3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2025; 16(2):498-505. doi:10.7150/jca.103945 This issue Cite
Research Paper
The association between the polymorphism of RAR related orphan receptor beta and the clinical manifestations of oral squamous cell carcinoma
1. Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan.
2. Department of Medical Research, Chung Shan Medical University Hospital, Taichung, Taiwan.
3. Institute of Oral Sciences, Chung Shan Medical University, Taichung, Taiwan.
4. Department of Dentistry, Chung Shan Medical University Hospital, Taichung, Taiwan.
5. Nobel Eye Institute, Taipei, Taiwan.
6. Department of Otorhinolaryngology, Head and Neck Surgery, Changhua Christian Hospital, Changhua, Taiwan.
7. Department of Post-Baccalaureate Medicine, College of Medicine, National Chung Hsing University, Taichung, Taiwan.
Received 2024-9-20; Accepted 2024-11-7; Published 2025-1-1
Abstract
Oral squamous cell carcinoma (OSCC) affects a substantial proportion of the Asian population and is influenced by various genetic risk factors. The RAR-related orphan receptor beta (RORB), a regulator of the circadian rhythm, has been implicated in certain neoplasms. Accordingly, this study investigated the association between RORB single-nucleotide polymorphisms and clinical manifestations of OSCC. A total of 1174 male patients without OSCC and 1254 male patients with OSCC were included in the study. Three RORB single-nucleotide polymorphism loci—rs3750420 (T/C), rs10781247 (A/G), and rs17611535 (C/T)—were genotyped using TaqMan allelic discrimination assays. RORB single-nucleotide polymorphism rs10781247 variants were significantly associated with moderate to poor cellular differentiation in patients with OSCC (p = 0.042). Additionally, among betel quid chewers with OSCC, rs10781247 variants were significantly associated with moderate to poor cell differentiation (p = 0.036). The rs3750420 variants were significantly associated with larger tumor size in individuals with buccal mucosa cancer (p = 0.036). An analysis of Cancer Genome Atlas data revealed that RORB mRNA levels were significantly higher in patients with head and neck squamous cell carcinoma compared with controls (p = 0.0002). Moreover, RORB mRNA levels were significantly higher in stage IV tumors than in stage III tumors (p = 0.0252). In conclusion, RORB single-nucleotide polymorphisms rs3750420 and rs10781247 are associated with adverse clinical characteristics in OSCC.
Keywords: single nucleotide polymorphism, RAR related orphan receptor beta, oral squamous cell carcinoma, betel quid chewing, buccal mucosa cancer, oral cancer
Introduction
Oral squamous cell carcinoma (OSCC) is a common malignancy worldwide that is characterized by epithelial dysplasia [1, 2]. In Asia, the prevalence of OSCC among men is approximately 21.2 per 100,000 [3]. The most common subtypes of OSCC include buccal cancer, mouth floor cancer, and tongue cancer. Hard palate cancer represents a relatively minor subset of OSCC [4, 5]. Current treatment modalities for OSCC are surgical excision, chemotherapy, radiotherapy, and targeted therapy [6]. The 5-year survival rate for OSCC is approximately 60%, but this declines to approximately 40% in stage IV cases [3, 7].
Several risk factors for OSCC have been identified [8, 9]. Cigarette smoking and betel quid chewing are two prominent lifestyle-related risk factors for OSCC, particularly in the Asian population [10]. Human papillomavirus is another risk factor, accounting for the majority of OSCC cases [11]. Regarding biomarkers, PD-L1 and interleukins have been implicated in OSCC development and can serve as potential indicators of the disease [12]. From a genetic perspective, mutations in the p53 gene are strongly associated OSCC pathogenesis [13]. However, other potential risk factors for OSCC remain to be elucidated and warrant further investigation.
RAR-related orphan receptor beta (RORB) is a protein-coding gene that regulates circadian rhythms in the human body [14]. RORB was previously demonstrated to be correlated with bipolar disorder [15]. Moreover, RORB expression was significantly correlated with alterations in the tumor immune microenvironment in head and neck squamous cell carcinoma [16]. In the context of neoplasms, RORB polymorphisms were associated with the development of breast cancer [17]. Evidence of the relationship between RORB polymorphisms and OSCC is limited. Because RAR-related orphan receptor alpha (RORA) has been implicated in the pathophysiology of OSCC [18], RORB, as part of the same receptor family, along with its single-nucleotide polymorphisms (SNPs), may also play a role in OSCC pathogenesis.
The present study evaluated the association between RORB SNPs and OSCC occurrence. Additionally, the relationship between RORB polymorphisms and OSCC across different populations was analyzed.
Materials and Methods
Ethics declarations
All procedures conducted in the present study adhered to the principles outlined in the Declaration of Helsinki (1964) and its subsequent amendments. The study protocol was reviewed and approved by the Institutional Review Board of Chung Shan Medical University Hospital (project identification code: CS1-21151). Written informed consent was obtained from all participants after a thorough explanation of the study objectives and procedures.
Medical data review and sample collection
The study included 1174 and 1254 male patients without and with OSCC, respectively. Basic demographic and clinical information, including age, OSCC stage, TNM stage, and degree of cell differentiation, were extracted from medical records held at Chung Shan Medical University Hospital. Only the most recent OSCC stage and TNM stage of each patient were used in the analysis. Regarding laboratory sample collection, venous blood samples were obtained from all participants. The blood samples were collected in ethylenediaminetetraacetic acid tubes, centrifuged, and stored in an experimental refrigerator at approximately -80°C, following protocols described elsewhere [19].
DNA analysis of RORB SNPs by using real-time PCR
The RORB SNPs examined in this study were rs3750420 (T/C), rs10781247 (A/G), and rs17611535 (C/T). These three RORB SNPs were selected because their minor allele frequencies exceeded 5% and because they were demonstrated to be associated with breast cancer and other disorders [14, 15, 20]. DNA extraction and analysis methods followed protocols established in other studies by the same authors [21]. Genomic DNA was isolated from leukocyte samples by using QIAamp DNA kits (Qiagen, Valencia, Valencia, CA, USA) following the manufacturer's instructions. Isolated DNA was stored at approximately -20°C until analysis. Genotyping of the three RORB SNPs was performed using the ABI StepOne Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The results were analyzed using SDS software version 3.0 (Applied Biosystems).
Bioinformatics analysis of RORB mRNA levels
To explore the association between RORB mRNA levels and clinical characteristics of OSCC, we used data from the Cancer Genome Atlas, a global genomic database [22]. In this analysis, patients with head and neck squamous cell carcinoma were stratified into subgroups on the basis of tumor presence and tumor stage.
Statistical analysis
Statistical analyses were conducted using SAS software version 9.4 (SAS Institute, NC, USA). Descriptive analyses were performed to summarize the demographic and tumor-related data of the two study groups. The chi-square test was employed to compare each group's demographic characteristics. A multiple logistic regression model was employed to examine the distribution frequency of each RORB SNP in each group after controlling for age, betel quid chewing, cigarette smoking, and alcohol consumption. Subsequently, the clinical characteristics of patients with OSCC with different genotypes of RORB SNP rs10781247 were analyzed using multiple logistic regression. The patients with OSCC were further stratified into betel quid chewers and non-betel quid chewers, and the association between RORB SNP genotypes and clinical characteristics of OSCC was reanalyzed using multiple logistic regression. A p value of <0.05 was considered significant.
Results
Baseline characteristics of each group
Baseline characteristics are presented in Table 1. The proportion of patients aged >60 years was significantly higher in the OSCC group than in the control group (p = 0.019). Additionally, the prevalence of betel quid chewing, cigarette smoking, and alcohol consumption was significantly greater in the OSCC group than in the control group (all p < 0.001). Overall, the OSCC group had a higher tumor stage and more advanced tumor T status than did the control group (Table 1).
The distributions of demographical characteristics in 1174 controls and 1254 male patients with oral squamous cell carcinoma.
| Variable | Control group (N=1174) | OSCC group (N=1254) | p value |
|---|---|---|---|
| Age (years) | |||
| < 60 | 761 (64.8%) | 755 (60.2%) | 0.019* |
| ≥ 60 | 413 (35.2%) | 499 (39.8%) | |
| Betel quid chewing | |||
| No | 978 (83.3%) | 398 (31.7%) | |
| Yes | 196 (16.7%) | 856 (68.3%) | < 0.001* |
| Cigarette smoking | |||
| No | 552 (47.0%) | 260 (20.7%) | |
| Yes | 622 (53.0%) | 994 (79.3%) | < 0.001* |
| Alcohol drinking | |||
| No | 942 (80.2%) | 773 (61.6%) | |
| Yes | 232 (19.8%) | 481 (38.4%) | < 0.001* |
| Stage | |||
| I+II | 566 (45.1%) | ||
| III+IV | 688 (54.9%) | ||
| Tumor T status | |||
| T1+T2 | 604 (48.2%) | ||
| T3+T4 | 650 (51.8%) | ||
| Lymph node status | |||
| N0 | 850 (67.8%) | ||
| N1+N2+N3 | 404 (32.2%) | ||
| Metastasis | |||
| M0 | 1247 (99.4%) | ||
| M1 | 7 (0.6%) | ||
| Cell differentiation | |||
| Well differentiated | 204 (16.3%) | ||
| Moderately or poorly differentiated | 1050 (83.7%) |
N: number, OSCC: oral squamous cell carcinoma
Mann-Whitney U test was used between healthy controls and patients with oral cancer. * p value < 0.05 as statistically significant.
Genotype frequencies of RORB SNPs in each group
In the multiple logistic regression analysis, the distributions of all three RORB SNPs exhibited no significant differences between the OSCC and control groups (all p > 0.05; Table 2). However, the RORB SNP rs10781247 variants were significantly associated with moderate to poor cell differentiation in the OSCC group (AOR: 1.403, 95% CI: 1.011 to 1.947, p = 0.042; Table 3). Additionally, the RORB SNP rs10781247 variants were significantly associated with moderate to poor cell differentiation in the patients with OSCC who were betel quid chewers (AOR: 1.486, 95% CI: 1.025 to 2.155, p = 0.036), whereas no significant association was observed between RORB SNPs and clinical characteristics of patients with OSCC who were nonbetel quid chewers (all p > 0.05; Table 4). Moreover, RORB SNP rs3750420 variants were significantly associated with larger tumor size in individuals with buccal mucosa cancer (AOR: 1.542, 95% CI: 1.027 to 2.315, p = 0.036). However, no significant association was observed between RORB SNP rs3750420 genotypes and clinical characteristics of OSCC in the participants with tongue cancer (all p > 0.05; Table 5).
Odds ratio and 95% confidence interval of oral squamous cell carcinoma associated with RORB genotypic frequencies.
| Variable | Controls (N=1174) (%) | Patients (N=1254) (%) | AOR (95% CI)# | p value |
|---|---|---|---|---|
| rs3750420 | ||||
| TT | 380 (32.4%) | 408 (32.5%) | 1.000 (reference) | |
| TC | 597 (50.9%) | 617 (49.2%) | 0.944 (0.765-1.166) | 0.593 |
| CC | 197 (16.7%) | 229 (18.3%) | 0.995 (0.753-1.314) | 0.970 |
| TC+CC | 794 (67.6%) | 846 (67.5%) | 0.957 (0.784-1.168) | 0.665 |
| rs10781247 | ||||
| AA | 312 (26.6%) | 322 (25.7%) | 1.000 (reference) | |
| AG | 598 (50.9%) | 650 (51.8%) | 1.194 (0.953-1.497) | 0.124 |
| GG | 264 (22.5%) | 282 (22.5%) | 1.148 (0.876-1.504) | 0.317 |
| AG+GG | 862 (73.4%) | 932 (74.3%) | 1.180 (0.952-1.462) | 0.130 |
| rs17611535 | ||||
| CC | 1004 (85.5%) | 1093 (87.2%) | 1.000 (reference) | |
| CT | 159 (13.5%) | 156 (12.4%) | 0.923 (0.699-1.219) | 0.573 |
| TT | 11 (1.0%) | 5 (0.4%) | 0.361 (0.106-1.226) | 0.102 |
| CT+TT | 170 (14.5%) | 161 (12.8%) | 0.884 (0.673-1.160) | 0.372 |
AOR: adjusted odds ratio, CI: confidence intervals, N: number
# The adjusted odds ratio with their 95% confidence intervals were estimated by multiple logistic regression models after controlling for age, betel quid chewing, cigarette smoking, and alcohol drinking.
Bioinformatics analysis of RORB expression in head and neck squamous cell carcinoma
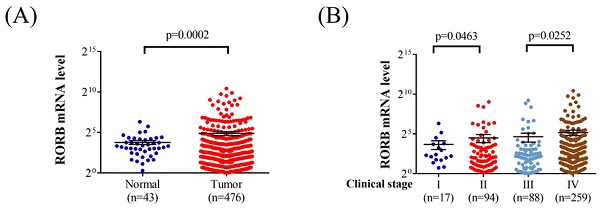
Analysis of Cancer Genome Atlas data revealed that RORB mRNA levels were significantly higher in patients with head and neck squamous cell carcinoma than in those with noncancerous tissues (p = 0.0002; Figure 1A). Furthermore, RORB mRNA levels were significantly higher in the stage II tumors than in stage I tumors (p = 0.0463). Moreover, the RORB mRNA levels were significantly higher in stage IV tumors than in stage III tumors (p = 0.0252; Figure 1B).
The Analysis of RORB mRNA by The Cancer Genome Atlas database. (A) The RORB mRNA level in head and neck squamous cell carcinoma patients and noncancerous tissues. (B) The RORB mRNA level in head and neck squamous cell carcinoma patients with different tumor stage.
Odds ratio and 95% confidence intervals of clinical statuses associated with genotypic frequencies of RORB rs10781247 in male oral squamous cell carcinoma patients.
| Variable | AA (N=322) | AG+GG (N=932) | AOR (95% CI)# | p value |
|---|---|---|---|---|
| Clinical Stage | ||||
| Stage I+II | 154 (47.8%) | 412 (44.2%) | 1.000 (reference) | 0.260 |
| Stage III+IV | 168 (52.2%) | 520 (55.8%) | 1.157 (0.897-1.492) | |
| Tumor size | ||||
| ≦ T2 | 161 (50.0%) | 443 (47.5%) | 1.000 (reference) | 0.445 |
| > T2 | 161 (50.0%) | 489 (52.5%) | 1.104 (0.857-1.422) | |
| Lymph node metastasis | ||||
| No | 221 (68.6%) | 629 (67.5%) | 1.000 (reference) | 0.705 |
| Yes | 101 (31.4%) | 303 (32.5%) | 1.054 (0.803-1.384) | |
| Metastasis | ||||
| M0 | 321 (99.7%) | 926 (99.4%) | 1.000 (reference) | 0.489 |
| M1 | 1 (0.3%) | 6 (0.6%) | 2.080 (0.249-17.342) | |
| Cell differentiated grade | ||||
| Well | 64 (19.9%) | 140 (15.0%) | 1.000 (reference) | 0.042* |
| Moderate or poor | 258 (80.1%) | 792 (85.0%) | 1.403 (1.011-1.947) |
AOR: adjusted odds ratio, CI: confidence intervals, N: number
# The adjusted odds ratio with their 95% confidence intervals were estimated by logistic regression models.
* p value < 0.05 as statistically significant.
Clinical statuses and genotypic frequencies of RORB rs10781247 in oral squamous cell carcinoma who are betel quid chewers or not betel quid chewers.
| Variable | Betel Quid Chewers (N=856) | Non-Betel Quid Chewers (N=398) | ||||||
|---|---|---|---|---|---|---|---|---|
| AA (N=230) | AG+GG (N=626) | AOR (95% CI)# | p value | AA (N=92) | AG+GG (N=306) | AOR (95% CI)# | p value | |
| Clinical Stage | ||||||||
| Stage I+II | 110 (47.8%) | 289 (46.2%) | 1.000 (reference) | 0.666 | 44 (47.8%) | 123 (40.2%) | 1.000 (reference) | 0.193 |
| Stage III+IV | 120 (52.2%) | 337 (53.8%) | 1.069 (0.790-1.447) | 48 (52.2%) | 183 (59.8%) | 1.364 (0.854-2.179) | ||
| Tumor size | ||||||||
| ≤ T2 | 111 (48.3%) | 310 (49.5%) | 1.000 (reference) | 0.744 | 50 (54.3%) | 133 (43.5%) | 1.000 (reference) | 0.066 |
| > T2 | 119 (51.7%) | 316 (50.5%) | 0.951 (0.703-1.287) | 42 (45.7%) | 173 (56.5%) | 1.549 (0.969-2.474) | ||
| Lymph node metastasis | ||||||||
| No | 159 (69.1%) | 426 (68.1%) | 1.000 (reference) | 0.763 | 62 (67.4%) | 203 (66.3%) | 1.000 (reference) | 0.851 |
| Yes | 71 (30.9%) | 200 (31.9%) | 1.051 (0.759-1.457) | 30 (32.6%) | 103 (33.7%) | 1.049 (0.638-1.722) | ||
| Metastasis | ||||||||
| M0 | 229 (99.6%) | 621 (99.2%) | 1.000 (reference) | 0.572 | 92 (100.0%) | 59 (99.7%) | 1.000 (reference) | --- |
| M1 | 1 (0.4%) | 5 (0.8%) | 1.844 (0.214-15.866) | 0 (0.0%) | 1 (0.3%) | --- | ||
| Cell differentiation | ||||||||
| Well | 53 (23.0%) | 105 (16.8%) | 1.000 (reference) | 0.036* | 11 (12.0%) | 35 (11.4%) | 1.000 (reference) | 0.891 |
| Moderate or poor | 177 (77.0%) | 521 (83.2%) | 1.486 (1.025-2.155) | 81 (88.0%) | 271 (88.6%) | 1.051 (0.511-2.164) | ||
AOR: adjusted odds ratio, CI: confidence intervals, N: number
# The adjusted odds ratio with their 95% confidence intervals were estimated by multiple logistic regression models after controlling for cigarette smoking, and alcohol drinking.
* p value < 0.05 as statistically significant.
Clinical statuses and genotypic frequencies of RORB rs3750420 in buccal mucosa cancer and tongue cancer
| Variable | Buccal mucosa cancer (N=433) | Tongue cancer (N=409) | ||||||
|---|---|---|---|---|---|---|---|---|
| TT (N=140) | TC+CC (N=293) | AOR (95% CI)# | p value | TT (N=130) | TC+CC (N=279) | AOR (95% CI)# | p value | |
| Clinical Stage | ||||||||
| Stage I+II | 69 (49.3%) | 133 (45.4%) | 1.000 (reference) | 0.448 | 62 (47.7%) | 126 (45.2%) | 1.000 (reference) | 0.632 |
| Stage III+IV | 71 (50.7%) | 160 (54.6%) | 1.169 (0.781-1.750) | 68 (52.3%) | 153 (54.8%) | 1.107 (0.729-1.680) | ||
| Tumor size | ||||||||
| ≤ T2 | 81 (57.9%) | 138 (51.8%) | 1.000 (reference) | 0.036* | 66 (50.8%) | 141 (50.5%) | 1.000 (reference) | 0.965 |
| > T2 | 59 (42.1%) | 155 (52.9%) | 1.542 (1.027-2.315) | 64 (49.2%) | 138 (49.5%) | 1.009 (0.666-1.530) | ||
| Lymph node metastasis | ||||||||
| No | 98 (70.0%) | 205 (70.0%) | 1.000 (reference) | 0.994 | 79 (60.8%) | 174 (62.4%) | 1.000 (reference) | 0.757 |
| Yes | 42 (30.0%) | 88 (30.0%) | 1.002 (0.645-1.554) | 51 (39.2%) | 105 (37.6%) | 0.935 (0.610-1.433) | ||
| Metastasis | ||||||||
| M0 | 139 (99.3%) | 292 (99.7%) | 1.000 (reference) | 0.592 | 130 (100.0%) | 277 (99.3%) | 1.000 (reference) | --- |
| M1 | 1 (0.7%) | 1 (0.3%) | 0.476 (0.030-7.667) | 0 (0.0%) | 2 (0.7%) | --- | ||
| Cell differentiation | ||||||||
| Well | 22 (15.7%) | 55 (18.8%) | 1.000 (reference) | 0.436 | 17 (13.1%) | 33 (11.8%) | 1.000 (reference) | 0.720 |
| Moderate or poor | 118 (84.3%) | 238 (81.2%) | 0.807 (0.469-1.386) | 113 (86.9%) | 246 (88.2%) | 1.121 (0.600-2.097) | ||
AOR: adjusted odds ratio, CI: confidence intervals, N: number
# The adjusted odds ratio with their 95% confidence intervals were estimated by multiple logistic regression models after controlling for cigarette smoking, and alcohol drinking.
* p value < 0.05 as statistically significant.
Discussion
In the present study, variants of the RORB SNP rs10781247 were associated with moderate to poor cell differentiation in both the overall OSCC group and the subgroup of patients with OSCC with a history of betel quid chewing. Moreover, variants of the RORB SNP rs3750420 were associated with a higher tumor T status in patients with buccal mucosa cancer. Furthermore, RORB mRNA levels were significantly higher in patients with head and neck squamous cell carcinoma, with the highest expression levels observed in those with advanced tumor stages.
The RORB gene has been implicated in various diseases [14, 15]. The RAR-related orphan receptor (ROR) family regulates the circadian rhythm and influences sleep patterns, with sleep duration affected by RORA and its SNP rs75981965 [23]. Additionally, the ROR family plays a role in the aging process, where both RORA and RORB independently contribute to the risk of cognitive aging [14]. In addition to circadian regulation, the ROR family is involved in the growth of specific cell types, such as epithelial cells and osteocytes [24], and research has demonstrated that RORA expression is suppressed during the development of OSCC [18]. The expression of RORB is also correlated with prognosis in breast cancer [25], with lower levels observed in the patients with endometrial cancer [24]. Zheng et al., reported that RORB expression was significantly associated with changes in the tumor immune microenvironment in head and neck squamous cell carcinoma [16]. Beyond gene expression, the genetic polymorphisms of RORB can influence disease development. For instance, the RORB SNP rs7867494 is associated with an increased incidence of breast cancer [17], and multiple RORB SNP variants are associated with the development of lung and prostate cancers [26]. In addition to the RORB gene family, several other genetic factors contribute to the development of OSCC [13, 27, 28]. The presence of CDKN2A was found to be significantly correlated with OSCC development [29], and mutations in the p53 gene are commonly observed in patients with OSCC [27]. Genetic polymorphisms, such as the interleukin-10 SNP-A592C, are more frequently observed in individuals with OSCC than in wild-type carriers [30], and the SNP rs1412115 A>G on chromosome 10 was demonstrated to increase the risk of OSCC in a Chinese population [31]. Additionally, GAS5 SNP rs145204276 (Ins/Del or Del/Del) variants are associated with a higher risk of moderate to poor cell differentiation in oral cancer [32]. Because both RORB and its SNPs have been demonstrated to regulate cancer growth [17, 24] and because RORA levels are significantly higher in individuals with OSCC [18], RORB SNP variants may be corrected with the clinical status of OSCC. This hypothesis is supported by the results of the present study.
In the present study, the distribution of RORB SNP rs10781247 variants was more commonly found in patients with OSCC with moderate to poor cell differentiation. Another study also reported that RORB SNP rs10781247 variants were more common in individuals with cognitive aging [14]. The association between RORB SNP rs10781247 variants and OSCC has not been extensively explored. To the best of our knowledge, this study provides preliminary evidence linking the distribution frequency of RORB SNP rs10781247 variants with cell differentiation in OSCC. Moreover, this study solely focused on male patients because male sex is a known risk factor for OSCC [33]. Several confounders, including age, betel quid chewing, cigarette smoking, and alcohol consumption, were adjusted in the multiple logistic regression model [3, 10]. Consequently, RORB SNP rs10781247 variants were independently associated with poorer cell differentiation in patients with OSCC. The RORB gene has been shown to increase the proliferation of retinal cells and neurons [24, 34], suggesting that RORB SNP rs10781247 variants may exert a similar effect on OSCC cells. Additionally, the OSCC subgroup with a history of betel quid chewing demonstrated a strong association between RORB SNP rs10781247 variants and moderate to poor cell differentiation. Betel quid chewing is a well-established risk factor for OSCC [35], and research has demonstrated that it contributes to a higher incidence of OSCC, particularly in buccal mucosa cancer [3]. Therefore, the synergic effect of betel quid chewing and RORB SNP rs10781247 variants may lead to worse differentiation of OSCC cells. By contrast, patients with OSCC with RORB SNP rs10781247 variants but no history of betel quid chewing did not exhibit an increased risk of poor cell differentiation during OSCC development.
In addition to the RORB SNP rs10781247 variants, a significantly higher distribution frequency of RORB SNP rs3750420 variants was observed in patients with buccal mucosa cancer and a higher tumor T status compared with individuals with tongue cancer. Although both buccal mucosa cancer and tongue cancer are types of OSCC, some differences exist between these two neoplasms [10]. The treatment approaches for buccal mucosa cancer and tongue cancer are also slightly different, with concurrent chemoradiotherapy providing additional benefits for tongue cancer [3]. Moreover, the 5-year distal metastasis rate for buccal mucosa cancer with nodal involvement is 30%, which is significantly higher than the 18% for tongue cancer with nodal invasion [36]. Genetically, buccal mucosa cancer has been associated with polymorphisms in the LINC00312 gene [37], whereas the stage of tongue cancer is correlated with the SNP rs9904341 of the survivin gene [38]. We speculated that the RORB SNP rs3750420 variants interact with genes involved in the development of buccal mucosa cancer, thereby influencing the clinical characteristics of this cancer type. Additionally, a study demonstrated that RORB SNP rs3750420 variants are associated with the development of breast cancer [20], which could also explain the larger tumor size and tumor progression observed in buccal mucosa cancer in patients with this particular variant.
In the bioinformatics analysis using the Cancer Genome Atlas database, individuals with head and neck squamous cell carcinoma exhibited significantly higher RORB mRNA expression compared with noncancerous tissues. Moreover, RORB mRNA expression was highest in stage IV head and neck squamous cell carcinoma and lowest in stage I head and neck squamous cell carcinoma. A study demonstrated that lower RORA expression is associated with OSCC development [18]. The bioinformatics analysis in the present study aligns with these findings, suggesting that the expression of the ROR gene family is correlated with OSCC development. Although OSCC does not fully represent head and neck squamous cell carcinoma [39], it constitutes a substantial proportion of cases within the broader head and neck squamous cell carcinoma population and ranks sixteenth in cancer incidence worldwide [10, 35, 40]. Additionally, the higher expression of RORB mRNA in advanced OSCC stages indicates that the RORB gene may shape the clinical characteristics of OSCC. By integrating the results from the bioinformatics analysis and the clinical data from patients with OSCC in this study, we hypothesize that genetic polymorphisms of RORB SNPs are attributable to variations in RORB mRNA expression, leading to higher levels of RORB mRNA in specific OSCC subgroups. However, further research is warranted to fully elucidate this hypothesis.
This study has several limitations. First, the study employed a case-control design rather than a prospective cohort design, limiting our ability to evaluate the progression and prognosis of OSCC. Second, the analysis did not include data on the dose and duration of betel quid chewing, cigarette smoking, and alcohol consumption because some patients did not provide detailed information regarding these habits. This absence of data may have compromised the completeness and accuracy of the analyses and results. Additionally, the population in the Cancer Genome Atlas database significantly differs from our study cohort, which may introduce considerable heterogeneity in the results. Finally, because nearly all the participants in this study were Han Taiwanese, the external validity of the findings may be limited, reducing the generalizability to other populations.
In conclusion, the presence of RORB SNP rs10781247 variants and RORB SNP rs3750420 variants was associated with poorer clinical characteristics of OSCC in specific populations. Furthermore, the RORB mRNA expression levels were significantly higher in patients with head and neck squamous cell carcinoma, particularly in those with advanced tumor stages. Consequently, more frequent follow-up is recommended for patients with OSCC with RORB SNP rs10781247 and rs3750420 variants for monitoring potential adverse tumor progression. Further large-scale, prospective studies are essential to investigate the correlation between RORB SNPs and therapeutic outcomes in OSCC.
Acknowledgements
This study was supported by grants from National Science and Technology Council, Taiwan (MOST-110-2314-B-371-004-MY3).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Lu HJ, Su CW, Su SC, Chang LC, Wu MF, Lin CW. et al. Prognostic impact of caspase-8 mutation in oral cavity squamous cell carcinoma. Oral Dis. 2024 Sep 17. Online ahead of print
2. Lin CW, Yang WE, Su CW, Lu HJ, Su SC, Yang SF. IGF2BP2 promotes cell invasion and epithelial-mesenchymal transition through Src-mediated upregulation of EREG in oral cancer. Int J Biol Sci. 2024;20:818-30
3. Howard A, Agrawal N, Gooi Z. Lip and Oral Cavity Squamous Cell Carcinoma. Hematol Oncol Clin North Am. 2021;35:895-911
4. Su SC, Chang LC, Lin CW, Chen MK, Yu CP, Chung WH. et al. Mutational signatures and mutagenic impacts associated with betel quid chewing in oral squamous cell carcinoma. Hum Genet. 2019;138:1379-89
5. Su SC, Yeh CM, Lin CW, Hsieh YH, Chuang CY, Tang CH. et al. A novel melatonin-regulated lncRNA suppresses TPA-induced oral cancer cell motility through replenishing PRUNE2 expression. J Pineal Res. 2021;71:e12760
6. Kitamura N, Sento S, Yoshizawa Y, Sasabe E, Kudo Y, Yamamoto T. Current Trends and Future Prospects of Molecular Targeted Therapy in Head and Neck Squamous Cell Carcinoma. Int J Mol Sci. 2020;22:240
7. Lu HJ, Peng CY, Tseng HC, Hsin CH, Chuang CY, Chen CC. et al. Preoperative prediction model to evaluate salvage surgery in patients with recurrent or second primary oral cavity squamous cell carcinoma. Oral Oncol. 2022;131:105951
8. Su SC, Chang LC, Huang HD, Peng CY, Chuang CY, Chen YT. et al. Oral microbial dysbiosis and its performance in predicting oral cancer. Carcinogenesis. 2021;42:127-35
9. Yang SF, Huang HD, Fan WL, Jong YJ, Chen MK, Huang CN. et al. Compositional and functional variations of oral microbiota associated with the mutational changes in oral cancer. Oral Oncol. 2018;77:1-8
10. Chamoli A, Gosavi AS, Shirwadkar UP, Wangdale KV, Behera SK, Kurrey NK. et al. Overview of oral cavity squamous cell carcinoma: Risk factors, mechanisms, and diagnostics. Oral Oncol. 2021;121:105451
11. Tumban E. A Current Update on Human Papillomavirus-Associated Head and Neck Cancers. Viruses. 2019;11:922
12. Nocini R, Vianini M, Girolami I, Calabrese L, Scarpa A, Martini M. et al. PD-L1 in oral squamous cell carcinoma: A key biomarker from the laboratory to the bedside. Clin Exp Dent Res. 2022;8:690-8
13. Pandya JA, Boaz K, Natarajan S, Manaktala N, Nandita KP, Lewis AJ. A correlation of immunohistochemical expression of TP53 and CDKN1A in oral epithelial dysplasia and oral squamous cell carcinoma. J Cancer Res Ther. 2018;14:666-70
14. Lin E, Kuo PH, Liu YL, Yang AC, Kao CF, Tsai SJ. Effects of circadian clock genes and environmental factors on cognitive aging in old adults in a Taiwanese population. Oncotarget. 2017;8:24088-98
15. Lai YC, Kao CF, Lu ML, Chen HC, Chen PY, Chen CH. et al. Investigation of associations between NR1D1, RORA and RORB genes and bipolar disorder. PLoS One. 2015;10:e0121245
16. Zheng J, Tang H, Yang Y, Yang K. Comprehensive analysis of the prognosis and biological significance of ROR(β) in head and neck squamous cell carcinoma. Environ Toxicol. 2024;39:487-508
17. Benna C, Helfrich-Förster C, Rajendran S, Monticelli H, Pilati P, Nitti D. et al. Genetic variation of clock genes and cancer risk: a field synopsis and meta-analysis. Oncotarget. 2017;8:23978-95
18. Zheng X, Wu K, Liao S, Pan Y, Sun Y, Chen X. et al. MicroRNA-transcription factor network analysis reveals miRNAs cooperatively suppress RORA in oral squamous cell carcinoma. Oncogenesis. 2018;7:79
19. Yeh JC, Chen YT, Chou YE, Su SC, Chang LC, Chen YL. et al. Interactive effects of CDKN2B-AS1 gene polymorphism and habitual risk factors on oral cancer. J Cell Mol Med. 2023;27:3395-403
20. Zienolddiny S, Haugen A, Lie JA, Kjuus H, Anmarkrud KH, Kjærheim K. Analysis of polymorphisms in the circadian-related genes and breast cancer risk in Norwegian nurses working night shifts. Breast Cancer Res. 2013;15:R53
21. Chen YT, Lin CW, Chou YE, Su SC, Chang LC, Lee CY. et al. Potential impact of ADAM-10 genetic variants with the clinical features of oral squamous cell carcinoma. J Cell Mol Med. 2023;27:1144-52
22. Su SC, Lin CW, Ju PC, Chang LC, Chuang CY, Liu YF. et al. Association of LINC00673 Genetic Variants with Progression of Oral Cancer. J Pers Med. 2021;11:468
23. Hou SJ, Tsai SJ, Kuo PH, Liu YL, Yang AC, Lin E. et al. An association study in the Taiwan Biobank reveals RORA as a novel locus for sleep duration in the Taiwanese Population. Sleep Med. 2020;73:70-5
24. Feng S, Xu S, Wen Z, Zhu Y. Retinoic acid-related orphan receptor RORβ, circadian rhythm abnormalities and tumorigenesis (Review). Int J Mol Med. 2015;35:1493-500
25. Wu F, Chen W, Kang X, Jin L, Bai J, Zhang H. et al. A seven-nuclear receptor-based prognostic signature in breast cancer. Clin Transl Oncol. 2021;23:1292-303
26. Mocellin S, Tropea S, Benna C, Rossi CR. Circadian pathway genetic variation and cancer risk: evidence from genome-wide association studies. BMC Med. 2018;16:20
27. Khurshid Z, Zafar MS, Khan RS, Najeeb S, Slowey PD, Rehman IU. Role of Salivary Biomarkers in Oral Cancer Detection. Adv Clin Chem. 2018;86:23-70
28. Gautam B, Pandi A, Girija ASS, Arumugam P, Priyadharsini VJ. Genetic Association of Toll-Like Receptor 4 (TLR4) Gene Polymorphism (rs4986790) With Oral Squamous Cell Carcinoma (OSCC): A Pilot Case-Control Study. Cureus. 2024;16:e56021
29. Solomon B, Young RJ, Rischin D. Head and neck squamous cell carcinoma: Genomics and emerging biomarkers for immunomodulatory cancer treatments. Semin Cancer Biol. 2018;52:228-40
30. Singh PK, Ahmad MK, Kumar V, Gupta R, Kohli M, Jain A. et al. Genetic polymorphism of interleukin-10 (-A592C) among oral cancer with squamous cell carcinoma. Arch Oral Biol. 2017;77:18-22
31. Ma L, Chen J, Song X, Yuan H, Wang Y, Wu Y. et al. Evidence that the genetic polymorphism rs1412115 on chromosome 10 is associated with risk for oral squamous cell carcinoma. Gene. 2015;560:137-9
32. Hsieh MH, Lu HJ, Lin CW, Lee CY, Yang SJ, Wu PH. et al. Genetic Variants of lncRNA GAS5 Are Associated with the Clinicopathologic Development of Oral Cancer. J Pers Med. 2021;11:348
33. Klasen C, Wuerdemann N, Rothbart P, Prinz J, Eckel HNC, Suchan M. et al. Sex-specific aspects in patients with oropharyngeal squamous cell carcinoma: a bicentric cohort study. BMC Cancer. 2023;23:1054
34. Liu H, Aramaki M, Fu Y, Forrest D. Retinoid-Related Orphan Receptor β and Transcriptional Control of Neuronal Differentiation. Curr Top Dev Biol. 2017;125:227-55
35. Barsouk A, Aluru JS, Rawla P, Saginala K, Barsouk A. Epidemiology, Risk Factors, and Prevention of Head and Neck Squamous Cell Carcinoma. Med Sci (Basel). 2023;11:42
36. Liao CT, Huang SF, Chen IH, Kang CJ, Lin CY, Fan KH. et al. Tongue and buccal mucosa carcinoma: is there a difference in outcome? Ann Surg Oncol. 2010;17:2984-91
37. Hsu HT, Lu YT, Chen YT, Hsing MT, Su CW, Su SC. et al. Impact of LINC00312 gene polymorphism coupled with habitual risks on buccal mucosa cancer. J Cancer. 2024;15:2354-60
38. Mostaan LV, Tabari A, Amiri P, Ashtiani MK, Mahdkhah A, Yazdani N. et al. Survivin gene polymorphism association with tongue squamous cell carcinoma. Genet Test Mol Biomarkers. 2013;17:74-7
39. Hayes RB, Ahn J, Fan X, Peters BA, Ma Y, Yang L. et al. Association of Oral Microbiome With Risk for Incident Head and Neck Squamous Cell Cancer. JAMA Oncol. 2018;4:358-65
40. Johnson DE, Burtness B, Leemans CR, Lui VWY, Bauman JE, Grandis JR. Head and neck squamous cell carcinoma. Nat Rev Dis Primers. 2020;6:92
Author contact
![]() Corresponding authors: Shun-Fa Yang, Ph.D. Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan; E-mail: ysfedu.tw. Mu-Kuan Chen, MD., PhD. Department of Otorhinolaryngology, Head and Neck Surgery, Changhua Christian Hospital, Changhua, Taiwan; E-mail: 53780org.tw.
Corresponding authors: Shun-Fa Yang, Ph.D. Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan; E-mail: ysfedu.tw. Mu-Kuan Chen, MD., PhD. Department of Otorhinolaryngology, Head and Neck Surgery, Changhua Christian Hospital, Changhua, Taiwan; E-mail: 53780org.tw.

 Global reach, higher impact
Global reach, higher impact