and spread [17-19]. Recent researches have shown SIGLEC1+ macrophages in CRC and hepatocellular carcinoma, suggesting their potential involvement in tumor progression [20, 21]. CD169 may be a specific marker of inflammatory macrophages with pro-inflammatory properties. The pro-inflammatory cytokine, type 1 interferon, induces strong expression of CD169 in macrophages of CRC[22]. Despite these advancements, the therapeutic potential of targeting CD169 in CRC remains largely unexplored. Current research has focused primarily on elucidating the mechanisms underlying CD169's functional roles, with limited investigations into the development of direct CD169-targeted therapies for CRC prevention or treatment [23, 24]. This gap highlights the need for further studies, particularly those aimed at translating our understanding of CD169's role in CRC into novel therapeutic strategies.
Traditional Chinese Medicine (TCM) that comprises multiple herbs with different active components, target sites, and pathway regulation activities can be a suitable treatment option against malignant diseases. Yiqi Huayu Jiedu Decoction (YHJD), a traditional polyherbal formulation, has been utilized in clinical practice since 1078 AD for addressing the pathogenesis of “spleen deficiency and stasis toxin” in CRC. [25]. According to Chinese Pharmacopoeia (2020 Edition), each 200 mL of YHJD consists of 15 g of Astragalus membranaceus (Fisch.), 15 g of Codonopsis pilosula (Nannf.), 10 g of Atractylodes macrocephala (Koidz.), 15 g of Poria Cocos (Schw.) (Wolf.), 15 g of Dioscorea opposita (Thunb.), 5 g of Prunus mume (Siebold & Zucc.), 10 g of Sparganium stoloniferum (Graebn.), 10 g of Curcuma phaeocaulis (Valeton.), 10 g of Agrimonia Pilosa (Ledeb.), 30 g of Patrinia villosa (Juss.), and 30 g of Glycyrrhiza uralensis (Fisch.), and all these herb names are available at “World Flora Online” (www.worldfloraonline.org). Some recent studies have shown YHJD inhibitory effects on CRC, indicating its potential for preventing and treating gastrointestinal CRC with satisfactory efficacy [25-27]. Zhou et al. indicates that YHJD can prohibit liver metastasis of CRC in vivo, especially through the PI3K-AKT signaling pathway and the nature killer (NK)-cell-led immune response [25]. However, the active ingredients and underlying molecular mechanisms require further thorough investigation. 5-Fluorouracil (5-FU) is a crucial fluoropyrimidine antimetabolite drug that exerts complex antitumor effects by interfering with a key enzyme in the DNA synthesis process - thymidylate synthase, and by being mistakenly incorporated into RNA and DNA, leading to DNA strand breaks. 5-FU plays a significant role in the treatment of colorectal cancer and is also widely used in the treatment of various other cancers, including breast cancer, head and neck cancer, ovarian cancer, gastrointestinal tumors, and basal cell carcinoma [28]. 5-Fluorouracil does show a limited overall response rate in monotherapy for colorectal cancer [29]. This generally implies that, although they may produce therapeutic effects in some patients, the response rate of fluoropyrimidine drugs as a monotherapy is not sufficient to make it the preferred treatment option for all patients with colorectal cancer. Therefore, fluoropyrimidine drugs often play an adjuvant role in the treatment of colorectal cancer. They are frequently combined with other chemotherapy drugs, targeted therapeutic drugs, or immunotherapy drugs to improve treatment efficacy and patient survival rates. This combined treatment regimen can utilize the synergistic effects between different drugs to attack multiple growth and survival pathways of tumor cells, thereby more effectively inhibiting tumor growth and spread [30].
Thus, herein, our objective was to assess the prognostic significance and potential mechanism of SIGLEC1 in CRC, and to explore the regulatory impact of YHJD on SIGLEC1, utilizing bioinformatics analyses for this purpose.
Materials and methods
Data source
To determine the expression pattern of SIGLEC1, we utilised The Cancer Genome Atlas (TCGA) database to select Primary Solid Tumor, Recurrent Solid Tumor, Solid Tissue Normal a total of 521 samples with expression profiles of 480 cancer tissue samples and 41 control samples, and excluded samples without survival information and duplicates. 430 cancer samples with Primary Solid Tumor and survival information were selected for survival analysis (Table S1). Meanwhile, the overall survival (OS) and clinic indicators were obtained from the TCGA-CRC. Subsequently, the CRC-related datasets, GSE39582 (annotation platform: GPL570) and GSE10972 (annotation platform: GPL6104), were collected from the Gene Expression Omnibus (GEO) database. The GSE39582 encompassed 566 colorectal cancer samples and 19 non-tumour colorectal mucosa samples. After excluding those lacking survival information and survival time data, a selection of 19 normal and 556 cancer samples was made to conduct a survival analysis (Table S2). Also, the GSE10972 included 24 primary tumors of CRC and 24 control samples. Of these, 430 CRC samples from TCGA-CRC (out of 463) and the 556 CRC samples from the GSE39582 dataset with available clinical information were analyzed for survival outcomes.
Characterization of the involvement of SIGLEC1 in CRC
To investigate the role of SIGLEC1 in CRC, the expression differences were compared between the CRC and normal controls in TCGA-CRC, GSE39582, and GSE10972 datasets by the Wilcoxon test (p < 0.05). Based on the median value of SIGLEC1 expression, the CRC patients with OS in TCGA-CRC and GSE39582 datasets were divided separately into low- and high-SIGLEC1 expression groups. Subsequently, K-M analysis and the log-rank test were employed to compare the survival rates of patients across these two expression groups using the survminer (v. 0.4.9) R package (https://CRAN.R-project.org/package=survminer). Then, the expression levels of SIGLEC1 in different clinical characteristics were compared by the Wilcoxon (two groups) and Kruskal-Wills tests (three groups or more), including age, race, sex, tumor stage, and p-TNM staging. Finally, we conducted univariate and multivariate Cox regression analyses to ascertain if SIGLEC1 expression served as an independent prognostic factor using the survminer (v. 0.4.9) R package (HR ≠ 1, p < 0.05). This was followed by the construction of a prognostic nomogram using the rms (v. 6.5-0) R package (https://CRAN.R-project.org/package=rms). Moreover, the calibration curve was applied to evaluate the predictive ability of the nomogram model.
Investigation of potential mechanisms of SIGLEC1 in CRC
As the aforementioned, we categorized the TCGA-CRC patients into low SIGLEC1 expression and high expression groups based on the median value of SIGLEC1 expression. Here, the differentially expressed genes (DEGs) were screened between the high- and low-SIGLEC1 expression groups via the limma (v 3.56.2) R package [31]. The log2 fold change (FC) values of each DEG were sorted from largest to smallest. Meanwhile, the and then Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways gene sets were downloaded from the Molecular Signatures Database (MSigDB, https://www.gsea-msigdb.org/gsea/msigdb) as background gene sets. Afterward, the Gene Set Enrichment Analysis (GSEA) was performed by the clusterProfiler (v 4.10.0) R package (|normalized enrichment score (NES)| > 1, p < 0.05) [32]. Additionally, we utilized the ESTIMATE algorithm to analyze and compare the immunological, stromal, and ESTIMATE scores between the low- and high-SIGLEC1 expression groups by the Wilcoxon test (p < 0.05). Through the single-sample GSEA (ssGSEA) algorithm of GSVA (v. 1.46.0) R package [33], the fraction of 29 immune cells was calculated, and the Wilcoxon test was used to compare the fraction differences between the high- and low-SIGLEC1 expression groups (p < 0.05). Simultaneously, the association between SIGLEC1 expression and immune-cell infiltration was studied using the Tumor IMmune Estimation Resource (TIMER, https://cistrome.shinyapps.io/timer/) database. Moreover, we employed immunologic signature gene sets as a reference to calculate the enrichment score (GSVA score) for each gene set in each sample of TCGA-CRC using the Gene Set Variation Analysis (GSVA) method. The correlation between SIGLEC1 expression and the obtained GSVA scores was then determined through Pearson analysis (p < 0.05 and |cor| ≥ 0.4). Except for the above analysis, Spearman correlation analysis was performed to find SIGLEC1-correlated genes with p-value < 0.05 and |cor| ≥ 0.6, and the functions of the obtained correlated genes were analyzed with the help of clusterProfiler (v 4.10.0) R package.
The effect of SIGLEC1 on the immune therapy of patients with CRC
To analyze the effect of SIGLEC1 on the immune therapy of patients with CRC, we identified the correlation between SIGLEC1 expression and inhibitory/coinhibitory immune genes [34] by the Spearman method. Additionally, we compared the TIDE score and the number of immunotherapeutic responders between the low- and high-SIGLEC1 expression groups and determined the correlation between SIGLEC1 expression and the TIDE score via the Wilcoxon test (p < 0.05).
Prediction of candidate drugs targeted to SIGLEC1 in CRC
SIGLEC1 was uploaded to the Comparative Toxicogenomics Database (CTD, https://ctdbase.org/), followed by candidate drug screening by upregulating or downregulating its expression. Next, the obtained drug-SIGLEC1 pairs were imported into Cytoscape to construct a drug-gene network.
Clinical samples
The tumor samples from all patients with CRC used in this study were provided by Jiangsu Provincial Hospital of Chinese Medicine, Affiliated Hospital of Nanjing University of Chinese Medicine (No.2022NL-110-02). From November 2021 to July 2022, surgically excised samples of tumor and para-cancerous tissue were gathered from 16 patients diagnosed with locally colorectal cancer (CRC) that were deemed resectable. The comprehensive clinicopathological profiles of all these patients are detailed in the supplementary in table S3. Treatment response was evaluated using Guidelines for Diagnosis and Treatment of Colorectal Cancer issued by the CSCO (Chinese Society of Clinical Oncology) and National Health Commission of the People's Republic of China [35]. Tumor and para-cancerous tissue samples of the patients were immediately treated at 4°C. Following the addition of Trizol, the samples were preserved in an ultra-low-temperature refrigerator at -80°C for later RNA or protein extraction. Simultaneously, patient data including the patient's admission number, gender, age, CRC type, and treatment stage, were collected.
Cell culture
Human colon cancer cells that could be used for the red fluorescence localization of nuclei (HCT-116-mkate2) were originally obtained from Cell Valley (Nanjing) Biotechnology Co., Ltd. The HCT-116-mkate2 cells were grown in a culture medium composed of 90% 1640 culture medium, 10% fetal bovine serum (FBS), and 1X antibiotics. They were maintained in an environment with 5% CO2 and O2 at a temperature of 37°C.
HCT-116-mkate2 xenograft model
Specific pathogen-free male BALB/c nude mice (5-6 weeks old, 20 g, SLAC, Shanghai, China) were used in the experiment. HCT-116-mkate2 cells, numbering 7.0 × 106, were suspended in 150 μL of Dulbecco's Modified Eagle Medium (DMEM) with 10% fetal bovine serum (FBS) and injected into the underarm regions of the hind limbs of the mice. The dimensions of the tumor were assessed every 2 days using a Vernier caliper.
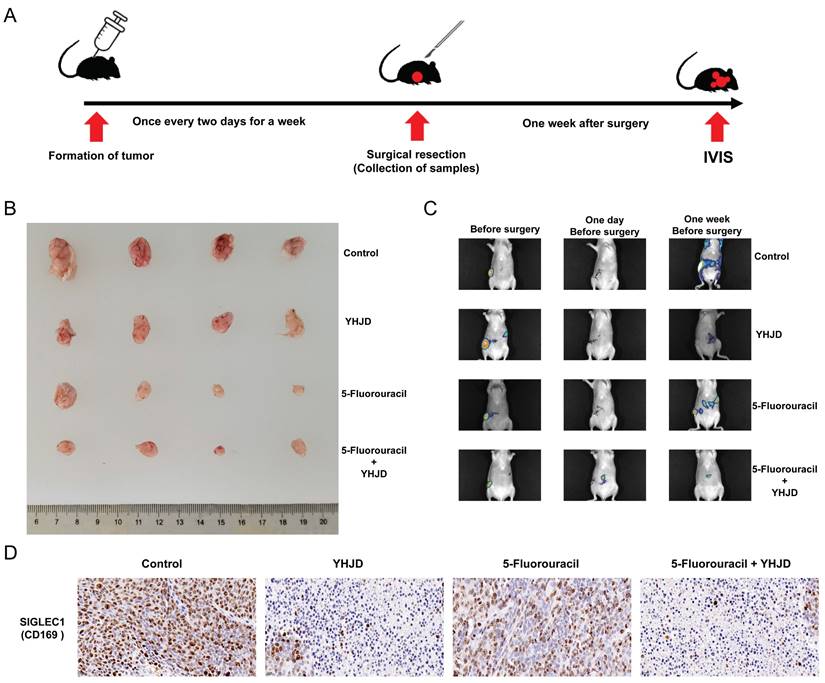
At the end of 14 days, mice bearing tumors of similar sizes were randomly allocated to various groups, including the control group, 5-Fluorouracil (5-Flu), YHJD, and 5-Flu+YHJD groups, with 4 mice in each group. YHJD (3.4 g/mL and 0.2 mL/20 g) and 5-Flu (30 mg/kg) were administered once every 2 days. The YHJD formulation was prepared in accordance with the Chinese Pharmacopoeia (2020 Edition) and a previously reported method [27], and the dose was selected with reference to the comparison of the reported growth inhibitory effects of YHJD on CRC in the mice [25]. During modeling and drug treatment, the mice in the control group were administered normal saline. After a week of treatment, the tumors of the tumor-bearing mice were surgically removed. The surgical incisions made on the mice were stitched up. Subsequently, YHJD was administered to them every two days over a period of one week, and all the mice were euthanized in the fifth week. The in vivo imaging system (IVIS) (Tanon ABL X5, Shanghai Tianneng Technology Co., Ltd.) was used to visualize the tumors on day 0 of modeling, the day before the surgery, and one week after the surgery.
Western blotting
Proteins were isolated using lysis buffer, incubated in SDS buffer, and then separated through SDS-polyacrylamide gel electrophoresis. Following this, they were transferred to a PVDF membrane for further electrophoresis. SIGLEC1/CD169 antibody (ab312840) and β-actin antibody (ab8226) were purchased from Abcam (Cambridge, MA, USA). The immunoreactive protein bands were visualized by means of the GE 600 RGB.
Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR)
RNA extraction from liver tissue and cell samples was carried out using RNAiso Plus reagent (TaKaRa Biotechnology Co., Ltd, Dalian, China), adhering to the protocol provided by the manufacturer. Gene expression quantification was carried out using SYBR green PCR mastermix on a BioRad real-time PCR detection system (Veenendaal, The Netherlands). GAPDH levels served as the normalization standard.
Immunohistochemical Staining
This method was executed in line with the procedures detailed in a previous report. The HRP-Polymer Conjugated anti-Mouse/Rabbit IgG complex was acquired from Maixin-Bio (Fuzhou, China), SIGLEC1/CD169 antibody(ab312840) and Ki67 antibody(ab15580) were purchased from Abcam (Cambridge, MA, USA). Analysis of the immunohistochemical sections was conducted using Mantra 1.01 software from Perkin Elmer (Waltham, MA, USA).
Hematoxylin-eosin staining
Tumor tissues from colorectal cancer (CRC) patients were processed as follows: they were stained with hematoxylin (Shanghai Ruji Biotechnology Development Co., LTD) and eosin (Beijing Solarbio Science & Technology Co., Ltd.), fixed in 4% paraformaldehyde (Sinopharm Co., LTD.), and then dehydrated in a gradient of ethanol (Sinopharm Co., LTD.). Following this, they were embedded in paraffin wax (Sinopharm Co., LTD.) and sectioned into 4-µm thick slices. These sections were subjected to hematoxylin staining for 5 minutes, followed by a 5-minute rinse in running water, and then stained with eosin for 2 minutes. The process concluded with routine dehydration, vitrification, and neutral resin mounting (Cida Biotechnology Co., LTD, Guangzhou). The OLYMPUS imaging system (DP74, Olympus Corporation, Japan) was used to capture images.
Statistical analysis
Statistical analyses were carried out using SPSS 19.0 software. The results were represented as the mean ± SD based on at least three independent experiments. In the experimental sections, to evaluate statistical differences between two groups, the unpaired Student's t-test was utilized. To make comparisons among multiple groups, we employed one-way ANOVA.
Results
Thirty-eight survival-related differentially expressed genes (DEGs) are identified in CRC
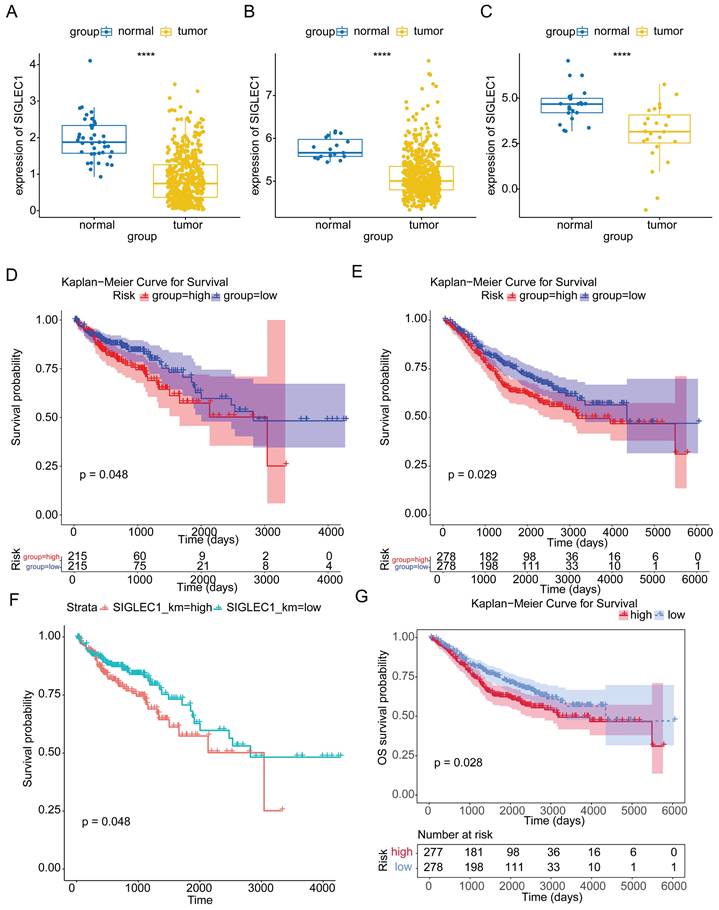
SIGLEC1 expression patterns were consistent among samples derived from TCGA, GSE39582, and GSE10972 cohorts. Gene expression in the tumor samples was markedly reduced compared to that in the control samples (p < 0.05) (Figure 1A to 1C). In patients with CRC, based on the median expression values of SIGLEC1, they were categorized into groups with low and high-SIGLEC1 expression. Notably, CRC patients exhibiting lower SIGLEC1 expression demonstrated improved survival compared to those with higher SIGLEC1 expression (p < 0.05), as observed in both TCGA and GSE39582 cohorts (Figures 1D and 1E). After eliminating the effects of gender and age (age is grouped according to the median value), although there was an individual in the GSE39582 data set with missing age information, it had no effect on the overall results (Figure 1F and G).
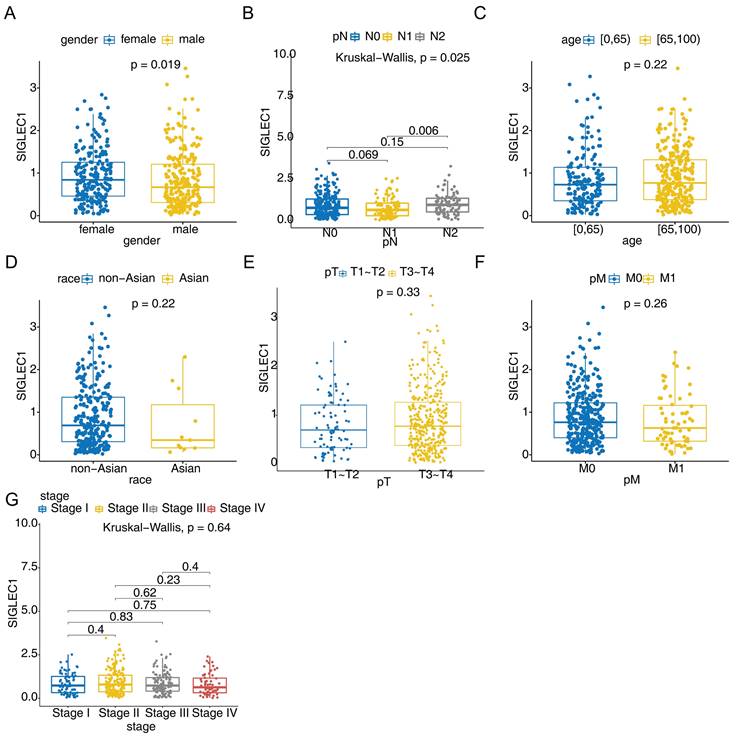
Additionally, the association between SIGLEC1 mRNA expression and the clinical characteristics of colon cancer patients in the TCGA cohort was examined. Among them, the SIGLEC1 expression of female CRC patients was higher than that of male (p < 0.05) (Figure 2A), and there was a significant difference between the N1 and N2 stage (p < 0.05) (Figure 2B). However, no such correlation was observed for age (Figure 2C), race (Figure 2D), T staging (Figure 2E), M staging (Figure 2F), and tumor stage (Figure 2G). These results indicated that SIGLEC1 expression was related to CRC clinicopathological features.
SIGLEC1 was an independent prognostic factor for CRC
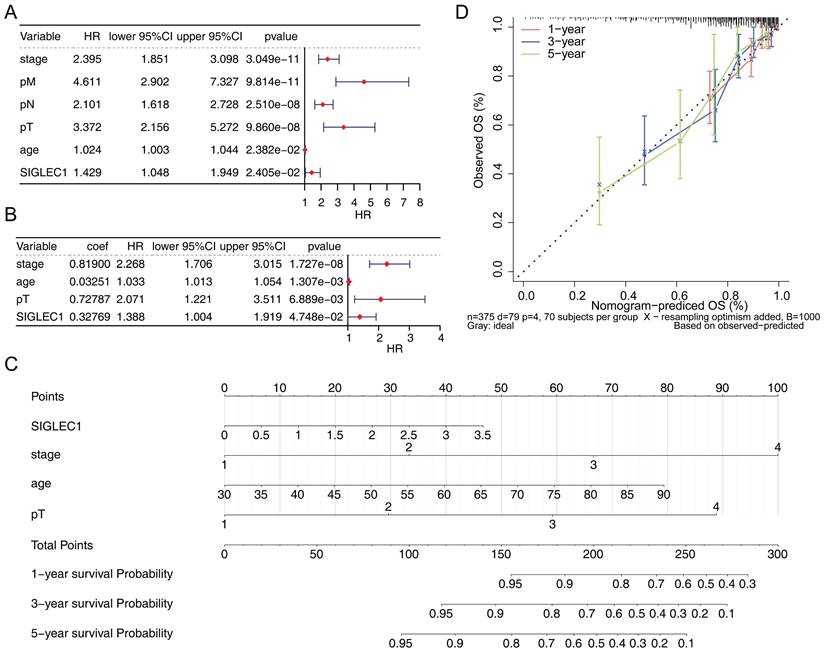
To investigate the independent prognostic factors affecting the prognosis of CRC, the univariate and multivariate Cox regression analyses were performed. Firstly, the outcomes from the univariate Cox regression analysis indicated that SIGLEC1 expression, tumor stage, T/N/M stage, and age were linked to the survival rate of patients with CRC (Figure 3A). Moreover, multivariate Cox regression analysis uncovered a notable association between SIGLEC1 expression and the survival rate of patients with CRC (Figure 3B). What's more, the age and pT were also independent prognostic factors for CRC. This suggested that SIGLEC1 served as an independent prognostic factor for CRC. In addition, we created a nomogram that incorporated independent prognostic factors such as SIGLEC1, tumor stage, age, and T stage, to forecast the 1-year, 3-year, and 5-year survival rates of CRC patients (Figure 3C). The calibration curves indicated a close alignment between the predicted and observed survival rates (Figure 3D), underscoring the nomogram's clinical utility in predicting CRC survival.
SIGLEC1 had a close relationship with the tumor immune microenvironment
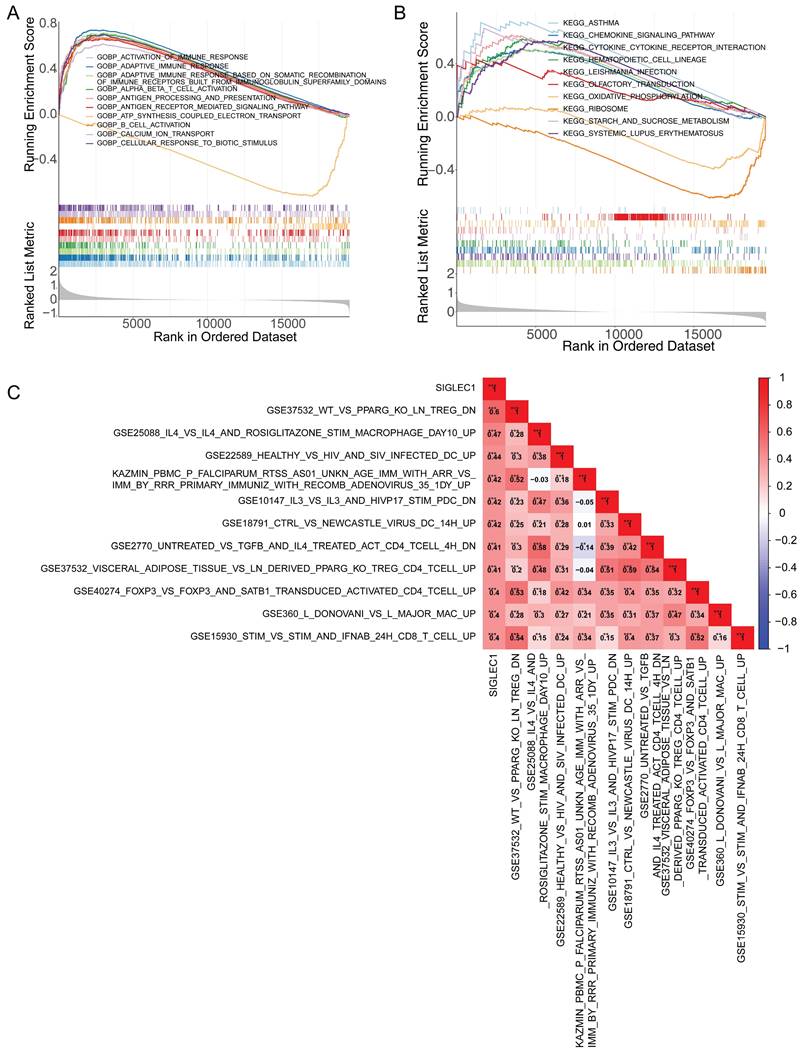
The GSEA demonstrated that immune-response-related GO terms, like “activation of immune response,” “adaptive immune response,” “antigen processing,” and “antigen expression” (Figure 4A) and KEGG pathways, such as cytokine signaling and cytokine-cytokine receptor interaction (Figure 4B) were notably enriched in the low- and high-SIGLEC1 expression groups, indicating a strong correlation between SIGLEC1 and immunity. Additionally, the GSVA results indicated that SIGLEC1 expression was positively associated with multiple immunologic signatures such as GSE37532_WT_VS_PPARG_KO_LN_TREG_DN, GSE25088_IL4_VS_IL4_AND_ROSIGLITAZONE_STIM_MACROPHAGE_DAY10_UP, GSE22589_HEALTHY_VS_HIV_AND_SIV_INFECTED_DC_UP, etc. (Figure 4C), which confirmed the close relationship between SIGLEC1 and immune responses.
Correlation between SIGLEC1 expression and overall survival in colon cancer. The expression of SIGLEC1 in TCGA-CRC, (A) GSE10972, and (B) GSE39582. Kaplan-Meier survival analysis of (D) TCGA-CRC and (E) GSE39582. (F-G) Kaplan-Meier survival analysis of (F) TCGA-CRC and (G) GSE39582. “****” represents p < 0.0001.
The clinicopathological features of SIGLEC1. These included (A) gender, (B) N stage, (C) age, (D) race, (E) T staging, (F) M staging, and (G) tumor stage.
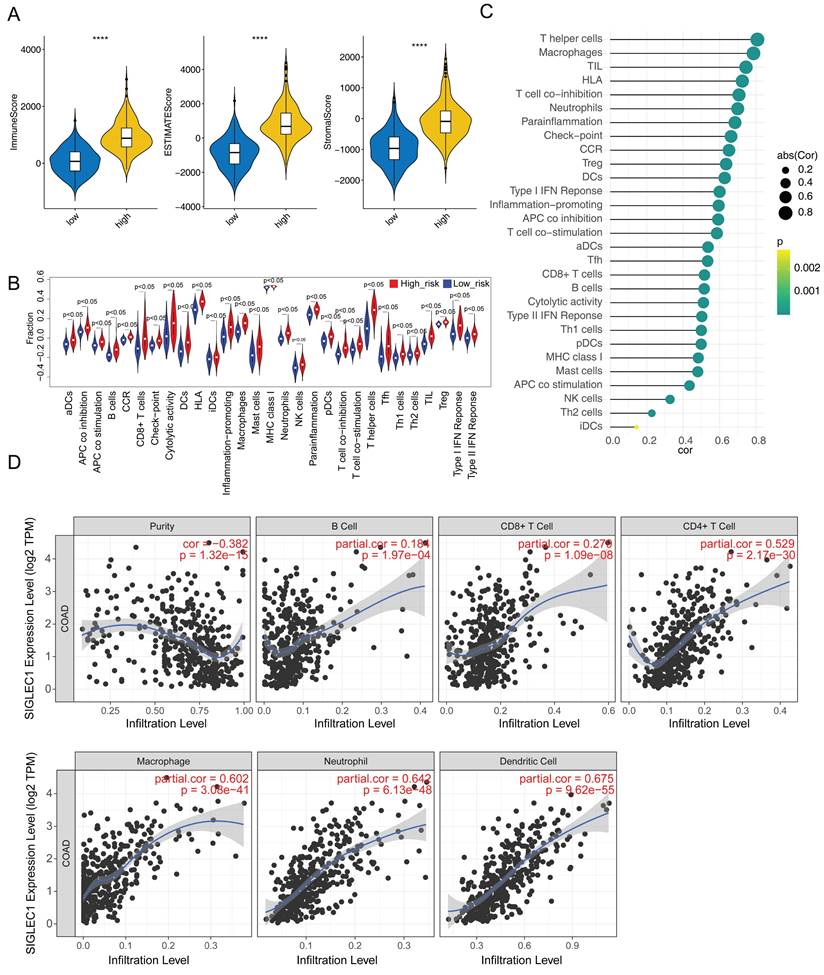
Additionally, we observed significantly increased immunological/stromal/ESTIMATE scores (Figure 5A) and immune cell and functional enrichment scores as well as enhanced pathways in the high SIGLEC1 expression group (Figure 5B). Moreover, SIGLEC1 expression was notably and positively associated with the 29 immune cells, functions, and pathways (Table S4 and Figure 5C). The TIMER results showed significant correlations between SIGLEC1 and immune-cell infiltration (Figure 5D).
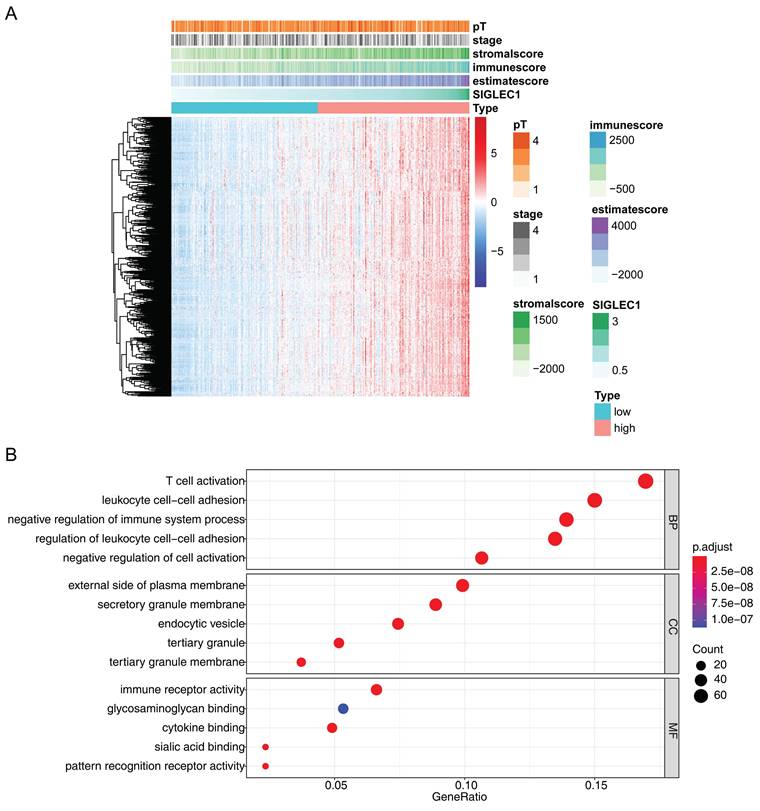
Additionally, 493 SIGLEC1 correlated genes (p < 0.05 and |cor| ≥ 0.6) were identified (Figure 6A), and their functions were notably enriched in immune-response-related GO terms, including “T-cell activation,” “leukocyte cell-cell adhesion,” and “immune receptor activity” (Figure 6B). Overall, these outcomes indicated that SIGLEC1 played a crucial part in the CRC tumor immune microenvironment.
SIGLEC1 affected immunotherapy of patients with CRC
Considering the above results, we investigated the role of SIGLEC1 in the immunotherapy of patients with CRC and found a positive correlation between SIGLEC1 and LAG3, CD38, PDCD1, KLRD1, TNFRSF4, FCRL4, CD200, and LGALS9 (Figure S2A), indicating that the patients with low SIGLEC1 expression might exhibit weaker anti-tumor immunity. The TIDE method predicts the reactivity of immune checkpoint inhibitors; thus, it can be used to evaluate disparities in immunotherapy sensitivity between high- and low-expression groups [34]. The present TIDE results showed that patients with high SIGLEC1 expression exhibited higher TIDE scores and were more sensitive to immunotherapy (Figure S2B). Furthermore, SIGLEC1 expression was positively correlated with the TIDE score (Figure S2B). Figure S2B showed a significant disparity in the number of responses between the groups with high and low expression. Next, we predicted 45 compounds that might regulate SIGLEC1 expression and constructed a drug-SIGLEC1 network (Figure S2C). Among the compounds predicted to upregulate the gene, bisphenol F (C000611646), estradiol (D004958), and genistein (D019833) were common active monomers in Chinese herbs. Moreover, the first-line antineoplastic drug, fluorouracil (D005472), was predicted to trigger SIGLEC1 downregulation, which was likely to further weaken the immune response. Thus, these findings have implications for anti-CRC therapy.
YHJD inhibited the development and metastasis of CRC via upregulated expression of SIGLEC1
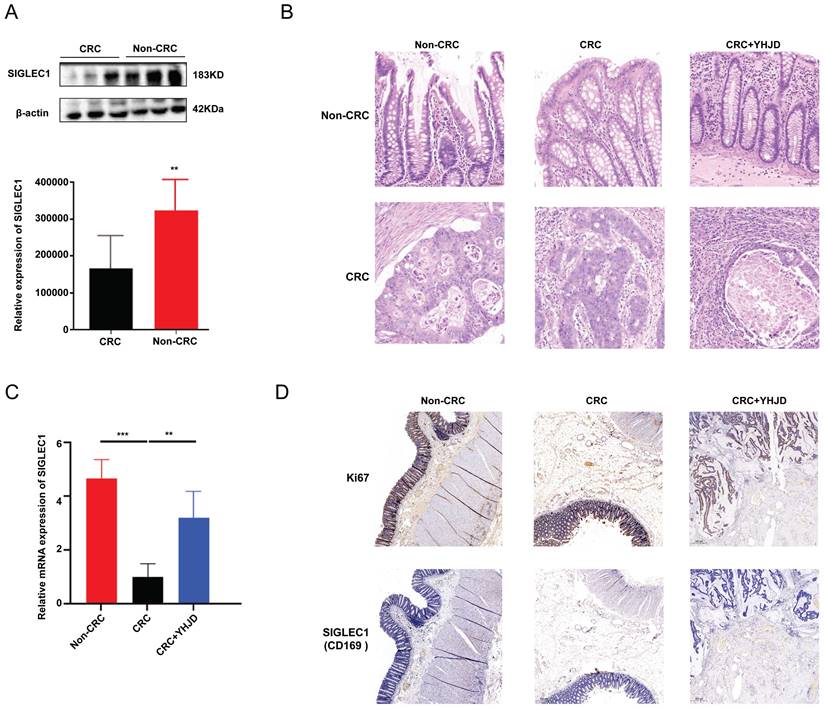
We surgically collected CRC tumors and paracancerous tissues from 16 patients. The Western blot analysis results demonstrated that SIGLEC1/CD169 expression was lower in the CRC samples than in the non-CRC samples (Figure 7A, B). The PCR results showed that YHJD increased SIGLEC1 mRNA levels in clinical samples obtained from the patients (Figure 7C). Relative to the non-YJHD treatment, YJHD treatment increased the expression of SIGLEC1/CD169, aligning with the findings from the database analysis (Figure 7D).
Independent prognostic analysis of CRC. The forest plot of (A) univariate and (B) multivariate cox analysis of the association between SIGLEC1 expression and the overall survival in CRC patients. (C) Nomogram and (D) calibration curve of an independent prognostic model.
Enrichment and Pearson's correlational analysis of SIGLEC1. (A) Enrichment results of SIGLEC1 in GO. (B) Enrichment results of SIGLEC1 in KEGG. (C) Pearson correlation of SIGLEC1 with immune-related pathways.
Analysis of ESTIMATE and ssGSEA. (A) The ESTIMATE algorithm was employed to assess the immune microenvironment in both the high and low expression groups. (B) The infiltration of immune cells in both the high and low expression groups was analyzed. (C) Relevance between SIGLEC1 and immune cells. (D) Association between SIGLEC1 expression and immune cells.
We performed an ectopic xenograft tumor assay to investigate the in vivo anticancer and metastasis inhibitory effects of YHJD. YHJD was administered every two days constantly after tumor formation in the inoculated cells (Figure 8A). A week following YHJD treatment, the tumor at the inoculation site was surgically excised. One week after the surgery, the tumors were detected using the IVIS, and the results indicated that the combined treatment of YHJD and 5-Fluorouracil (5-Flu) substantially suppressed tumor growth (Figure 8B) and metastasis, in comparison to the control group (Figure 8C). Immunohistochemistry assays were conducted to delve deeper into the effects of YHJD on CRC in vivo (Figure. 8D). When YHJD was administered alone, the expression of SIGLEC1/CD169 was significantly upregulated. However, when combined with 5-Flu, this upregulation was even more pronounced. In contrast, the use of 5-Flu alone only induced a modest and non-significant upregulation of SIGLEC1/CD169. Consequently, it is evident that 5-Flu acts as an effective adjuvant, enhancing the ability of YHJD to upregulate SIGLEC1/CD169 expression. These findings indicate that YHJD plays a key role in treating CRC by upregulating SIGLEC1/CD169 expression and inhibiting tumor metastasis.
Expression analysis of genes significantly strongly associated with and SIGLEC1. (A) Heat map and (B) enrichment analysis.
In clinical terms, the SIGLEC1 expression in CRC and the effect of YHJD. (A) The protein expression of SIGLEC1 was assayed by Western blotting, respectively. (B) Exploration of the infiltration of CRC using HE experiments. (C) The mRNA expression of SIGLEC1 were assayed by RT-PCR, respectively. (D) Proteins expressions in the tumor were assessed by immunohistochemistry. Scale bar = 100 µm. (**p < 0.01, ***p < 0.001, n > 4).
Discussion
Herein, we first investigated SIGLEC1 differential expression in both colon tumors and normal tissues, and the results showed significantly downregulated SIGLEC1 expression in the tumor tissues than that in the normal tissues. Simultaneously, patients with colon cancer with lower SIGLEC1 expression exhibited lower survival rates. The univariate and multivariate analyses confirmed the strong correlation between SIGLEC1 and the survival of patients with CRC. To further validate these findings, we selected 10 patients with CRC and collected adjacent cancer tissues as a control. The PCR and western blotting results showed significantly low SIGLEC1 expression in the patients with CRC compared with controls, which aligned with the results of the database analysis (figure 7A).
Additionally, we identified a positive correlation between SIGLEC1 and various immunological characteristics of CRC. We observed that upregulated SIGLEC1 expression was related to increased immunological scores, indicating improved immune responses and prolonged OS rates in the patients. The study found that the high-density infiltration of CD169-positive (CD169+) macrophages in the sinusoids of regional lymph nodes (RLN) is closely related to the increase of cytotoxic CD8 T cells in tumor tissues. The high-density infiltration of CD169+ macrophages is significantly associated with favorable overall survival (OS) in patients with colorectal cancer (CRC) [22]. The significant increase in the number of SIGLEC1-binding cells observed in CRC mouse models is directly related to the disease state, revealing mechanism of CRC-induced changes in the immune microenvironment. As a key molecule for immune regulation, the increased expression level of SIGLEC1 may indicate more complex immune escape mechanisms and worse prognosis. Therefore, SIGLEC1 is expected to become an important biomarker for prognosis assessment in CRC patients and serve as a potential therapeutic target [36]. A previous study has shown that SIGLEC1 is predominantly abundant in immune process signaling pathways in patients with CRC [37], which is consistent with the present findings. Interestingly, a lower expression of SIGLEC1 was observed in CRC patients, while a high-SIGLEC1 expression group had a better prognosis. Here, we hypothesize that SIGLEC1 showed inhibitory effects in the transition of normal cells in the colorectal mucosa to early CRC tumors. In tumor cells of CRC, SIGLEC1 can further prevent the progression of tumors by promoting the activity of immune cells. We found a significant positive correlation between SIGLEC1 expression and inhibitory/co-inhibitory immune genes, which further emphasizes the potential role of SIGLEC1 in CRC immune evasion and immunotherapy resistance. This discovery provides new insights into the mechanisms of CRC immune evasion and may guide the development of novel immunotherapies targeting these mechanisms. In particular, we found that the expression levels of male CRC were much lower, therefore, these individuals were more likely to develop early CRC. These findings were consistent with the consensus that males were more likely to develop CRC [38]. These results suggest that SIGLEC1 may be a viable candidate for cancer immunity research and a target for cancer immunotherapy.
In vivo, the anticancer effects of YHJD on CRC. (A) Experimental procedures for a CRC xenograft model. (B) Images of separated tumors from the control, YHJD only, 5-Flu only, and 5-Flu with the YHJD groups. (C) Fluorescence imaging was used to detect tumor growth and metastasis before and after the surgery. (D) Immunohistochemical methods were employed to detect the expression of proteins in tumors. (Scale bar = 20 µm. **p < 0.01, ***p < 0.001, n = 4).
Lately, there has been an increasing emphasis on the role of the tumor microenvironment in the emergence and progression of cancer, especially in colon cancer. Studies have revealed that immune-cell infiltration can serve as a promising prognostic and therapeutic biomarker [39-42]. Figure 5 shows a strong positive correlation between SIGLEC1 expression and T helper (Th) cells, macrophages, and tumor-infiltrating lymphocytes (TIL). Similarly, the presence of Th17 cells and interleukin (IL)-17 was probably associated with the progression of different malignancies such as CRC, lung cancer, breast cancer, ovarian cancer, and gastric cancer [43-46]. A notable reduction in Th17 cells was detected in CRC patients, which aligned with the decreased expression of SIGLEC1 [47]. Additionally, colon cancer cells can stimulate IL-1β production from THP1 macrophages, activating the Wnt signaling pathway and promoting cancer cell growth [48]. Moreover, the presence of TILs has been recognized as a prognostic indicator for survival in patients with CRC. indicating that increasing TIL density is associated with improved survival rates [49]. Additionally, the DEGs were enriched in the differentiation and chemokine signaling pathway of Th17 cells, which play a vital role in colon cancer development and advancement by producing and releasing cytokines [50-52].
The role of SIGLEC1 was notably enriched in GO terms related to immune response, such as “T-cell activation,” “leukocyte cell adhesion,” and “immune receptor activity” (Figure 6). T-cell activation encompasses both the stimulation and inhibition of checkpoint signals. Elevated T-cell counts lead to an increased number of T cells, including cytotoxic lymphocytes and Th1 cells, which are related to improved survival rates in patients with CRC [53]. The levels of activated leukocyte adhesion molecules in CRC tissues are higher than those in the normal mucosa, which is an independent prognostic factor for CRC [54]. Additionally, immune receptors can suppress tumor growth by decreasing colonic tumorigenesis and key tumor-promoting signals. Therefore, their activity is critical for carcinogenesis and progression [55].
Among the predicted upregulated molecules, we focused on bisphenol F, genistein, and estradiol (Figure S2C). Bisphenol F is present in many TCM plants [56]. Estradiol and genistein are active monomers commonly found in legumes and other TCM herbs [57].
YHJD is a TCM formulation developed based on colon cancer etiology, “spleen deficiency and blood stasis.” It originates from the classic prescription “Gui Shao Liujun Decoction” in the Song Dynasty's “Taiping Imperial Prescription” (1078 AD). A prior study has provided confirmation that YHJD effectively inhibits epithelial-mesenchymal transition and stem-cell formation in CRC by modulating the hsa-miR-374a-3p/Wnt3/β-catenin signaling pathway [27]. Furthermore, YHJD can inhibit liver metastasis of colorectal cancer (CRC) in vivo, and its therapeutic effect is closely related to the regulation of multiple targets and effector processes, especially the PI3K-AKT signaling pathway and the immune response dominated by natural killer (NK) cells [25]. Figure 8 displays a strong correlation between YHJD and SIGLEC1 in clinical and animal CRC samples, indicating that YHJD inhibits SIGLEC1 expression and restricts tumor growth and metastasis. Therefore, this study provides a basis for further studies on the effects and underlying mechanisms of YHJD. Currently, most studies focus on the functions of SIGLEC1 itself and its interactions with other immune molecules, rather than the regulatory mechanism of YHJD on SIGLEC1. Our findings not only uncover the pivotal role of YHJD in immunomodulation but also open up novel avenues for further elucidating its antitumor mechanisms. We plan to utilize high-throughput sequencing, proteomics, and other technologies in future studies to further uncover the underlying molecular mechanisms. Through in vitro cell experiments and in vivo animal models, we will verify the effects of YHJD upregulating SIGLEC1 expression on immune cell function and how this effect promotes anti-tumor immune responses. Additionally, we can combine preclinical research and clinical trial data to evaluate the potential clinical application value of YHJD as an anti-tumor drug or immune modulator.
In investigating the correlation between SIGLEC1 and tumors, despite meticulous efforts to select representative and rigorously screened samples, tumor heterogeneity remains a complex and largely uncontrollable factor. This intratumoral heterogeneity can lead to varying SIGLEC1 expression levels across different regions or cells, potentially influencing the composition and function of the tumor microenvironment, thereby complicating and obscuring the relationship between SIGLEC1 and its microenvironment. Additionally, variations among samples from diverse patient populations, stemming from factors such as genetic backgrounds, environmental exposures, and treatment histories, can also contribute to differences in SIGLEC1 expression levels. To better mitigate the influence of tumor heterogeneity and sample source variations on results, future endeavors should integrate multiple technologies and methodologies (e.g., genomics, transcriptomics, proteomics) to comprehensively analyze tumor heterogeneity. Experimental validation and clinical data comparison will be employed to multidimensionally verify and assess analytical outcomes. While we strive to manage confounding factors, analytical methods inherently possess assumptions and limitations that may not fully eliminate confounding effects. Moreover, complex interactions among confounding factors may be difficult to fully capture and explain within current analytical frameworks. Consequently, more advanced statistical approaches like machine learning and causal inference are being considered to enhance the control of confounding factors and improve the accuracy of results. Larger-scale, multi-center studies will be conducted to gather broader and more comprehensive data, facilitating better control of confounding factors and validating the universality of research findings. Furthermore, more detailed subgroup analyses and exploration of potential biological mechanisms can be undertaken to deepen our understanding.
Herein, SIGLEC1 has been acknowledged as a standalone prognostic factor for CRC by multiple analyses including bioinformatics analysis, clinical sample verification, and corresponding animal experiments. Our study demonstrated that SIGLEC1 not only serves as an independent prognostic factor but also displays a strong correlation with immunological parameters and immune cell infiltration within the tumor microenvironment. Importantly, our findings extend beyond mere correlations by uncovering a potential role for SIGLEC1 in modulating immunotherapeutic responsiveness, an area that has been understudied in CRC. Furthermore, our investigation represents a pioneering effort in exploring the therapeutic potential of TCM, specifically YHJD, in modulating SIGLEC1 expression and thereby influencing CRC progression.
Conclusion
Our study offers a fresh perspective on CRC prognosis and therapy, with implications that extend beyond SIGLEC1 itself to the broader field of cancer immunology and TCM-based anticancer therapies. Nevertheless, additional research is required to gain a comprehensive understanding of the impact of YHJD on the tumor microenvironment and immune response.
Abbreviations
BSA: Bovine serum albumin; COPD: Chronic obstructive pulmonary disease; CRC: Colorectal cancer; CTLs: Cytotoxic lymphocytes; DEGs: Differentially expressed genes; GEO: Gene Expression Omnibus; GO: Gene Ontology; GSEA: Gene set enrichment analysis; GSVA: Gene set variation analysis; IVIS: In vivo imaging system; KEGG: Kyoto Encyclopedia of Genes and Genomes; K-M: Kaplan-Meier; M stage: Metastasis stage; N stage: Node stage; OS: Overall survival; SIGLEC1: Sialic acid binding immunoglobulin-like lectin 1; ssGSEA: Single-sample gene set enrichment analysis; T stage: Tumor stage; TCGA: The Cancer Genome Atlas; TCM: Traditional Chinese Medicine; TIDE: Time-series Dense Encoder; YHJD: Yiqi Huayu Jiedu decoction.
Supplementary Material
Supplementary figure and table legends.
Supplementary tables.
Acknowledgements
Funding
This study was supported by National Natural Science Foundation of China (82304595, 82304854), National Natural Science Foundation of China by Nanjing University of Chinese Medicine (XPT82304595, XPT82304854), Natural Science Foundation of Jiangsu Province (BK20220465), Project of Key Laboratory of National Health Commission (JSHD2022039), Science and Technology Development Project of Jiangsu Province Traditional Chinese Medicine (YB201916).
Data availability statement
Data will be made available on request.
Ethics statement
In vivo experiments comply with the National Institutes of Health guide for the Care and Use of Laboratory Animal, which were approved by the Ethics Committee Nanjing University of Chinese Medicine (2022DW-36-02).
Clinical research follows the guidelines of the Helsinki Declaration and the Tokyo Declaration approved by the Ethics Committee of the Affiliated Hospital of Nanjing University of Chinese Medicine (2022NL-110-02).
Author contributions
Yejin Zhu: Conceptualization, Funding acquisitions, and methodology. Xinping Wang: Writing, Data management, and surgical operation. Qianyi Wang: Surgical operation, and methodology. Shanfan Shi: Writing, Data management, and methodology. Jun He: Data management and Project administration. Chao Jiang: Conceptualization, Funding acquisitions, and Validation.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Baran B, Mert Ozupek N, Yerli Tetik N, Acar E, Bekcioglu O, Baskin Y. Difference Between Left-Sided and Right-Sided Colorectal Cancer: A Focused Review of Literature. Gastroenterology Res. 2018;11:264-73
2. Xi Y, Xu P. Global colorectal cancer burden in 2020 and projections to 2040. Transl Oncol. 2021;14:101174
3. Cho YA, Lee J, Oh JH, Chang HJ, Sohn DK, Shin A. et al. Genetic Risk Score, Combined Lifestyle Factors and Risk of Colorectal Cancer. Cancer Res Treat. 2019;51:1033-40
4. Holtedahl K, Borgquist L, Donker GA, Buntinx F, Weller D, Campbell C. et al. Symptoms and signs of colorectal cancer, with differences between proximal and distal colon cancer: a prospective cohort study of diagnostic accuracy in primary care. BMC Fam Pract. 2021;22:148
5. Hugen N, van de Velde CJH, de Wilt JHW, Nagtegaal ID. Metastatic pattern in colorectal cancer is strongly influenced by histological subtype. Ann Oncol. 2014;25:651-7
6. Chu E. An update on the current and emerging targeted agents in metastatic colorectal cancer. Clin Colorectal Cancer. 2012;11:1-13
7. Asano K, Kikuchi K, Tanaka M. CD169 macrophages regulate immune responses toward particulate materials in the circulating fluid. J Biochem. 2018;164:77-85
8. Fraschilla I, Pillai S. Viewing Siglecs through the lens of tumor immunology. Immunol Rev. 2017;276:178-91
9. Stanczak MA, Laubli H. Siglec receptors as new immune checkpoints in cancer. Mol Aspects Med. 2023;90:101112
10. van Houtum EJH, Bull C, Cornelissen LAM, Adema GJ. Siglec Signaling in the Tumor Microenvironment. Front Immunol. 2021;12:790317
11. Uchil PD, Pi R, Haugh KA, Ladinsky MS, Ventura JD, Barrett BS. et al. A Protective Role for the Lectin CD169/Siglec-1 against a Pathogenic Murine Retrovirus. Cell Host Microbe. 2019;25:87-100 e10
12. Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7:255-66
13. Souza de Lima D, Nunes VCL, Ogusku MM, Sadahiro A, Pontillo A, Alencar BC. Polymorphisms in SIGLEC1 contribute to susceptibility to pulmonary active tuberculosis possibly through the modulation of IL-1ss. Infect Genet Evol. 2017;55:313-7
14. Bukvic BK, Blekic M, Simpson A, Marinho S, Curtin JA, Hankinson J. et al. Asthma severity, polymorphisms in 20p13 and their interaction with tobacco smoke exposure. Pediatr Allergy Immunol. 2013;24:10-8
15. Ferreira RC, Guo H, Coulson RM, Smyth DJ, Pekalski ML, Burren OS. et al. A type I interferon transcriptional signature precedes autoimmunity in children genetically at risk for type 1 diabetes. Diabetes. 2014;63:2538-50
16. Biesen R, Demir C, Barkhudarova F, Grun JR, Steinbrich-Zollner M, Backhaus M. et al. Sialic acid-binding Ig-like lectin 1 expression in inflammatory and resident monocytes is a potential biomarker for monitoring disease activity and success of therapy in systemic lupus erythematosus. Arthritis Rheum. 2008;58:1136-45
17. O'Neill AS, van den Berg TK, Mullen GE. Sialoadhesin - a macrophage-restricted marker of immunoregulation and inflammation. Immunology. 2013;138:198-207
18. Komohara Y, Ohnishi K, Takeya M. Possible functions of CD169-positive sinus macrophages in lymph nodes in anti-tumor immune responses. Cancer Sci. 2017;108:290-5
19. Saito Y, Ohnishi K, Miyashita A, Nakahara S, Fujiwara Y, Horlad H. et al. Prognostic Significance of CD169+ Lymph Node Sinus Macrophages in Patients with Malignant Melanoma. Cancer Immunol Res. 2015;3:1356-63
20. Li C, Luo X, Lin Y, Tang X, Ling L, Wang L. et al. A Higher Frequency of CD14+ CD169+ Monocytes/Macrophages in Patients with Colorectal Cancer. PLoS One. 2015;10:e0141817
21. Zhang Y, Li JQ, Jiang ZZ, Li L, Wu Y, Zheng L. CD169 identifies an anti-tumour macrophage subpopulation in human hepatocellular carcinoma. J Pathol. 2016;239:231-41
22. Ohnishi K, Komohara Y, Saito Y, Miyamoto Y, Watanabe M, Baba H. et al. CD169-positive macrophages in regional lymph nodes are associated with a favorable prognosis in patients with colorectal carcinoma. Cancer Sci. 2013;104:1237-44
23. Hou X, Chen G, Zhao Y. Research progress on CD169-positive macrophages in tumors. Am J Transl Res. 2021;13:8589-97
24. Zhang J, Xu J, Zhang RX, Zhang Y, Ou QJ, Li JQ. et al. CD169 identifies an activated CD8(+) T cell subset in regional lymph nodes that predicts favorable prognosis in colorectal cancer patients. Oncoimmunology. 2016;5:e1177690
25. Zhou JY, Wu C, Shen Z, Liu S, Zou X, Qian J. et al. Yiqi Huayu Jiedu Decoction inhibits liver metastasis of colorectal cancer via enhancing natural killer cells function. J Ethnopharmacol. 2024;318:116915
26. Xu XQ. Clinical efficacy of Yiqi Qushi Recipe in treating myasthenia gravis and observation of its immunomodulatory effects. Zhongguo Zhong Xi Yi Jie He Za Zhi. 2013;33:177-9
27. Zhuang Y, Zhou J, Liu S, Wang Q, Qian J, Zou X. et al. Yiqi Jianpi Huayu Jiedu Decoction Inhibits Metastasis of Colon Adenocarcinoma by Reversing Hsa-miR-374a-3p/Wnt3/beta-Catenin-Mediated Epithelial-Mesenchymal Transition and Cellular Plasticity. Front Oncol. 2022;12:904911
28. Thorn CF, Marsh S, Carrillo MW, McLeod HL, Klein TE, Altman RB. PharmGKB summary: fluoropyrimidine pathways. Pharmacogenet Genomics. 2011;21:237-42
29. Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3:330-8
30. Fekete JT, Gyorffy B. New Transcriptomic Biomarkers of 5-Fluorouracil Resistance. Int J Mol Sci. 2023;24:1508
31. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47
32. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16:284-7
33. Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7
34. Bao Y, Jiang A, Dong K, Gan X, Gong W, Wu Z. et al. DDX39 as a predictor of clinical prognosis and immune checkpoint therapy efficacy in patients with clear cell renal cell carcinoma. Int J Biol Sci. 2021;17:3158-72
35. Diagnosis Treatment Guidelines For Colorectal Cancer Working Group C. Chinese Society of Clinical Oncology (CSCO) diagnosis and treatment guidelines for colorectal cancer 2018 (English version). Chin J Cancer Res. 2019;31:117-34
36. Farago A, Zvara A, Tiszlavicz L, Hunyadi-Gulyas E, Darula Z, Hegedus Z. et al. Lectin-Based Immunophenotyping and Whole Proteomic Profiling of CT-26 Colon Carcinoma Murine Model. Int J Mol Sci. 2024;25:4022
37. Pan S, Tang T, Wu Y, Zhang L, Song Z, Yu S. Identification and Validation of Immune-Related Prognostic Genes in the Tumor Microenvironment of Colon Adenocarcinoma. Front Genet. 2021;12:778153
38. Baraibar I, Ros J, Saoudi N, Salvà F, García A, Castells MR. et al. Sex and gender perspectives in colorectal cancer. ESMO Open. 2023;8:101204
39. Baghban R, Roshangar L, Jahanban-Esfahlan R, Seidi K, Ebrahimi-Kalan A, Jaymand M. et al. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun Signal. 2020;18:59
40. Whiteside TL. The tumor microenvironment and its role in promoting tumor growth. Oncogene. 2008;27:5904-12
41. Tauriello DVF, Batlle E. Targeting the Microenvironment in Advanced Colorectal Cancer. Trends Cancer. 2016;2:495-504
42. Pedrosa L, Esposito F, Thomson TM, Maurel J. The Tumor Microenvironment in Colorectal Cancer Therapy. Cancers (Basel). 2019;11:1172
43. Chen X, Wan J, Liu J, Xie W, Diao X, Xu J. et al. Increased IL-17-producing cells correlate with poor survival and lymphangiogenesis in NSCLC patients. Lung Cancer. 2010;69:348-54
44. Zhu X, Mulcahy LA, Mohammed RA, Lee AH, Franks HA, Kilpatrick L. et al. IL-17 expression by breast-cancer-associated macrophages: IL-17 promotes invasiveness of breast cancer cell lines. Breast Cancer Res. 2008;10:R95
45. Miyahara Y, Odunsi K, Chen W, Peng G, Matsuzaki J, Wang RF. Generation and regulation of human CD4+ IL-17-producing T cells in ovarian cancer. Proc Natl Acad Sci U S A. 2008;105:15505-10
46. Zhuang Y, Peng LS, Zhao YL, Shi Y, Mao XH, Chen W. et al. CD8(+) T cells that produce interleukin-17 regulate myeloid-derived suppressor cells and are associated with survival time of patients with gastric cancer. Gastroenterology. 2012;143:951-62 e8
47. Li F, Zhou J, Li Z, Zhang L. Screening of immunosuppressive cells from colorectal adenocarcinoma and identification of prognostic markers. Biosci Rep. 2021;41:BSR20203496
48. Kaler P, Godasi BN, Augenlicht L, Klampfer L. The NF-kappaB/AKT-dependent Induction of Wnt Signaling in Colon Cancer Cells by Macrophages and IL-1beta. Cancer Microenviron. 2009;2:69-80
49. Tong Y, Peng M, Li J, Niu Y. Comprehensive Analyses of Stromal-Immune Score-Related Competing Endogenous RNA Networks In Colon Adenocarcinoma. Dis Markers. 2022;2022:4235305
50. Donnelly RP, Sheikh F, Dickensheets H, Savan R, Young HA, Walter MR. Interleukin-26: an IL-10-related cytokine produced by Th17 cells. Cytokine Growth Factor Rev. 2010;21:393-401
51. Fridman WH, Galon J, Pages F, Tartour E, Sautes-Fridman C, Kroemer G. Prognostic and predictive impact of intra- and peritumoral immune infiltrates. Cancer Res. 2011;71:5601-5
52. Korn T, Oukka M, Kuchroo V, Bettelli E. Th17 cells: effector T cells with inflammatory properties. Semin Immunol. 2007;19:362-71
53. Fridman WH, Galon J, Dieu-Nosjean MC, Cremer I, Fisson S, Damotte D. et al. Immune infiltration in human cancer: prognostic significance and disease control. Curr Top Microbiol Immunol. 2011;344:1-24
54. Zhang Y, Qian C, Jing L, Ren J, Guan Y. Meta-analysis indicating that high ALCAM expression predicts poor prognosis in colorectal cancer. Oncotarget. 2017;8:48272-81
55. Fletcher R, Wang YJ, Schoen RE, Finn OJ, Yu J, Zhang L. Colorectal cancer prevention: Immune modulation taking the stage. Biochim Biophys Acta Rev Cancer. 2018;1869:138-48
56. Huang T, Danaher LA, Bruschweiler BJ, Kass GEN, Merten C. Naturally occurring bisphenol F in plants used in traditional medicine. Arch Toxicol. 2019;93:1485-90
57. Wei TT, Chandy M, Nishiga M, Zhang A, Kumar KK, Thomas D. et al. Cannabinoid receptor 1 antagonist genistein attenuates marijuana-induced vascular inflammation. Cell. 2022;185:2387-9
Author contact
![]() Corresponding authors: E-mail addresses: Yejin Zhu (yejin_zhuedu.cn), Jun He (hejedu.cn) and Chao Jang (doctorjccom).
Corresponding authors: E-mail addresses: Yejin Zhu (yejin_zhuedu.cn), Jun He (hejedu.cn) and Chao Jang (doctorjccom).

 Global reach, higher impact
Global reach, higher impact