Impact Factor
ISSN: 1837-9664
J Cancer 2025; 16(2):479-485. doi:10.7150/jca.102801 This issue Cite
Research Paper
MiR-196a2 rs11614913 C Allele is Associated with Increased Wilms Tumor Susceptibility in Chinese Children
1. Department of General Surgery, Jiangxi Provincial Children's Hospital, Nanchang 330006, Jiangxi, China.
2. Department of Pathology, Anhui Provincial Children's Hospital, Hefei 230051, Anhui, China.
3. Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, Guangzhou 510623, Guangdong, China.
4. Department of Pathology, Children's Hospital of Nanjing Medical University, Nanjing 210008, Jiangsu, China.
5. Department of Pediatric Surgery, The Affiliated Hospital of Qingdao University, Qingdao 266000, Shandong, China.
6. Department of Hematology, The Key Laboratory of Pediatric Hematology and Oncology Diseases of Wenzhou, The Second Affiliated Hospital and Yuying Children's Hospital of Wenzhou Medical University, Wenzhou 325027, Zhejiang, China.
7. Department of Pediatric Surgery, Shengli Clinical Medical College of Fujian Medical University, Fuzhou 350001, Fujian, China.
8. Department of Clinical Laboratory, Biobank, Harbin Medical University Cancer Hospital, Harbin 150040, Heilongjiang, China.
Received 2024-8-25; Accepted 2024-11-10; Published 2025-1-1
Abstract
Wilms tumor, also known as nephroblastoma, is the most common kidney cancer in children. The miR-196a2 rs11614913 T>C polymorphism has been identified as a susceptibility locus in various adult cancers. However, it is unclear whether this polymorphism also increases the risk of pediatric cancer. In this study, we examined the genotypes of miR-196a2 rs11614913 T>C in 416 pediatric patients with Wilms tumor and 936 age- and gender-matched healthy controls of Chinese Han ethnicity using the TaqMan technology. The association between rs11614913 T>C polymorphism and the susceptibility to Wilms tumor was analyzed by univariable and multivariable logistic regression analyses, and the odds ratio (OR) and 95% confidence interval (CI) were calculated. Overall analysis indicated that the miR-196a2 rs11614913 T>C was associated with an increased risk of Wilms tumor in the homozygous (adjusted OR (AOR)=1.58, 95% CI=1.14-2.21, P=0.007), additive (AOR=1.26, 95% CI=1.06-1.49, P=0.007), dominant (AOR=1.33, 95% CI=1.02-1.74, P=0.034), and recessive (AOR=1.38, 95% CI=1.05-1.81, P=0.023) models. Stratification analysis further demonstrated that the association was dependent on age and tumor stages rather than gender. Furthermore, miR-196a2 rs11614913 T>C was significantly associated with the disease risk only in children older than 18 months and those with III+IV stage tumors. The expression quantitative trait locus (eQTL) analysis showed that rs11614913 T>C was associated with decreased expression of HOXC-AS1 and increased expression of GPR84 and HOXC8. Our results provide evidence that miR-196a2 rs11614913 T>C may confer genetic susceptibility to Wilms tumor. These findings warrant validation in larger cohorts and across different ethnicities.
Keywords: Wilms tumor, susceptibility, miR-196a2, rs11614913, polymorphism
Introduction
Wilms tumor, also referred to as nephroblastoma, is a pediatric kidney cancer stemming from the immature cells in the developing kidney. The age-standardized incidence rates (ASR) of Wilms tumor have been varying from 9.1 to 9.8 cases per million in North America and Europe and around 4.3 in East Asia annually [1, 2]. It is the most frequently diagnosed childhood kidney cancer [3]. The prognosis for Wilms tumor is generally optimistic as a result of early diagnosis and multimodality regimes, including surgery, radiation, and chemotherapy [4]. However, relapse, late effects of treatment, as well as a subgroup of high-risk patients with a survival rate of 50% or lower remain essential long-term concerns. Current research focuses on improving the management of relapsed or refractory tumors, as well as minimizing the adverse effects of therapy in survivors.
Genetic risk factors for Wilms tumor development have been reported, such as syndromes and genetic abnormalities. Common predisposing syndromes include WAGR, Denys-Drash, and Beckwith-Wiedemann syndromes [3]. Moreover, mutations of WT1, CTNNB1, ASXL1, MAP3K4, and WTX genes and loss of H19-IGF2 imprinting can help explain a subset of patients [5]. Besides, many Wilms tumor-predisposing loci have also been identified, such as single nucleotide polymorphisms (SNPs) in the RAN, RANBP2, miR34b/c, TP53, TRMT6, YTHDF2, and WTAP genes [6-14]. However, more studies are warranted to understand pediatric renal tumors better, dissect more risk factors, and develop more risk-adapted therapies.
Apart from SNPs of protein-encoding genes, genetic variants in non-coding RNA genes are also nonnegligible. MicroRNAs, evolutionarily conserved small non-coding RNAs, impede the translation or stability of mRNAs by binding to the target transcript's 3' untranslated region (UTR) and then negatively regulate gene expression. MicroRNAs approximately regulate more than 30% of gene expression whose abnormal expression is tightly related to various diseases, including cancer [15, 16]. A genome-wide suppression of miRNA maturation leads to cellular transformation and carcinogenesis [17]. miRNAs are involved in the pathogenesis of Wilms tumor [18]. Alterations in miRNA expression profiles have been observed in Wilms tumor tissues compared to normal kidney tissues, suggesting that miRNAs play a significant role in the tumor's biology [19, 20]. miR-378c, miR-204, and miR-483-5p are among the miRNAs previously reported to be dysregulated in Wilms tumor [21, 22]. These miRNAs are involved in key pathways that promote tumorigenesis, such as cell cycle regulation, apoptosis evasion, and angiogenesis, underscoring the critical role of miRNA dysregulation in the disease [20, 23, 24]. SNPs within miRNAs (e.g., seed region) or nearby microRNA binding sites of functional genes can compromise or strengthen a microRNA function, thereby interfering with the miRNA-targeted mRNAs, such as altered expression of tumor suppressors, oncoproteins, or genes-related to drug sensitivity. Moreover, SNPs in pri- and pre-microRNAs may also modulate the processing and maturation of microRNAs [25, 26]. These variations can modulate individual susceptibility to various cancers. Studies have identified several miR-SNPs associated with altered cancer risk, such as miR-499 rs3746444, miR-146a rs2910164, and miR-423 rs6505162 [11, 27, 28]. The association of miR-196a2 SNPs with disease risk has been investigated in a broad of cancers, including breast cancer [29], esophageal carcinoma [30], non-small cell lung cancer (NSCLC) [31], glioma [32], colorectal cancer (CRC) [33]. However, its roles in pediatric solid cancer are much less investigated [34].
This study aimed to interrogate whether the miR-196a2 rs11614913 T>C polymorphism is associated with Wilms tumor susceptibility in 416 cases and 936 matched healthy controls. The impacts of the rs11614913 polymorphism on the expression of neighboring genes were also investigated using expression quantitative traits locus (eQTL) analysis.
Methods and Materials
Study population
This Chinese Han children study cohort consisted of 416 cases and 936 controls. To meet inclusion criteria, cases were diagnosed with Wilms tumor based on histopathological confirmation and had not received any prior chemotherapy, radiotherapy, or targeted therapy before sample collection. Cases were excluded if there was a simultaneous presence of other malignancies or significant comorbid conditions. Healthy controls should be free of any history of malignancies or other chronic diseases, as well as genetic conditions associated with increased Wilms tumor risk. Children with abnormal clinical examination findings or laboratory tests suggesting underlying diseases were excluded. Cases meeting the inclusion criteria were recruited from various medical centers in East China, situated in Anhui, Jiangsu, Zhejiang, Shandong, Fujian, and Jiangxi provinces. Accordingly, 936 age- and gender-matched healthy controls were enrolled from the participating hospitals during the same periods. We conducted this study in accordance with the Declaration of Helsinki. The study protocol was reviewed and proved by the Ethics Committee of Guangzhou Women and Children Medical Center (No. 202016601). Parents or equally legal guardians of all participants were informed and signed the written consent. Venous blood was then withdrawn from each subject prior to any medical intervention.
SNP selection and genotyping
We chose the miR-196a2 SNP rs11614913 (T>C) for this study based on a literature review [26, 29-35]. We first obtain genomic DNA from the above-mentioned blood samples using the Genomic DNA kit (Tian Gen Biotech Co. Ltd., Beijing, China). Then, we used a spectrophotometer to measure the DNA concentration of each sample, and then adjusted it to match the desired concentration level. The SNP of interest was genotyped with TaqMan real-time polymerase chain reaction (PCR) in 384-well plates, as reported in published studies [36-38].
Statistical analysis
A chi-square goodness-of-fit test was used to check whether the genotype frequency distribution of the miR-196a2 rs11614913 (T>C) polymorphism is consistent with Hardy-Weinberg equilibrium. Logistic regression was employed to estimate the strength of the association between the SNP and Wilms tumor susceptibility. The odds ratio (OR) and 95% confidence interval (CI) of the associations were calculated under different genetic models, including heterozygous, homozygous, additive, dominant, and recessive models. All models used age, gender, and clinical stage as covariates. Statistical analyses were executed utilizing SAS v10.0 (SAS Institute Inc., Cary, NC) and we recognized results with a P-value of less than 0.05 (two-sided) as statistically significant.
Expression quantitative traits locus (eQTL) analysis
An eQTL is considered to be a genetic variant (a locus) or region that influences the level of activity of a particular gene. The Genotype-Tissue Expression (GTEx) project contains a comprehensive database of eQTLs for various types of tissues and cell types [39, 40]. In this study, we identified genes that could be potentially affected by the miR-196a2 SNP rs11614913 T>C, using the GTEx Portal (https://gtexportal.org/home/).
Results
Participant characteristics
The baseline characteristics of all subjects are provided in Table S1. The median age of cases and controls were 34.09±26.35 and 33.87±30.88 months, respectively. In summary, 29.81% of patients had stage I disease, 34.86% stage II, 18.51% stage III, and 9.13% had stage IV. Tumor stage information was not available for 7.69% of patients. The distribution frequencies of age, gender, and tumor clinical stages are displayed in Table S1.
Association study
Different types of genetic inheritance patterns were employed to test the association between the rs11614913 T>C polymorphism and susceptibility to Wilms tumor (Table 1). Overall analysis indicated that the miR-196a2 rs11614913 T>C was associated with an increased risk of Wilms tumor under homozygous (adjusted OR (AOR)=1.58, 95% CI=1.14-2.21, P=0.007), a model used to identify genetic factors that have a substantial effect on disease risk (Table 1). We also detected a significant association between the rs11614913 and the risk of Wilms tumor under dominant (AOR=1.33, 95% CI=1.02-1.74, P=0.034) and recessive (AOR=1.38, 95% CI=1.05-1.81, P=0.023) models. In addition, this SNP was also shown to elevate Wilms tumor risk under the additive (AOR=1.26, 95% CI=1.06-1.49, P=0.007), indicating that the effect of the rs11614913 on the disease risk increases linearly with the number of C alleles. The stratification analysis indicated that the link between Wilms tumor susceptibility and miR-196a2 rs11614913 T>C was influenced by age and clinical stage, with no differences observed by gender (Table 2). A significant association was only detected in children > 18 months of age (AOR=1.45, 95% CI=1.03-2.04, P=0.032) and subgroup with tumors in the III+IV stage (AOR=1.92, 95% CI=1.17-3.16, P=0.01). These results suggest that older children with rs11614913 C allele are likely to have Wilms tumor. Moreover, children with this risk allele tended to develop advanced tumors (III+IV stage).
Association of miR-196a2 rs11614913 T>C polymorphism with Wilms tumor susceptibility
| Genotype | Cases (N=415) | Controls (N=936) | P a | Crude OR (95% CI) | P | Adjusted OR (95% CI) b | P b |
|---|---|---|---|---|---|---|---|
| rs11614913 (HWE=0.351) | |||||||
| TT | 98 (23.61) | 273 (29.17) | 1.00 | 1.00 | |||
| TC | 212 (51.08) | 478 (51.07) | 1.24 (0.93-1.64) | 0.141 | 1.24 (0.93-1.64) | 0.139 | |
| CC | 105 (25.30) | 185 (19.76) | 1.58 (1.13-2.21) | 0.007 | 1.58 (1.14-2.21) | 0.007 | |
| Additive | 0.007 | 1.26 (1.06-1.49) | 0.007 | 1.26 (1.06-1.49) | 0.007 | ||
| Dominant | 317 (76.39) | 663 (70.83) | 0.035 | 1.33 (1.02-1.74) | 0.035 | 1.33 (1.02-1.74) | 0.034 |
| TT/TC | 310 (74.70) | 751 (80.24) | 1.00 | 1.00 | |||
| CC | 105 (25.30) | 185 (19.76) | 0.022 | 1.38 (1.05-1.81) | 0.023 | 1.38 (1.05-1.81) | 0.023 |
OR, odds ratio; CI, confidence interval.
a χ2 test for genotype distributions between Wilms tumor patients and controls.
b Adjusted for age and gender.
Stratification analysis for the association of miR-196a2 rs11614913 T>C polymorphism with Wilms tumor susceptibility
| Variables | rs11614913 (cases/controls) | Crude OR | P | Adjusted OR a | P a | |
|---|---|---|---|---|---|---|
| TT | TC/CC | (95% CI) | (95% CI) | |||
| Age, month | ||||||
| ≤18 | 37/116 | 105/288 | 1.14 (0.74-1.76) | 0.545 | 1.15 (0.74-1.77) | 0.538 |
| >18 | 61/157 | 212/375 | 1.46 (1.04-2.05) | 0.031 | 1.45 (1.03-2.04) | 0.032 |
| Gender | ||||||
| Female | 45/122 | 139/283 | 1.33 (0.90-1.98) | 0.158 | 1.33 (0.89-1.98) | 0.162 |
| Male | 53/151 | 178/380 | 1.34 (0.93-1.91) | 0.116 | 1.34 (0.94-1.93) | 0.108 |
| Clinical stage | ||||||
| I+II | 68/273 | 201/663 | 1.22 (0.89-1.66) | 0.213 | 1.21 (0.89-1.65) | 0.226 |
| III+IV | 21/273 | 94/663 | 1.84 (1.13-3.02) | 0.015 | 1.92 (1.17-3.16) | 0.010 |
a Adjusted for age and gender, omitting the corresponding stratify factor.
OR, odds ratio; CI, confidence interval.
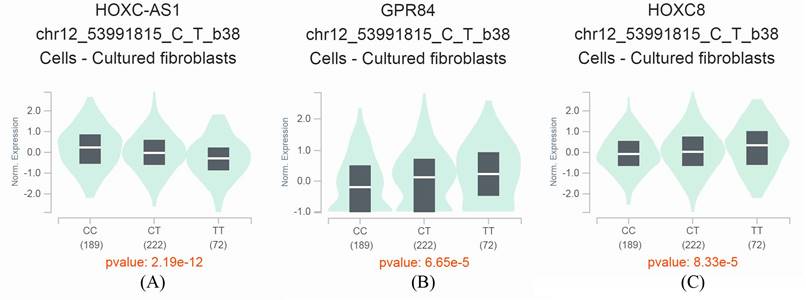
GTEx Portal analysis shows that the miR-196a2 rs11614913 T>C variant affects the expression of neighboring genes. A. HOXC cluster antisense RNA 1 (HOXC-AS1). B. inflammation-related G protein-coupled receptor EX33/G-protein coupled receptor 84 (GPR84). C. a transcription factor homeobox protein HOX-C8 (HOXC8).
Gene tissue expression analysis
By taking advantage of the GTEx Portal, we observed that the miR-196a2 rs11614913 T>C was associated with expression levels of several neighboring genes, including HOXC cluster antisense RNA 1 (HOXC-AS1), inflammation-related G protein-coupled receptor EX33/G-protein coupled receptor 84 (GPR84), and a transcription factor homeobox protein HOX-C8 (HOXC8). With an increase in C alleles, the HOXC-AS1 expression levels decrease, and the levels of GPR84 and HOXC8 increase significantly (Figure 1A-C). These results suggest that the miR-196a2 rs11614913 is an eQTL that may have impacts on the expression of these essential genes.
Discussion
In the present study, we identified the rs11614913 T>C in the miR-196a2 as a medium penetrant SNP in Wilms tumor, showing a moderate correlation with the risk of developing the disease. Overall analysis indicated that the rs11614913 T>C significantly conferred an increased susceptibility to Wilms tumor. The association was reliant on age and clinical stage rather than gender.
The rs11614913 polymorphism in the 3p mature miRNA region of hsa-mir-196a2 is a functional miRNA gene polymorphism. This SNP impacts the interaction between the mature hsa-mir-196a2-3p and its target mRNA. For instance, LSP1 is a predicated target gene of hsa-mir-196a2. Compared to hsa-mir-196a2 expression plasmids (T allele), the plasmids carrying rs11614913 C could significantly suppress the expression of LSP1 in Chinese hamster ovary (CHO), 293T, and A549 cell lines transfected with LSP1 3′UTR luciferase reporter plasmids, which was also validated with mature hsa-mir-196a2-3p miRNAs (C or U allele) [26]. Several pieces of evidence have shown that the rs11614913 was associated with the expression of mature miR-196a2 [26, 35]. The study on the correlation between the rs11614913 genotype and phenotype in 23 human lung cancer tissue samples revealed that tissues with the rs11614913 CC allele showed a substantial change in mature hsa-mir-196a expression. However, no alterations were detected in the precursor levels, indicating a potentially enhanced processing of pre-miRNA into its mature format [26]. Similarly, analyzing 50 CRC tissues showed that the expression level of mature miR-196a2 was significantly increased in CRC tumor tissues with rs11614913 TT compared to those with CC genotypes [35]. Recently, Yueh reported a significant genotype-phenotype of serum miR-196a2 in healthy controls, with the rs11614913 CC homozygotes showing significantly higher levels of serum miR-196a2 than CT heterozygous and TT homozygous patients [33].
The frequency of the T allele of the rs11614913 shows notable variation across various ethnic groups: approximately 18.8% in Africans, 39.4% in Europeans, 41.1% in Mexicans, 30.7% in South Asians, and 54.8% in East Asians.[33]. Consistently, our controls had a T allele frequency of 58.59, close to East Asians (54.8%). The T allele represents the major allele in East Asians but is the minor allele in all other ethnicities. The clinical implications of the rs11614913 have been extensively investigated in adult cancers. The study conducted by Hu et al. revealed that individuals with NSCLC who have the CC homozygote genotype of rs11614913 had poorer survival outcomes than those with the combined CT heterozygote and TT homozygote genotypes [26]. A systematic meta-analysis indicated that the rs11614913 (miRNA-196a2) was significantly associated with breast cancer susceptibility in Asia and South America based on data collected from Brazil, China, India, Iran, and Saudi Arabia [29]. Liu et al. observed no association between the miR-196a2 rs11614913 and susceptibility to esophageal squamous cell cancer (ESCC) in a study population of 829 cases and 1522 controls of Chinese Han nationality [30]. However, after adjustment for age, sex, smoking, drinking status, and body mass index (BMI), this SNP seemed to be associated with reduced ESCC susceptibility in women and in the subgroups aged 63 years old or beyond [30]. Yueh et al. found that the miR-196a2 rs11614913 CC genotype was associated with an increased CRC risk by 2.04 folds in Taiwan [33]. So far, no association between the SNP and renal cancer risk has been reported. Regarding pediatric cancer, our results in a previous study with 312 cases and 762 healthy controls suggested that the miR-196a2 rs11614913 might not be a causal genetic variant for neuroblastoma susceptibility in Eastern Chinese children [34]. Taken together, the association of rs11614913 with cancer risk may be tissue- and ethnic-specific. However, the sampling bias or other factors should be ruled out before the conclusion.
Finally, eQTL analysis demonstrated the potential association of the rs11614913 with HOXC-AS1, GPR84, and HOXC8. HOXC8 is a member of the homeobox family of genes that are highly conserved across species and encode a group of transcription factors responsible for regulating morphogenesis. Four HOX gene clusters, HOXA, HOXB, HOXC, and HOXD, have been characterized in mammals. The gene clusters are positioned on distinct chromosomes and comprise a series of 9 to 11 genes aligned consecutively. Interestingly, miR-196 was reported to be encoded at three paralogous locations within the HOX clusters A, B, and C in mammals. This microRNA exhibits significant evolutionary conservation and has great complementarity to the transcripts of HOXB8, HOXC8, and HOXD8. Moreover, human HOXB8, HOXC8, HOXD8, and HOXA7 were predicted to be targets of miR-196, followed by experimental validation [41].
Our findings have important clinical relevance. The identification of genetic variants associated with Wilms tumor susceptibility, such as the miR-196a2 rs11614913 C allele, facilitates the stratifying of children based on their genetic risk. Incorporating genetic testing for the rs11614913 C allele into routine screening panels for at-risk populations could improve the identification of high-risk individuals, guiding surveillance strategies, allowing for more targeted monitoring of those at greatest risk and potentially leading to earlier detection of Wilms tumor. Early identification of at-risk individuals through genetic markers can facilitate closer clinical follow-up and timely diagnostic evaluations, such as ultrasound screening for kidney abnormalities. Detecting Wilms tumor at an earlier stage significantly improves the prognosis and survival outcomes for affected children. In clinical practice, the presence of the miR-196a2 rs11614913 C allele could be used as part of a genetic risk assessment tool, prompting early imaging or other diagnostic measures in children with known genetic susceptibility. This approach aligns with precision medicine, where early intervention is tailored based on individual genetic profiles.
The limitations of this study were as follows. First, due to the rarity of Wilms tumor, the sample size of the case-control study was relatively moderate. Second, we investigated only one SNP in the study, and more functional SNPs in the hsa-mir-196a gene should be studied to explore the effect of gene-gene interactions. Third, to establish the role of the rs11614913 in the etiology of Wilms tumor, tumor tissue sample should be collected to determine the linkage between mature hsa-mir-196a expression and the rs11614913 genotypes (TT, TC, and CC). Fourth, the functional implications should be confirmed in Wilm's tumor cell lines. Fifth, environmental exposures, such as prenatal exposures, parental smoking, or other environmental influences, could also confound genetic associations. Due to the retrospective nature of our study and limitations in exposure data collection, these factors were unavailable. Finally, multiple statistical tests could increase the risk of type I errors, leading to false-positive findings. Replication in independent cohorts is a powerful method to strengthen the validity of the results and reduce concerns about false positives due to multiple comparisons. Therefore, our findings warrant validation in larger, well-designed case-control studies involving different ethnicities.
In conclusion, we demonstrated that the rs11614913 T>C is a moderate penetrant SNP associated with enhanced Wilms tumor predisposition in Chinese children. This risk genetic variant may have the potential to contribute to the Wilms tumor screening in the future.
Supplementary Material
Supplementary table.
Acknowledgements
This study was funded by grants from the Youth Medical Innovation and Practice Research Program of Guangzhou (No: 2023QNYXYB010), the Natural Science Foundation of Zhejiang Province (No: LGF21H260012), the Project of Jiangxi Province Key Laboratory of Child Development (No: 2024SSY06191), Jiangxi Provincial Natural Science Foundation (No:20242BAB25444), and the Jiangxi Provincial Health Technology Project (202410446).
Data availability statement
All the data are available upon request.
Ethical statement
We conducted this study in accordance with the Declaration of Helsinki. The study protocol was reviewed and proved by the Ethics Committee of Guangzhou Women and Children Medical Center (No. 202016601).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bao PP, Li K, Wu CX, Huang ZZ, Wang CF, Xiang YM. et al. [Recent incidences and trends of childhood malignant solid tumors in Shanghai, 2002-2010]. Zhonghua Er Ke Za Zhi. 2013;51:288-94
2. Libes J, Hol J, Neto JCA, Vallance KL, Tinteren HV, Benedetti DJ. et al. Pediatric renal tumor epidemiology: Global perspectives, progress, and challenges. Pediatr Blood Cancer. 2023;70(Suppl 2):e30343
3. Spreafico F, Fernandez CV, Brok J, Nakata K, Vujanic G, Geller JI. et al. Wilms tumour. Nat Rev Dis Primers. 2021;7:75
4. Ortiz MV, Koenig C, Armstrong AE, Brok J, de Camargo B, Mavinkurve-Groothuis AMC. et al. Advances in the clinical management of high-risk Wilms tumors. Pediatr Blood Cancer. 2023;70(Suppl 2):e30342
5. Gadd S, Huff V, Walz AL, Ooms A, Armstrong AE, Gerhard DS. et al. A Children's Oncology Group and TARGET initiative exploring the genetic landscape of Wilms tumor. Nat Genet. 2017;49:1487-94
6. Chang X, Zhu J, Hua RX, Deng C, Zhang J, Cheng J. et al. TRMT6 gene rs236110 C > A polymorphism increases the risk of Wilms tumor. Gene. 2023;882:147646
7. Huang X, Zhao J, Fu W, Zhu J, Lou S, Tian X. et al. The association of RAN and RANBP2 gene polymerphisms with Wilms tumor risk in Chinese children. J Cancer. 2020;11:804-9
8. Wang J, Lou S, Huang X, Mo Y, Wang Z, Zhu J. et al. The association of miR34b/c and TP53 gene polymorphisms with Wilms tumor risk in Chinese children. Biosci Rep. 2020;40(2):BSR20194202
9. Wang Z, Zhuo Z, Li L, Hua RX, Li L, Zhang J. et al. The contribution of YTHDF2 gene rs3738067 A>G to the Wilms tumor susceptibility. J Cancer. 2021;12:6165-9
10. Zhu J, Fu W, Jia W, Xia H, Liu GC, He J. Association between NER Pathway Gene Polymorphisms and Wilms Tumor Risk. Mol Ther Nucleic Acids. 2018;12:854-60
11. Fu W, Li L, Xiong SW, Zhang T, Jia W, Zhu J. et al. miR-423 rs6505162 C>A polymorphism contributes to decreased Wilms tumor risk. J Cancer. 2018;9:2460-5
12. Zhu J, Jia W, Wu C, Fu W, Xia H, Liu G. et al. Base Excision Repair Gene Polymorphisms and Wilms Tumor Susceptibility. EBioMedicine. 2018;33:88-93
13. Zhuo Z, Hua RX, Zhang H, Lin H, Fu W, Zhu J. et al. METTL14 gene polymorphisms decrease Wilms tumor susceptibility in Chinese children. BMC Cancer. 2021;21:1294
14. Ma L, Hua RX, Lin H, Zhu J, Fu W, Lin A. et al. The contribution of WTAP gene variants to Wilms tumor susceptibility. Gene. 2020;754:144839
15. Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259-69
16. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857-66
17. Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673-7
18. Alfaifi J. miRNAs Role in Wilms tumor pathogenesis: Signaling pathways interplay. Pathol Res Pract. 2024;256:155254
19. Csok A, Micsik T, Magyar Z, Tornoczky T, Kuthi L, Nishi Y. et al. Alterations of miRNA Expression in Diffuse Hyperplastic Perilobar Nephroblastomatosis: Mapping the Way to Understanding Wilms' Tumor Development and Differential Diagnosis. Int J Mol Sci. 2023;24(10):8793
20. Cerqueira DM, Tayeb M, Ho J. MicroRNAs in kidney development and disease. JCI Insight. 2022;7(9):e158277
21. Orgen Calli A, Issin G, Yilmaz I, Ince D, Tural E, Guzelis I. et al. The association of miR-204 and mir-483 5p expression with clinicopathological features of Wilms tumor: Could this provide foresight? Jpn J Clin Oncol. 2023;53:1170-6
22. Yu Q, Zheng B, Ji X, Li P, Guo Z. miR-378c suppresses Wilms tumor development via negatively regulating CAMKK2. Acta Biochim Biophys Sin (Shanghai). 2021;53:739-47
23. Liang XL, Wang YL, Wang PR. MiR-200a with CDC7 as a direct target declines cell viability and promotes cell apoptosis in Wilm's tumor via Wnt/beta-catenin signaling pathway. Mol Cell Biochem. 2021;476:2409-20
24. Luo X, Dong J, He X, Shen L, Long C, Liu F. et al. MiR-155-5p exerts tumor-suppressing functions in Wilms tumor by targeting IGF2 via the PI3K signaling pathway. Biomed Pharmacother. 2020;125:109880
25. Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2008;105:7269-74
26. Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L. et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;118:2600-8
27. Torabi M, Khafaei M, Jahanbin B, Sadeghi M. Assessment of the relationship between miR-499C/T (rs3746444) polymorphism and lung carcinoma in Iranian population; a case-control study. Afr Health Sci. 2023;23:301-7
28. Wang BR, Chang WS, Liao CH, Wang YC, Gu J, Bau DT. et al. Impacts of Mir146a Genotypes on Bladder Cancer Risk in Taiwan. Biomedicines. 2023;11(5):1396
29. Arancibia T, Morales-Pison S, Maldonado E, Jara L. Association between single-nucleotide polymorphisms in miRNA and breast cancer risk: an updated review. Biol Res. 2021;54:26
30. Liu C, Gao W, Shi Y, Lv L, Tang W. Association between miR-146a rs2910164, miR-196a2 rs11614913, and miR-499 rs3746444 polymorphisms and the risk of esophageal carcinoma: A case-control study. Cancer Med. 2022;11:3949-59
31. Qiu H, Xie Z, Tang W, Liu C, Wang Y, Gu H. et al. Association between microRNA-146a, -499a and -196a-2 SNPs and non-small cell lung cancer: a case-control study involving 2249 subjects. Biosci Rep. 2021;41(2):BSR20201158
32. Yang S, Zheng Y, Zhou L, Jin J, Deng Y, Yao J. et al. miR-499 rs3746444 and miR-196a-2 rs11614913 Are Associated with the Risk of Glioma, but Not the Prognosis. Mol Ther Nucleic Acids. 2020;22:340-51
33. Yueh TC, Wang YC, Chin YT, Hung YC, Mong MC, Yang YC. et al. Impact of Mir196a-2 Genotypes on Colorectal Cancer Risk in Taiwan. Int J Mol Sci. 2023;24(14):11613
34. Zhou C, Tang Y, Zhu J, He L, Li J, Wang Y. et al. Association of miR-146a, miR-149 and miR-196a2 polymorphisms with neuroblastoma risk in Eastern Chinese population: a three-center case-control study. Biosci Rep. 2019;39(6):BSR20181907
35. Zhan JF, Chen LH, Chen ZX, Yuan YW, Xie GZ, Sun AM. et al. A functional variant in microRNA-196a2 is associated with susceptibility of colorectal cancer in a Chinese population. Arch Med Res. 2011;42:144-8
36. Li L, Zhu J, Lu T, Liu W, Tang J, Zhang J. et al. Association of miR-34b/c rs4938723 and TP53 Arg72Pro Polymorphisms with Neuroblastoma Susceptibility: Evidence from Seven Centers. Transl Oncol. 2019;12:1282-8
37. Hua RX, Zhuo Z, Zhu J, Zhang SD, Xue WQ, Li XZ. et al. LIG3 gene polymorphisms and risk of gastric cancer in a Southern Chinese population. Gene. 2019;705:90-4
38. Chen H, Duan F, Wang M, Zhu J, Zhang J, Cheng J. et al. Polymorphisms in METTL3 gene and hepatoblastoma risk in Chinese children: A seven-center case-control study. Gene. 2021;800:145834
39. Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580-5
40. Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648-60
41. Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594-6
Author contact
![]() Corresponding authors: Shouhua Zhang, Department of General Surgery, Jiangxi Provincial Children's Hospital, Nanchang 330006, Jiangxi, China, Email: zshouhua416com; or Jinhong Zhu, Department of Clinical Laboratory, Biobank, Harbin Medical University Cancer Hospital, Harbin 150040, Heilongjiang, China, zhujinhongedu.
Corresponding authors: Shouhua Zhang, Department of General Surgery, Jiangxi Provincial Children's Hospital, Nanchang 330006, Jiangxi, China, Email: zshouhua416com; or Jinhong Zhu, Department of Clinical Laboratory, Biobank, Harbin Medical University Cancer Hospital, Harbin 150040, Heilongjiang, China, zhujinhongedu.

 Global reach, higher impact
Global reach, higher impact