Impact Factor
ISSN: 1837-9664
J Cancer 2025; 16(3):888-897. doi:10.7150/jca.100610 This issue Cite
Research Paper
Elucidating the Role of Estrogen Effects in Leukemia: Insights from Single-Cell RNA Sequencing and Mendelian Randomization
1. College of First Clinical Medicine, Shandong University of Traditional Chinese Medicine, Jinan, 250014, China.
2. State Key Laboratory of Quality Research in Chinese Medicine, and Faculty of Chinese Medicine, Macau University of Science and Technology, Avenida Wai Long, Taipa, 999078, Macau, China.
3. College of Traditional Chinese Medicine, Shandong Second Medical University, Weifang, 261000, China.
4. Department of Oncology, Weifang Traditional Chinese Hospital, Weifang, 261000, China.
#These authors have contributed equally to this work and are co-first authors.
Received 2024-7-7; Accepted 2024-10-28; Published 2025-1-1
Abstract
Background: Epidemiological studies have confirmed the potential role of estrogen effects in influencing the development and outcome of leukemia. Estrogen effects are increasingly attracting research interest for their potential antitumor effects beyond gynecological tumors. However, their causal relationship remains unclear.
Methods: In a novel approach, this study integrates single-cell RNA sequencing (scRNA-seq) with Mendelian randomization (MR) to explore the relationship between estrogen (and its receptor) and leukemia (and its related proteins). This integration showcases the uniqueness of our methodology and provides a new perspective for understanding the molecular relationship between them. Secondary analyses using genetic risk scores (GRS) were performed to further verify the robustness of the results.
Results: Our scRNA-seq analysis identified 14 BMMC mononuclear cell subsets, and the result showed that the estrogen receptor was implicated in leukemia. The MR results showed that there was a relationship between estradiol and leukemia inhibitory factor (β = 0.0621; P = 0.0229), and leukemia inhibitory factor receptor (β = 0.0665; P = 0.0218). The result of GRS analysis verified the MR analysis.
Conclusions: While both scRNA-seq and MR have yielded intriguing results, inconsistencies between these methodologies hint at a more elaborate underlying mechanism. The observed discrepancies underscore the complexity of the estrogen effects-leukemia relationship, suggesting that elucidating these interactions demands larger cohorts and enhanced sequencing depth in future studies. This research paves the way for a more nuanced understanding of the role of estrogen effects in leukemia and sets the stage for targeted therapeutic interventions.
Keywords: Estrogen, Leukemia, Single-cell RNA sequencing, Mendelian randomization, Genetic risk score
Introduction
Leukemia constitutes a collection of lethal blood-related malignancies characterized by the transformation of hemopoietic progenitors and the diffuse infiltration of the bone marrow, including the bone marrow and the lymphatic system. In recent years, leukemia has become one of the highest mortalities among all cancers, accounting for 2.8% of all new cancer cases and 3.4% of new cancer deaths [1-3]. Different genetic perturbations drive leukemia to exist in a variety of subtypes, some of which are more prevalent in children, while cases of other types of leukemia affect adults [4,5]. Previous studies have recognized a number of etiological and pathogenetic mechanisms, such as genetic disorders, metabolic abnormalities, and environmental factors, which contribute to excessive proliferation, differentiation blockade, and resistance to apoptosis of leukemia cells [6]. However, the primary cause of most cases is unexplained.
Given the rising incidence of hematopoietic malignancies and the challenges associated with their treatment, investigating the risk and protective factors for blood cancers, particularly leukemias, is crucial. Epidemiological studies have revealed that males, encompassing both adults and children, exhibit a heightened susceptibility to the incidence and mortality rates associated with leukemia, which suggests a potential role of estrogen effects in influencing the development and outcome of leukemia [7-9]. Previous studies have confirmed that targeting the estrogenic effects of estrogen receptors α and β holds potential as a therapeutic strategy for leukemia [10-12].
Leukemia inhibitory factor (LIF) is a pleiotropic cytokine of the interleukin-6 superfamily. LIF was initially discovered as a factor to induce the differentiation of myeloid leukemia cells and thus inhibit their proliferation [13]. LIF affects multiple types of leukemia cells through its functional receptor, the leukemia inhibitory factor receptor (LIFR)[14]. LIF displays a wide variety of important functions in a cell-, tissue-, and context-dependent manner in many physiological and pathological processes, including regulating cell proliferation, pluripotent stem cell self-renewal, tissue and organ development and regeneration, inflammation, infection, immune response, and metabolism. LIF and its receptor, LIFR, are proteins deeply implicated in the pathogenesis of leukemia. The meticulous regulation of them is paramount, and any perturbation can precipitate the cascade of events leading to leukemia development and its relentless progression [15-18]. Their influence on such transformative cellular dynamics positions them as keystones in the intricate mosaic of leukemia biology.
Recent technological breakthroughs in single-cell RNA sequencing (scRNA-seq) have revolutionized our ability to dissect the cellular subpopulations within tumors. This innovative approach has been instrumental in the field of cancer research, providing detailed insights into how specific cell subsets contribute to cancer initiation and progression [19,20]. Concurrently, epidemiological studies have increasingly utilized Mendelian randomization (MR) to assess the causal impact of risk factors [21,22]. The sophistication of MR methods has made them a preferred tool for deducing the genetic underpinnings of associations between risk factors and complex diseases, thereby enhancing our comprehension of disease pathogenesis. Furthermore, the genetic risk score (GRS) serves as a valuable metric, aggregating genetic variants to quantify the overall genetic risk [23,24]. This strategy not only consolidates the findings from MR analysis but also bolsters the biological validity of our research outcomes. Despite these methodological advancements, comprehensive investigations into the connection between estrogen effects and leukemia are scarce, and the causal links and regulatory mechanisms between them remain obscure.
Although a handful of studies have begun to explore the potential of these techniques—using scRNA-seq to reveal cellular heterogeneity and MR to establish causal relationships [25,26], in the current landscape of leukemia research, this combination is still a novel approach. In this groundbreaking study, we harness the power of scRNA-seq to illuminate the cellular and molecular heterogeneity of estrogen receptor in leukemia with unprecedented clarity. By integrating MR analysis, which leverages the natural randomization of genes during meiosis, we provide robust evidence for the causal association between estrogen and leukemia risk. This sophisticated combination of methodologies offers new insights into the intricate relationship between estrogen effects and leukemia, presenting potential avenues for the development of targeted preventive and therapeutic interventions that could enhance leukemia management.
Materials and Methods
Study design
Our investigation into the relationship between estrogen effects and leukemia was conducted with utmost rigor. We combined two robust research approaches: first, we rigorously analyzed the relationship between estrogen receptor (ESR1) and leukemia using scRNA-seq. Given the central role of ESR1 in mediating the effects of estrogen, we employed ESR1 expression as a surrogate for estrogen receptor activity within the leukemia cellular microenvironment. This involved a meticulous mapping of single cells and a thorough analysis of the expression differences of related genes. Then, a two-sample MR analysis, known for its statistical power, was conducted to explore the bidirectional causality between estrogen and leukemia. To further strengthen the validity of our findings, we performed a GRS analysis. The flow diagram summarizing the methodology of the study is depicted in Figure 1.
Acquisition of scRNA-seq data
Four samples for scRNA-seq were obtained from the Gene Expression Omnibus (GEO) database (accession number GSE139369), including two normal Bone marrow mononuclear cells (BMMC) samples and two BMMC samples from leukemia patients. Unlike the original study, we focused on estrogen receptors and leukemia-related proteins, ensuring the study's novelty of our findings.
scRNA-seq analysis
We first set up the Seurat object (version 5.0.2) to begin the scRNA-seq analysis. Count data were normalized after quality control. Then, we identify highly variable features for the selection and scale of the data. Then, we perform linear dimensional reduction to reduce the complexity of the dataset while preserving its structure. The number of principal components involved was determined with the aid of the ElbowPlot function in Seurat. We eliminated batch effects between samples based on the top 2000 highly variable genes obtained, and Harmony was then applied. Subsequently, we run non-linear dimensional reduction techniques such as UMAP to visualize the data in a lower-dimensional space. After that, we identified cluster biomarkers by finding differentially expressed features within each cluster. Only genes that were both enriched and expressed in a minimum of 25% of the cells within at least one cell type and exhibited a log fold change exceeding 0.25, were deemed eligible for inclusion in the study. Finally, we use the “GPTCelltype” R package to assign cell type. Compared with the existing automatic and manual annotations, this annotation method has high accuracy and robustness [27], and we use “cellmarker2.0” to verify the result of its annotation. This thoroughness ensures the validity and reliability of our results.
Differential expression analysis
We investigated the intergroup expression differences of ESR1, LIF, and LIFR, and analyzed their expression across different cell types. Additionally, we constructed pseudotime trajectory plots and illustrated the proportions of various cell types between the two groups.
Study design of our study. (A) Overview of the study. (B) Hypothesis of MR analysis. (C) Flow chart of scRNA-seq. (D) Flow chart of MR analysis.
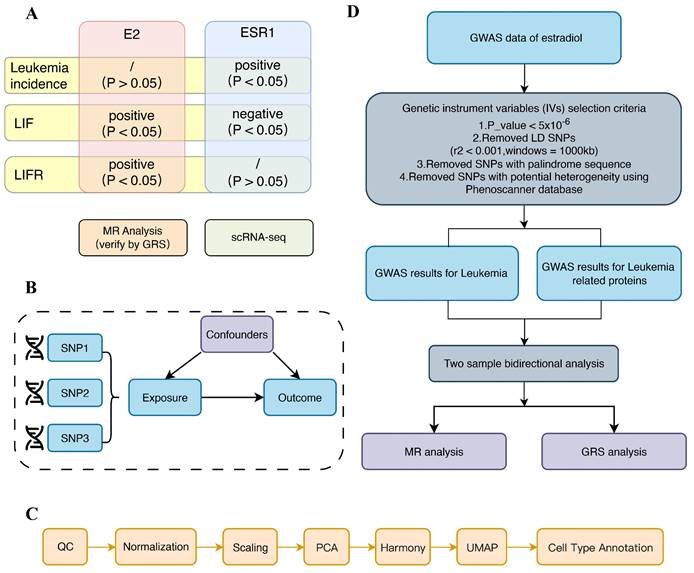
Source data of MR
The source study of exposure (estradiol) was from a generalized linear mixed model analysis [28], which applied fast GWA or fast GWA GLMM to individuals of European ancestry in the UK Biobank. They analyzed 456,348 individuals and up to 11,842,647 variants.
Data for the outcomes were obtained from two sources: the GWAS data for leukemia incidence were retrieved from the IEU OpenGWAS project dataset “ieu-b-4914”, while the genetic instruments for assessing the levels of LIF and LIFR were sourced from the INTERVAL study.
Instrumental variable selection
Genetic IVs for estradiol, leukemia incidence, LIF and LIFR were constructed according to the following criteria [29]: Only SNPs with a P value less than the genome-wide significance level of 5 × 10-6, indicating an association with disease in the respective GWAS study, were retained. Firstly, we applied a genome-wide significance threshold of P < 5 × 10-8 to filter out SNPs closely associated with estradiol, leukemia, LIF, and LIFR. However, the results indicated that the number of obtainable SNPs was limited. In such a scenario, conducting MR analysis could lead to low statistical power and weak instrument problems, resulting in biased parameter estimates [30]. Therefore, we used a more liberal criterion of P < 5 × 10-6 to identify SNPs significantly associated with exposure. Then SNPs with an r2 > 0.001 within a 10,000 kb range of the most significant SNPs were eliminated. Allelic directions of SNPs associated with exposure and outcomes were aligned while incompatible SNPs were removed. IVs associated with potential confounding factors were further eliminated using PhenoScanner [31]. Finally, to enhance the accuracy of our analysis, we applied criterion “F > 10” to filter out weak SNPs [32].
MR analyses
MR analysis is conducted utilizing the “TwoSampleMR” R package (version 0.5.6) of the RStudio (version 4.3.2). Inverse variance weighting (IVW) was employed as the primary method of MR analysis, which is the most commonly used and mainstream method for MR analysis, uses a meta-analysis approach to combine ratio estimates of SNPs in an inverse variance weighted way and obtain an estimate of the effect of risk factors on outcomes [33,34]. It ignores the presence of the intercept term in the regression and takes the inverse of the result variance as the weight for the fit, assuming that all the SNPs turn out to be valid instrumental variables and are completely independent of each other. The weighted median method incorporates weight for each value [35]. A minimum of 50% of the IVs is required to be valid to obtain a robust estimate. This method tolerates more invalid IVs [36]. MR-Egger regression accounts for the existence of an intercept term. It assumes that the instrument exposure and instrument outcome associations are independent-that is, the instrument strength is independent of the assumption of direct effect [37]. MR-Egger regression method can provide a weighted linear regression of the outcome coefficients on the exposure coefficients and detect some violations of the standard instrumental variable assumptions and provide a non-violation-prone effect estimate [38].
Sensitivity analysis of MR
We employed sensitivity analysis methods to assess the sensitivity of MR results, including the heterogeneity test, pleiotropy test, and leave-one-out sensitivity tests. Cochran's Q test and Rucker's Q test were used to detect the heterogeneity. If the P value of Cochran's Q test was less than 0.05, the final MR result referred to a multiplicative random-effects model of IVW [39]. The intercept of the MR-Egger analysis results was used to test the horizontal pleiotropy [40]. Additionally, the MR PRESSO method could simultaneously identify outliers and detect horizontal pleiotropy [41]. We cross-validated these two horizontal pleiotropy tests to provide more robust results or correct for any pleiotropy. Finally, we performed a leave-one-out analysis and created a funnel plot to examine whether individual SNPs introduced biases into the MR results [42].
GRS
To validate the above MR results, we conducted a secondary analysis using the GRS method, utilizing R (version 4.3.2) with the "gtx" R package (version 0.0.8 for Windows), whose "grs.summary" module contains the GRS function. The “grs.summary” module merely used single SNP association summarized data obtained from the results of the GWAS analysis, which was similar to a method that regresses an outcome onto an additive GRS [43,44].
Results
Comparison of Tumor Microenvironment (TME) in leukemia and normal samples
To compare TME between leukemia and normal samples, we collected four BMMC samples for scRNA-seq data. After quality control, 22595 cells were retained for subsequent analysis, including 10010 cells from the leukemia sample and 12585 cells from the normal sample.
Characterization of BMMC in normal and leukemia samples. (A) Expression of marker genes for BMMC clusters. (B) UMAP plot of BMMC, colored by cluster. (C) UMAP plot of BMMC, colored by cell type.
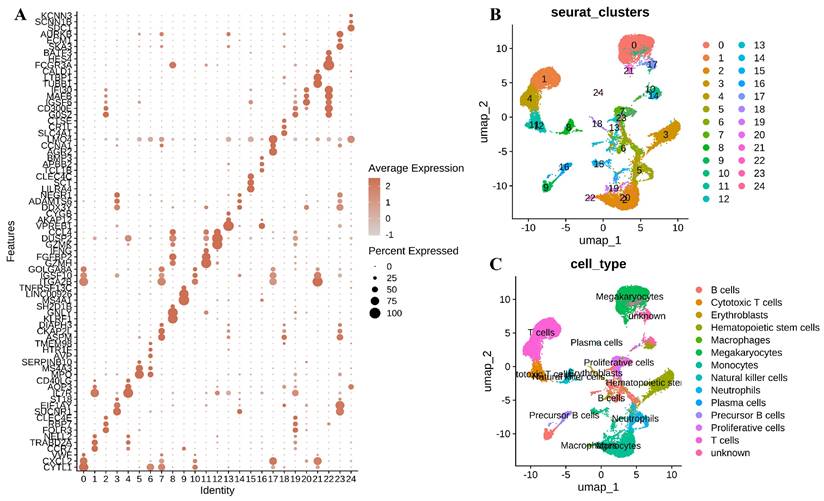
We visualized the marker genes across these single-cell subpopulations within different cell clusters using a bubble plot (Figure 2A). UMAP revealed 25 distinctive cellular clusters within the BMMC context (Figure 2B). These clusters were further categorized into 14 cell types based on the expression patterns of marker genes associated with various cell lineages, including Megakaryocytes, T cells, Monocytes, Hematopoietic stem cells, Neutrophils, B cells, Proliferative cells, Natural killer cells, Cytotoxic T cells, Precursor B cells, Erythroblasts, Macrophages, and Plasma cells (Figure 2C).
Heterogeneity of gene expression in BMMC
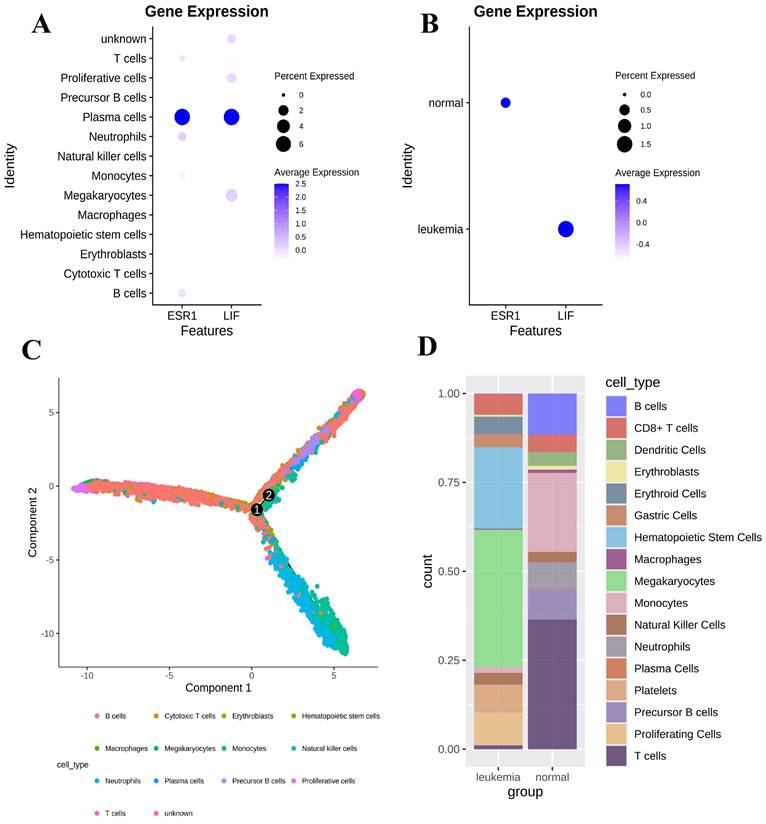
Cell clustering results showed that the ESR1 gene and LIF were mainly expressed in plasma cells (Figure 3A). We found that the expression of the ESR1 gene was higher in the normal group compared to the leukemia group, while the expression of the LIF gene was lower in the normal group compared to the leukemia group (P < 0.05). The LIFR gene was not detected in either group (Figure 3B). According to the scRNA-seq analysis, ESR1 expression was negatively correlated with leukemia, whereas LIF expression was positively correlated with leukemia. Then we performed a pseudotime analysis using the “monocle2" R package to better understand the progression of leukemia in the BMMC (Figure 3C). Furthermore, the cell proportion plot revealed substantial heterogeneity in the proportions of different cell types between the disease and normal groups (Figure 3D).
MR results
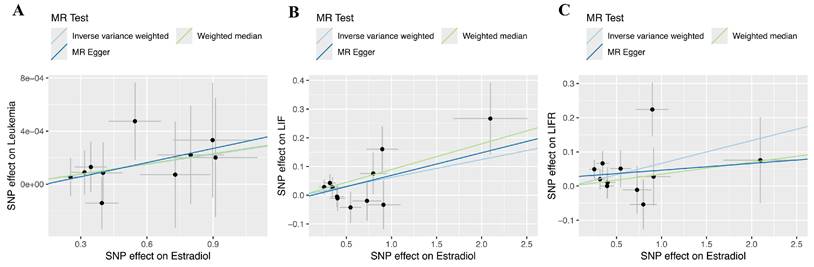
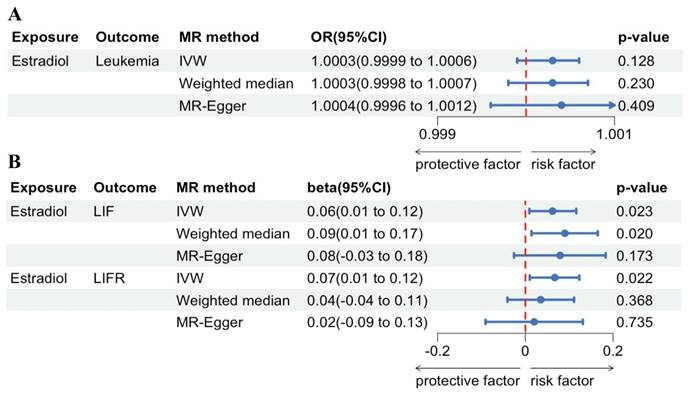
Regarding forward-direction MR, the estradiol to leukemia incidence MR study showed no significant association (IVW: OR = 1.0003, 95% CI = 0.9999-1.0005, P = 0.1278; Weighted Median: OR = 1.0003, 95% CI = 0.9998-1.0007, P = 0.2412; MR-Egger: OR = 1.0004, 95% CI = 0.9996-1.0012, P = 0.4092). In the estradiol to LIF MR study, IVW analysis and weighted median methods revealed a significant association, but MR-Egger showed no significant association (IVW: β = 0.0621, 95% CI = 0.0086-0.1156, P = 0.0229; Weighted Median: β = 0.0896, 95% CI = 0.0167-0.1625, P = 0.0161; MR-Egger: β = 0.0787, 95% CI = -0.0255-0.1828, P = 0.1728). In the estradiol to LIFR MR study, IVW analysis revealed a significant association, but weighted median methods and MR-Egger showed no significant association (IVW: β = 0.0665, 95% CI = 0.0097-0.1234, P = 0.0218; Weighted Median: β = 0.0349, 95% CI = -0.0411-0.1108, P = 0.3685; MR-Egger: β = 0.0198, 95% CI = -0.0913-0.1309, P = 0.7353). Scatter plots and forest plots are presented in Figure 4 and Figure 5.
Heterogeneity of gene expression in BMMC. (A) Gene expression differences between cell types. (B) Gene expression differences between normal and leukemia groups. (C) Pseudotime analysis of BMMC. (D) Proportions of cell clusters in the normal and leukemia groups.
Regarding reverse-direction MR, the IVW results of the estradiol to leukemia incidence were insignificant (P = 0.3852). The IVW results of the estradiol to LIF were insignificant (P = 0.8823). The results of the IVW analysis for the association between estradiol and LIFR were also not statistically significant (P = 0.0845).
MR sensitivity analysis
Cochran's Q test and Rucker's Q test showed there was no heterogeneity. The intercept of the MR Egger analysis and the MR PRESSO method indicated that there was no horizontal pleiotropy, too. The F-statistics for each SNP was greater than 10, indicating that there were no weak IVs. Using Phenoscanner, we manually removed SNPs related to confounding factors, such as viral infections, immune abnormalities [45], chemical exposure [46], ionizing radiation exposure [47], and alcohol consumption [48]. The results of the leave-one-out analysis and the funnel plot are provided in Supplemental Figure S1-S2. Detailed information about the SNPs used in the MR analysis can be found in Supplementary Table S1-S4.
Scatterplots of the genetic IVs association between estradiol and leukemia and its associated proteins. (A) Leukemia incidence. (B) LIF. (C) LIFR.
MR analysis forest plot of estradiol on leukemia and its associated proteins. (A) Leukemia incidence. (B) LIF and LIFR.
The results of the GRS
| Exposure | Outcome | OR/β (95%CI) | P value | Exposure | Outcome | OR/β (95%CI) | P value |
|---|---|---|---|---|---|---|---|
| Estradiol | leukemia | 1.0003 (0.9999-1.0006) | 0.1278 | leukemia | Estradiol | -8.7674(-43.4587, 25.9239) | 0.6204 |
| Estradiol | LIF | 0.0621(0.0086, 0.1156) | 0.0229 | LIF | Estradiol | -0.0183(-0.2605, 0.2239) | 0.8823 |
| Estradiol | LIFR | 0.0665(0.0130, 0.1201) | 0.0148 | LIFR | Estradiol | 0.1977(-0.0269, 0.4224) | 0.0845 |
GRS analysis
The results of GRS analysis confirmed the causal relationship obtained by MR analysis, and the specific GRS analysis values are shown in Table 1.
Discussion
Previous studies and epidemiological evidence have suggested a possible association between estrogen effects and leukemia [7-9]. However, studies with higher levels of evidence are lacking, and little is known about the causal relationship and molecular mechanisms. In this thesis, we complemented these two approaches with scRNA-seq and MR analysis studies to explore the relationship between estrogen effects and leukemia.
Estrogen receptors are not limited to females but are present in all vertebrates, where they are involved in various physiological and pathological states [49]. The effects of estrogen are influenced by both estrogen and estrogen receptors [50]. Either an increase in estrogen or an increase in estrogen receptors potentiates the effects of estrogen.
Initially, we aimed to explore the relationship between estrogen, estrogen receptors, and leukemia using scRNA-seq analysis. While scRNA-seq is excellent at dissecting transcriptional heterogeneity and identifying cell types within complex tissues, it has limitations. Specifically, it only studies gene expression correlations within local tissues and cannot capture interactions between different tissues. Therefore, while we can use scRNA-seq to investigate estrogen receptor relationships in leukemia cells, we cannot assess the effects of estrogen, which is primarily secreted by the ovary [51], on leukemia using this method.
Our study takes a novel approach by using Mendelian randomization combined with scRNA-seq to delve deeper into the relationship between estrogen and leukemia. This method allows us to observe the direct effect of estrogen on leukemia, unrestricted by local tissue conditions. The use of MR analysis effectively mitigates potential biases, such as confounders and reverse causation, thereby enhancing causal inference [52]. By combining scRNA-seq and the Mendelian randomization method, we effectively address the inherent limitations of each approach, providing a comprehensive exploration of the correlation between estrogen (and its receptors) and leukemia (and its related proteins). ScRNA-seq offers detailed molecular information at the cellular level, while Mendelian randomization provides causal insights into relationships between genetic variants and complex traits or diseases.
As previous studies have shown, leukemia is a highly heterogeneous disease, with significant differences between patient and normal samples [53]. We specifically chose to study mixed-phenotype acute leukemias, which present with features of multiple hematopoietic lineages and can demonstrate the common characteristics of myeloid leukemia and lymphoid leukemia to a certain extent [54,55]. This choice of mixed leukemia for our single-cell data set is crucial for the context and relevance of our research.
As shown in Figure 1, we explored the relationship between estrogen (E2, ESR1) and leukemia (leukemia incidence, LIF, LIFR) separately. ScRNA-seq shows a significant association between estradiol receptors and leukemia, and MR analysis suggests a causal relationship between estrogen and leukemia. While both methods yielded significant findings, they also presented some inconsistencies, indicating a more complex relationship between estrogen effects and leukemia. Possibly due to the high heterogeneity of leukemia. The Mendelian randomization study, with its larger sample size, and the scRNA-seq, which was limited to four samples, may have introduced a degree of chance, leading to the observed disparities. Besides, because the Mendelian randomized dataset is primarily derived from European populations, this is a source of potential heterogeneity, which may also limit the generality of our results.
In addition to affecting LIF and LIFR, estrogen also affects the actual clinical pathogenesis of leukemia through more complex molecular actions and cellular signaling pathways. The action of cytokines is usually regulated by a complex regulatory network, including their own regulation, the expression of ligands and receptors, and the activation state of signaling pathways [56]. Cytokines can influence tumor behavior and reprogram the tumor microenvironment [57]. The cytokine network is intricate, with over a hundred cytokines that share receptor components and signal transduction pathways, creating complex interactions [58]. Therefore, even if the expression level of LIF and LIFR themselves changes, it is possible that the association with leukemia pathogenesis may change due to the influence of other factors.
According to the genetic central dogma, from genes to proteins, there are two processes, transcription, and translation, and there are complex regulations such as epigenetics [59,60]. Methods of scATAC-seq and CITE-seq are promising approaches and deserve to be included in future studies to comprehensively investigate the relationship between estrogen effects and leukemia [61-63]. At the same time, due to the strong heterogeneity of leukemia, this has led to the existing studies reporting that there is a relationship between estrogen effects and leukemia. Thus, although both methods showed a significant association between them, as for clinical incidence, only scRNA-seq showed an association, whereas MR analysis studies did not show a significant causal relationship.
Our study investigates the possible involvement of estrogen (and its receptor, ESR1) and leukemia (and its related proteins, LIF and LIFR). This research aims to explore whether estrogen effects might have antitumor effects in leukemia. By understanding the role of estrogen effects, we hope to guide treatment strategies and possibly improve outcomes for certain patients. For instance, secondary leukemia following breast cancer is a common occurrence [64]. It is a question worth studying whether breast cancer patients treated with estrogen therapy may experience changes in their risk of developing leukemia.
At present, the scRNA-seq and MR datasets are still limited, and the depth of scRNA-seq is not enough. We look forward to larger samples and deeper sequencing results to further understand the relationship between estrogen effects and leukemia.
Abbreviations
GWAS: genome-wide association studies; GRS: genetic risk score; IVs: instrumental variables; IVW: inverse-variance weighted; LIF: leukemia inhibitory factor; LIFR: leukemia inhibitory factor receptor; Mcl-1: myeloid cell leukemia 1; MR: mendelian randomization; MR-PRESSO: mendelian randomization pleiotropy residual sum and outlier; SNP: single nucleotide polymorphism; scRNA-seq: single-cell RNA sequencing; BMMC: bone marrow mononuclear cells; E2: estradiol.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
We thank the study participants and research staff for their contributions to this study.
The Graphical Abstract was created in BioRender. Zhao, J. (2024) https://BioRender.com/t32j490.
Funding
This work was supported by funds from the National Natural Science Foundation of China (Grant Number: 82174222) and the Natural Science Foundation of Shandong Province (Grant Number: ZR2021LZY015).
Availability of data and materials
All data are publicly available.
Author contributions
Changgang Sun and Qibiao Wu conceptualized the study. Jiahan Zhao and Yang Yu curated the data. Jiahan Zhao, Yang Yu, Cun Liu, Ruijuan Liu, Mengxuan Sun, and Jing Zhuang developed the methodology. Jiahan Zhao and Yang Yu wrote the original draft. Changgang Sun and Qibiao Wu reviewed and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Li B, Tang H, Cheng Z, Zhang Y, Xiang H. The Current Situation and Future Trend of Leukemia Mortality by Sex and Area in China. Front Public Health. 2020;8:598215
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-49
3. Du M, Chen W, Liu K, Wang L, Hu Y, Mao Y, Sun X, Luo Y, Shi J, Shao K, Huang H, Ye D. The Global Burden of Leukemia and Its Attributable Factors in 204 Countries and Territories: Findings from the Global Burden of Disease 2019 Study and Projections to 2030. J Oncol. 2022;2022:1612702
4. Bispo JAB, Pinheiro PS, Kobetz EK. Epidemiology and Etiology of Leukemia and Lymphoma. Cold Spring Harb Perspect Med. 2020;10:a034819
5. Lin L, Yan L, Liu Y, Yuan F, Li H, Ni J. Incidence and death in 29 cancer groups in 2017 and trend analysis from 1990 to 2017 from the Global Burden of Disease Study. J Hematol OncolJ Hematol Oncol. 2019;12:96
6. Egan G, Schimmer AD. Contribution of metabolic abnormalities to acute myeloid leukemia pathogenesis. Trends Cell Biol. 2023;33:455-62
7. Lin K, Jia H, Cao M, Xu T, Chen Z, Song X, Miao Y, Yao T, Dong C, Shao J, Guo H, Hu Y, Yan Y. Epidemiological characteristics of leukemia in China, 2005-2017: a log-linear regression and age-period-cohort analysis. BMC Public Health. 2023;23:1647
8. Amini M, Sharma R, Jani C. Gender differences in leukemia outcomes based on health care expenditures using estimates from the GLOBOCAN 2020. Arch Public Health Arch Belg Sante Publique. 2023;81:151
9. Sztolsztener K, Żywno H, Hodun K, Konończuk K, Muszyńska-Rosłan K, Latoch E. Apolipoproteins-New Biomarkers of Overweight and Obesity among Childhood Acute Lymphoblastic Leukemia Survivors? Int J Mol Sci. 2022;23:10634
10. Li Q, Kopecky KJ, Mohan A, Willman CL, Appelbaum FR, Weick JK, Issa JP. Estrogen receptor methylation is associated with improved survival in adult acute myeloid leukemia. Clin Cancer Res Off J Am Assoc Cancer Res. 1999;5:1077-84
11. Rosen ST, Maciorowski Z, Wittlin F, Epstein AL, Gordon LI, Kies MS, Kucuk O, Kwaan HC, Vriesendorp H, Winter JN. Estrogen receptor analysis in chronic lymphocytic leukemia. Blood. 1983;62:996-9
12. Rota S-G, Roma A, Dude I, Ma C, Stevens R, MacEachern J, Graczyk J, Espiritu SMG, Rao PN, Minden MD, Kreinin E, Hess DA, Doxey AC. et al. Estrogen Receptor β Is a Novel Target in Acute Myeloid Leukemia. Mol Cancer Ther. 2017;16:2618-26
13. Wang J, Chang C-Y, Yang X, Zhou F, Liu J, Feng Z, Hu W. Leukemia inhibitory factor, a double-edged sword with therapeutic implications in human diseases. Mol Ther J Am Soc Gene Ther. 2023;31:331-43
14. Sun Q, Gao G, Xiong J, Wu Q, Liu H. Leukemia inhibitory factor receptor α-chain: a potential method for acute promyeloid leukemia therapy. Med Hypotheses. 2012;79:864-6
15. Mittal P, Singh S, Sinha R, Shrivastava A, Singh A, Singh IK. Myeloid cell leukemia 1 (MCL-1): Structural characteristics and application in cancer therapy. Int J Biol Macromol. 2021;187:999-1018
16. Zhang C, Liu J, Wang J, Hu W, Feng Z. The emerging role of leukemia inhibitory factor in cancer and therapy. Pharmacol Ther. 2021;221:107754
17. Viswanadhapalli S, Dileep KV, Zhang KYJ, Nair HB, Vadlamudi RK. Targeting LIF/LIFR signaling in cancer. Genes Dis. 2022;9:973-80
18. Kadia TM, Kantarjian HM, Konopleva M. Myeloid cell leukemia-1 dependence in acute myeloid leukemia: a novel approach to patient therapy. Oncotarget. 2019;10:1250-65
19. Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol. 2018;18:35-45
20. Morris JA, Sun JS, Sanjana NE. Next-generation forward genetic screens: uniting high-throughput perturbations with single-cell analysis. Trends Genet. Elsevier. 2024;40:118-33
21. Burgess S, Butterworth A, Thompson SG. Mendelian Randomization Analysis With Multiple Genetic Variants Using Summarized Data. Genet Epidemiol. 2013;37:658-65
22. Jin P, Xing Y, Xiao B, Wei Y, Yan K, Zhao J, Tian W. Diabetes and intervertebral disc degenera-tion: A Mendelian randomization study. Front Endocrinol (Lausanne). 2023;14:1100874
23. Belsky DW, Moffitt TE, Sugden K, Williams B, Houts R, McCarthy J, Caspi A. Development and Evaluation of a Genetic Risk Score for Obesity. Biodemography Soc Biol. Routledge. 2013;59:85-100
24. Cornelis MC, Qi L, Zhang C, Kraft P, Manson J, Cai T, Hunter DJ, Hu FB. Joint Effects of Common Genetic Variants on the Risk for Type 2 Diabetes in U.S. Men and Women of European Ancestry. Ann Intern Med. American College of Physicians. 2009;150:541-50
25. Li H, Jiang X, Xiao Y, Zhang Y, Zhang W, Doherty M, Nestor J, Li C, Ye J, Sha T, Lyu H, Wei J, Zeng C, Lei G. Combining single-cell RNA sequencing and population-based studies reveals hand osteoarthritis-associated chondrocyte subpopulations and pathways. Bone Res. 2023Nov2;11(1):58
26. Qian X, Zheng Y, Xu L, Liu Z, Chen M, Tong F, Fan P, Chen Z, Dong N, Zhang C, Liu J. Deciphering the role of CX3CL1-CX3CR1 in aortic aneurysm pathogenesis: insights from Mendelian randomization and transcriptomic analyses. Front Immunol. 2024;15:1383607
27. Hou W, Ji Z. Assessing GPT-4 for cell type annotation in single-cell RNA-seq analysis. Nat Methods. 2024Aug;21(8):1462-1465
28. Jiang L, Zheng Z, Fang H, Yang J. A generalized linear mixed model association tool for biobank-scale data. Nat Genet. 2021;53:1616-21
29. Chen L, Yang H, Li H, He C, Yang L, Lv G. Insights into modifiable risk factors of cholelithiasis: A Mendelian randomization study. Hepatol Baltim Md. 2022;75:785-96
30. Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol. 2011;40:740-52
31. Kamat MA, Blackshaw JA, Young R, Surendran P, Burgess S, Danesh J, Butterworth AS, Staley JR. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinforma Oxf Engl. 2019;35:4851-3
32. Burgess S, Thompson SG. Use of allele scores as instrumental variables for Mendelian randomi-zation. Int J Epidemiol. 2013;42:1134-44
33. Bowden J, Del Greco M F, Minelli C, Davey Smith G, Sheehan N, Thompson J. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat Med. 2017;36:1783-802
34. Burgess S, Scott RA, Timpson NJ, Davey Smith G, Thompson SG, EPIC- InterAct Consortium. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol. 2015;30:543-52
35. Burgess S, Thompson SG. Improving bias and coverage in instrumental variable analysis with weak instruments for continuous and binary outcomes. Stat Med. 2012;31(15):1582-1600
36. Li C, Niu M, Guo Z, Liu P, Zheng Y, Liu D, Yang S, Wang W, Li Y, Hou H. A Mild Causal Relationship Between Tea Consumption and Obesity in General Population: A Two-Sample Mendelian Randomization Study. Front Genet. 2022;13:795049
37. Bowden J. Misconceptions on the use of MR-Egger regression and the evaluation of the InSIDE assumption. Int J Epidemiol. 2017;46:2097-9
38. Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32:377-89
39. Higgins JPT, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557-60
40. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512-25
41. Verbanck M, Chen C-Y, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693-8
42. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R, Tan VY, Yarmolinsky J, Shihab HA. et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife. 2018;7:e34408
43. Luo Q, Wen Z, Li Y, Chen Z, Long X, Bai Y, Huang S, Yan Y, Lin R, Mo Z. Assessment Causality in Associations Between Serum Uric Acid and Risk of Schizophrenia: A Two-Sample Bidirectional Mendelian Randomization Study. Clin Epidemiol. 2020;12:223-33
44. Liu Y, Xiao Z, Ye K, Xu L, Zhang Y. Smoking, alcohol consumption, diabetes, body mass index, and peptic ulcer risk: A two-sample Mendelian randomization study. Front Genet. 2022;13:992080
45. Rutella S, Vadakekolathu J, Mazziotta F, Reeder S, Yau T-O, Mukhopadhyay R, Dickins B, Altmann H, Kramer M, Knaus HA, Blazar BR, Radojcic V, Zeidner JF. et al. Immune dysfunction signatures predict outcomes and define checkpoint blockade-unresponsive microenvironments in acute myeloid leukemia. J Clin Invest. 2022;132:e159579
46. Beane Freeman LE, Blair A, Lubin JH, Stewart PA, Hayes RB, Hoover RN, Hauptmann M. Mortality from lymphohematopoietic malignancies among workers in formaldehyde industries: the National Cancer Institute Cohort. J Natl Cancer Inst. 2009;101:751-61
47. Metz-Flamant C, Samson E, Caër-Lorho S, Acker A, Laurier D. Leukemia risk associated with chronic external exposure to ionizing radiation in a French cohort of nuclear workers. Radiat Res. 2012;178:489-98
48. Engen PA, Green SJ, Voigt RM, Forsyth CB, Keshavarzian A. The Gastrointestinal Microbiome: Alcohol Effects on the Composition of Intestinal Microbiota. Alcohol Res Curr Rev. 2015;37:223-36
49. Amenyogbe E, Chen G, Wang Z, Lu X, Lin M, Lin AY. A Review on Sex Steroid Hormone Estrogen Receptors in Mammals and Fish. Int J Endocrinol. Hindawi. 2020;2020:e5386193
50. Dietrich W, Haitel A, Huber JC, Reiter WJ. Expression of estrogen receptors in human corpus cavernosum and male urethra. J Histochem Cytochem Off J Histochem Soc. 2004;52:355-60
51. Zhao H, Zhou L, Shangguan AJ, Bulun SE. Aromatase expression and regulation in breast and endometrial cancer. J Mol Endocrinol. Bioscientifica Ltd. 2016;57:R19-33
52. Rasooly D, Peloso GM. Two-Sample Multivariable Mendelian Randomization Analysis Using R. Curr Protoc. 2021;1:e335
53. Granja JM, Klemm S, McGinnis LM, Kathiria AS, Mezger A, Corces MR, Parks B, Gars E, Liedtke M, Zheng GXY, Chang HY, Majeti R, Greenleaf WJ. Single-cell multiomic analysis identifies regulatory programs in mixed-phenotype acute leukemia. Nat Biotechnol. NIH Public Access. 2019;37:1458
54. Matutes E, Pickl WF, Van't Veer M, Morilla R, Swansbury J, Strobl H. et al. Mixed-phenotype acute leukemia: clinical and laboratory features and outcome in 100 patients defined according to the WHO 2008 classification. Blood. 2011;117:3163-71
55. Horvat NP, Logothetis CN, Zhang L, Yun S, Sweet K. Gilteritinib Combined with Azacitidine as Salvage Therapy for B/Myeloid Mixed Phenotype Acute Leukemia. Cureus. 2022;14:e23618
56. Fairfax BP, Humburg P, Makino S, Naranbhai V, Wong D, Lau E, Jostins L, Plant K, Andrews R, McGee C, Knight JC. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science. 2014;343:1246949
57. Chen W, Qin Y, Liu S. Cytokines, breast cancer stem cells (BCSCs) and chemoresistance. Clin Transl Med. 2018;7:e27
58. Morel PA, Lee REC, Faeder JR. Demystifying the cytokine network: Mathematical models point the way. Cytokine. 2017;98:115-23
59. Ross SA, Dwyer J, Umar A, Kagan J, Verma M, Van Bemmel DM, Dunn BK. Introduction: diet, epigenetic events and cancer prevention. Nutr Rev. 2008;66:S1-6
60. Guo Y, Ren G, Zhang K, Li Z, Miao Y, Guo H. Leaf senescence: progression, regulation, and application. Mol Hortic. 2021;1:5
61. Zhang Z, Yang C, Zhang X. scDART: integrating unmatched scRNA-seq and scATAC-seq data and learning cross-modality relationship simultaneously. Genome Biol. 2022;23:139
62. Hu K, Liu H, Lawson ND, Zhu LJ. scATACpipe: A nextflow pipeline for comprehensive and repro-ducible analyses of single cell ATAC-seq data. Front Cell Dev Biol. 2022;10:981859
63. Huang Y, Li Y, Liu Y, Jing R, Li M. A Multiple Comprehensive Analysis of scATAC-seq Based on Auto-Encoder and Matrix Decomposition. Symmetry. Multidisciplinary Digital Publishing Institute. 2021;13:1467
64. Jabagi MJ, Goncalves A, Vey N, Le Tri T, Zureik M, Dray-Spira R. Risk of Hematologic Malignant Neoplasms after Postoperative Treatment of Breast Cancer. Cancers. 2019;11:1463
Author contact
![]() Corresponding authors: Changgang Sun, M.D. PhD. College of Traditional Chinese Medicine, Shandong Second Medical University, Weifang, 261000, China; Department of Oncology, Weifang Traditional Chinese Hospital, Weifang, 261000, China. Email: scgdoctorcom. Qibiao Wu, M.D. PhD. State Key Laboratory of Quality Research in Chinese Medicine, and Faculty of Chinese Medicine, Macau University of Science and Technology, Avenida Wai Long, Taipa, 999078, Macau, China. Email: qbwuedu.mo.
Corresponding authors: Changgang Sun, M.D. PhD. College of Traditional Chinese Medicine, Shandong Second Medical University, Weifang, 261000, China; Department of Oncology, Weifang Traditional Chinese Hospital, Weifang, 261000, China. Email: scgdoctorcom. Qibiao Wu, M.D. PhD. State Key Laboratory of Quality Research in Chinese Medicine, and Faculty of Chinese Medicine, Macau University of Science and Technology, Avenida Wai Long, Taipa, 999078, Macau, China. Email: qbwuedu.mo.

 Global reach, higher impact
Global reach, higher impact