Impact Factor
ISSN: 1837-9664
J Cancer 2025; 16(3):952-968. doi:10.7150/jca.103708 This issue Cite
Research Paper
Sensitivity of renal cell carcinoma to cuproptosis: a bioinformatics analysis and experimental verification
1. Laboratory of Stem Cell Regulation with Chinese Medicine and Its Application, School of Pharmacy, Hunan University of Chinese Medicine, Changsha 410208, China
2. Department of Urology, The First Hospital of Hunan University of Chinese Medicine, Changsha 410007, China
3. Institutional Key Laboratory of Vascular Biology and Translational Medicine in Hunan Province, Changsha, China
4. Hunan Province Engineering Research Center of Bioactive Substance Discovery of Traditional Chinese Medicine, Hunan University of Chinese Medicine, Changsha, China.
Received 2024-9-15; Accepted 2024-12-10; Published 2025-1-1
Abstract
Purpose: Targeting cuproptosis is considered as a promising therapeutic strategy for the prevention of tumors. However, the potential role of cuproptosis and its related genes in clear cell renal cell carcinoma (ccRCC) remains elusive. The present study aims to explore the sensitivity of ccRCC to cuproptosis and its underlying mechanism.
Methods: Cuproptosis differential genes (CDGs) were extracted using the GSE53757 and GSE66272 datasets. A comprehensive analysis of the role of CDGs was conducted through multiple public databases and experiments.
Results: It was found that cuproptosis inducer elesclomol significantly induced cell death in 786-O and A498 cells. FDX and DLAT exhibited significantly low expression, which were independent prognostic factors for poor survival, and had a strong positive correlation in ccRCC patients. Functional analysis of differentially expressed genes positively or negatively correlated with both FDX1 and DLAT indicated that acetyl-CoA biosynthetic process and acetyl-CoA metabolic process were remarkably affected. In ccRCC patients, the methylation levels and sites of FDX1 and DLAT genes were dramatically correlated with overall survival (OS). The expressions of FDX1 and DLAT were closely related to immune infiltration and immune checkpoints. Docking results indicated that mitotane, adicicol and dihydrolipoic acid might be potential drug targets for FDX1 and DLAT.
Conclusions: Overall, the present study demonstrates the sensitivity of ccRCC to cuproptosis, and targeting the combination of FDX1 and DLAT may be a novel therapeutic strategy to induce cuproptosis in ccRCC.
Keywords: Cuproptosis, Clear cell renal cell carcinoma, FDX1, DLAT
Introduction
Clear cell renal cell carcinoma (ccRCC), a major histological subtype of RCCs, accounts for 70%-80% of all renal cell carcinoma cases[1, 2]. At present, early-stage ccRCC can usually be treated effectively by surgery, but the treatment of advanced ccRCC still faces severe clinical challenges[3]. Although early detection and timely treatment are beneficial for ccRCC patients, clinically successful ccRCC-specific markers are still lacking[4, 5]. As such, there is an urgent require to identify sensitive molecular biomarkers and therapeutic strategies for ccRCC therapy.
Programmed cell death (PCD) is a type of gene-regulated cell death that not only plays an important role in fundamental biological processes but also participates in a variety of pathological processes[6]. An increasing number of PCD types, such as apoptosis, necroptosis, autophagy, ferroptosis and pyroptosis, have been implicated in the pathological and physiological processes of various diseases, especially tumors[7, 8]. A typical example is that targeting autophagy to treat tumors has brought enormous clinical benefits to cancer patients[9, 10]. For example, gefitinib, lapatinib and erlotinib are common clinical tyrosine kinase inhibitors that target autophagy to treat breast cancer, chronic myeloid leukemia, and gastrointestinal stromal tumors[11, 12]. However, resistance to autophagy is largely responsible for the failure of these therapeutic strategies[13]. Different forms of PCD can be substituted for each other in the treatment of tumors[14]. Therefore, finding novel forms of PCD is crucial for therapy-resistant tumors. More recently, a novel regulated form of cell death, termed cuproptosis, has attracted considerable academic interest, referring to copper ionophore-induced (including elesclomol, disulfiram and NSC319726) binding of copper to the lipoylated tricarboxylic acid (TCA) cycle proteins, ultimately stimulating cell death[15]. The progression of ccRCC is closely related to the reprogramming of the TCA cycle based on metabolomics analysis[16], which regulates energy metabolism through the TCA cycle, allowing tumor cells to survive under nutrient-depleted conditions and escape the immune system[17, 18]. Although a previous study has analyzed the prognostic role of cuproptosis-related genes in ccRCC[19], these theories lacked experimental verification. In particular, the sensitivity of ccRCC to cuproptosis still lacks a large amount of reliable experimental data, and the relationship between cuproptosis related genes and methylation and immune microenvironment remains unclear.
Herein, we sought to find the potential functions of the most critical cuproptosis-related genes in ccRCC on the basis of public databases. Additionally, the protein expression of cuproptosis-related genes was verified in our collected ccRCC clinical samples. Furthermore, the sensitivity of ccRCC to cuproptosis was examined and its potential target genes were evaluated in vitro experiments.
Methods and Materials
Cuproptosis-related genes and ccRCC differential genes collection and preprocessing
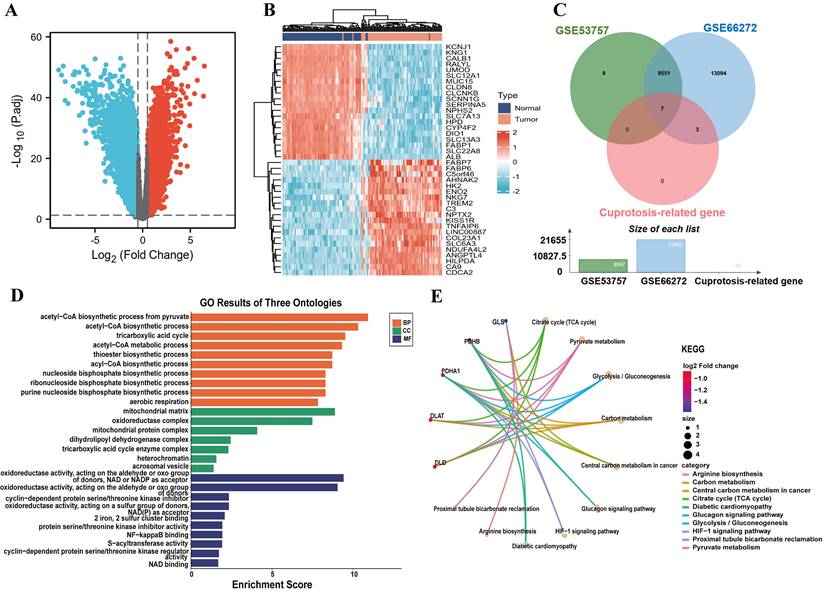
The 10 cuproptosis-related genes were obtained from a previously study[15]. The GSE53757 and GSE66272 datasets are the commonly utilized expression profiles for analyzing differentially expressed genes in ccRCC. Compared with other ccRCC datasets, GSE53757 and GSE66272 contain a relatively large number of samples. Thus, we chose the GSE53757 and GSE66272 cohorts for the differentially analysis. The GSE53757 and GSE66272 datasets was downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/), which includestumor samples and matched adjacent paracancerous tissues from 99 patients with ccRCC. A total of 99 ccRCC tumor tissue and adjunct nontumor samples were analyzed by GEO2R, an online analysis tool, to acquire the differential expression of genes. The differentially expressed genes were selected under the following criteria: |log2 (fold-change) | > 0.5 and P < 0.05. Subsequently, cuproptosis-related genes and ccRCC differential genes were intersected to identify CDGs. A venn diagram of differentially expressed genes was created using the online tool Venny2.1.0, and the heatmap and volcano plot of differential expression genes about the gene expression profiles in the GSE53757 and GSE66272 databases were drawn by R software (version 4.0.3; R Foundation).
The expression, mapping and prognosis of cuproptosis differential genes
Following that, the mRNA expression profile data of CDGs in tumor tissues and different clinical stages of ccRCC, such as pathologic state and OS events, were analyzed using RNA-seq gene expression data and clinical information downloaded from the TCGA database. The protein expression and localization of CDGs were explored by the Human Protein Atlas (HPA, https://www.proteinatlas.org/). The R software packages "Survival" and "Survminer" perform the Kaplan-Meier (KM) survival curve analysis. The univariate Cox regression analysis was used to calculate the relationship between CDGs expression and the ccRCC patient's OS. A multivariate analysis was done to determine whether the CDGs were independent prognostic factors for ccRCC patient survival, and their diagnostic values were assessed with a receiver operating characteristic (ROC) curve analysis.
Co-expression network and functional enrichment analysis of CDGs
The genes co-expressed with FDX and DLAT in ccRCC were acquired from LinkedOmics (http://linkedomics.org/login.php), and screened according to the Pearson correlation coefficient (|cor| > 0.3, P < 0.05). Heatmaps of the top 50 negative or positive FDX and DLAT expression correlated genes were plotted by the “pheatmap” R package. To demonstrate the biological function of CDGs and the top 50 genes most positively or negatively correlated with FDX and DLAT, gene ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed on the Metascape website (http://metascape.org/gp/index).
Gene mutation and methylation of ferredoxin 1 (FDX1) and dihydrolipoyl transacetylase (DLAT)
The FDX1 and DLAT gene mutations in ccRCC were evaluated using cBioPortal (http://www.cbioportal.org/). To further investigate the effect of DNA methylation status of FDX1 and DLAT on ccRCC, MEXPRESS visualized expression, DNA methylation, and clinical parameters according to TCGA data[20]. Then, the relative DNA methylation site data for FDX1 and DLAT was evaluated using an online tool called MethSurv (https://biit.cs.ut.ee/methsurv/). In addition, the survival analysis and prognostic value of all methylation sites were explored.
Analysis of immune infiltration and immune checkpoints
The molecular characterization of tumor immune interactions in ccRCC was investigated using the single-sample GSEA (ssGSEA) approach from the “GSVA” R package and the Tumor Immune Estimation Resource (TIMER) database (http://timer.cistrome.org/). Furthermore, we explored the effect of low or high FDX and DLAT expression on immune cell infiltration. Immune checkpoints were identified using the TISIDB database (http://cis.hku.hk/TISIDB/).
Drug targets screening and validation
The candidate drugs targeting the FDX1 and DLAT were acquired from DrugBank database (https://go.drugbank.com/). To validate the drug targets of FDX1 and DLAT, molecular docking was performed with AutoDock vina 1.1.2. The structures of FDX1 (PDB: 3P1M) and DLAT (PDB: 1Y8N) were obtained from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Databank (https://www.rcsb.org/). Their X-ray crystal structures: FDX1: Resolution: 2.54 Å, R-Value Free: 0.241, R-Value Work: 0.204, R-Value Observed: 0.205[21]; DLAT: Resolution: 2.60 Å, R-Value Free: 0.248, R-Value Work: 0.210, R-Value Observed: 0.211[22]. The results are analyzed and visualized by using PyMOL.
Cell lines and clinical samples
Human ccRCC cell lines 786-O and A498 were purchased from the American Type Culture Collection (Rockville, MD, USA) and maintained at 37°C in a humidified, 5% CO2 controlled atmosphere in RPMI medium 1640 and MEM medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin, respectively.
Tumor and adjacent non-tumorous tissues were taken from five patients with ccRCC who underwent surgical treatment in the First Hospital of Hunan University of Chinese Medicine. All procedures involving human participants in this study complied with the Declaration of Helsinki (2013 revision). This study was approved by the Ethics Committee of Hunan University of Chinese medicine (Ethics number: HN-LL-GZR-2022-22). Before surgery, all patients with ccRCC provided informed consent and received no further special therapy. When we conducted experiments using samples, patients provided oral informed consent due to age, cognitive ability, or other health conditions.
CCK-8 assay
The CCK-8 assay was performed to explore elesclomol-induced cuproptosis in ccRCC. Cells at 5000 cells/well were seeded in 96-well plates and co-treated with different concentrations of elesclomol (0-100 nM) and 1µM CuCl2, incubated for 12 h at 37℃. Meanwhile, cells treated with elesclomol alone act as a control. CCK-8 solution (5 mg/mL) was added to each well at 10 µL. After 4 h of incubation. The absorbance of each well was detected by using an enzyme-labeled instrument (Bio-Rad Laboratories, Hercules, CA, USA) at a wavelength of 490 nm.
Western blot assay
Total protein from cells and tumor tissue from ccRCC patients were extracted with RIPA lysis buffer containing proteinase inhibitor (Beyotime, Shanghai, China). Proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and treated with primary antibodies overnight at 4°C, which were washed and incubated with the peroxidase-conjugated anti-rabbit secondary antibody at room temperature for 1 h. Chemiluminescent detection was carried out utilizing ECL reagents with a ChemiDoc MP imaging system (Millipore Corporation). The following primary and secondary antibodies were used: antibodies for β-actin (1:2,000), FDX1 (1:2,000), DLAT (1:2,000) and peroxidase-conjugated anti-rabbit (1:5000). All antibodies were purchased from Abcam.
Statistics
SPSS20.0 was used for statistical analysis of all data; and the results were presented as mean ± standard deviation (mean ± SD). The significant difference was analyzed by one-way analysis of variance or unpaired Student's t-test, and P < 0.05 was considered statistically significant. All data shown are representative of at least three independent experiments.
Results
Cuproptosis differential genes involved in ccRCC collection and functional enrichment analysis
We downloaded the GSE53757 and GSE66272 microarray expression profiling datasets from the GEO database and obtained 8551 differential genes at |log2 (fold-change) | > 0.5 and P < 0.05 (Fig. 1A, 1C). In addition, cuproptosis-related genes were obtained including 10 genes from a previous study[15] and intersected them with GSE53757 and GSE66272 to identify cuproptosis differential genes in ccRCC. The results indicated six down-regulated genes and only one up-regulated genes (Fig. 1C and Table S1). The Top 20 up- and down-regulated genes from ccRCC patients were exhibited in the heatmap (Fig. 1B). As we identified, functional and pathway enrichment analysis of seven cuproptosis differential genes was carried out using the online tool Metascape. The results of functional analysis revealed that acetyl-CoA biosynthetic process, acetyl-CoA metabolism process, acyl-CoA biosynthetic process, mitochondrial matrix, S-acyltransferase activity and Fe2+, 2 sulfur cluster binding were dramatically triggered in the gene sets (Fig. 1D). The activation of the citrate cycle (TCA cycle) signaling pathway, pyruvate metabolism, glycolysis/gluconeogenesis and carbon metabolism were all regulated by PDHB, PDHA1, DLAT and DLD; among them, PDHB and PDHA1 were involved in the glucagon signaling pathway, HIF-1 signaling pathway and diabetic cardiomyopathy (Fig. 1E).
Validation and mapping of cuproptosis differential gene expression
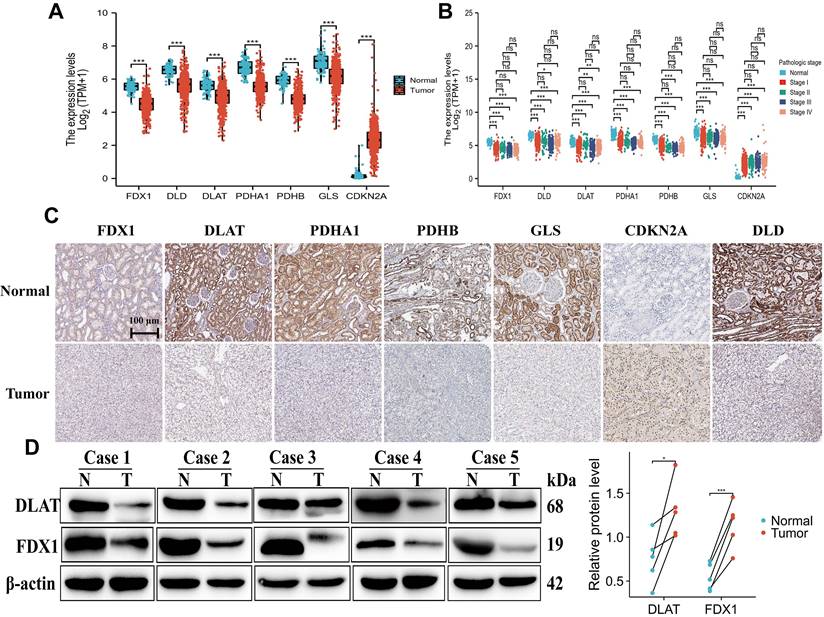
We further analyzed the mRNA expression of CDGs in ccRCC through the TCGA database (Unmatched renal clear cell carcinoma consisted of 72 paracancer tissues and 537 tumor samples). The results demonstrated that FDX1, DLD, DLAT, PDHA1, GLS and PDHB exhibited significantly lower expression in ccRCC tissues than normal tissues. Interestingly, only CDKN2A showed higher expression in ccRCC tissues than normal tissues (Fig. 2A). In addition, we found that the expressions of FDX1, DLD, DLAT, PDHA1, GLS and PDHB were significantly down-regulated in pathologic state, whereas CDKN2A was up-regulated as compared to comparable normal tissues (Fig. 2B). The results from the HPA database showed that protein expression levels of CDGs had also been consistent with mRNA expression in ccRCC (Fig. 2C). Based on the above results, we validated the protein expression of FDX1 and DLAT in five matched normal and ccRCC clinical samples, and found that their protein expressions were poorly expressed in ccRCC tissues compared to adjacent tissues (Fig. 2D). The results illustrated that cuproptosis differential genes were abnormally expressed in ccRCC patients in comparison to those of normal tissues, which might be predictive of poor prognosis and is related to disease stage progression in ccRCC patients.
Effects of cuproptosis differential genes on ccRCC patient prognosis
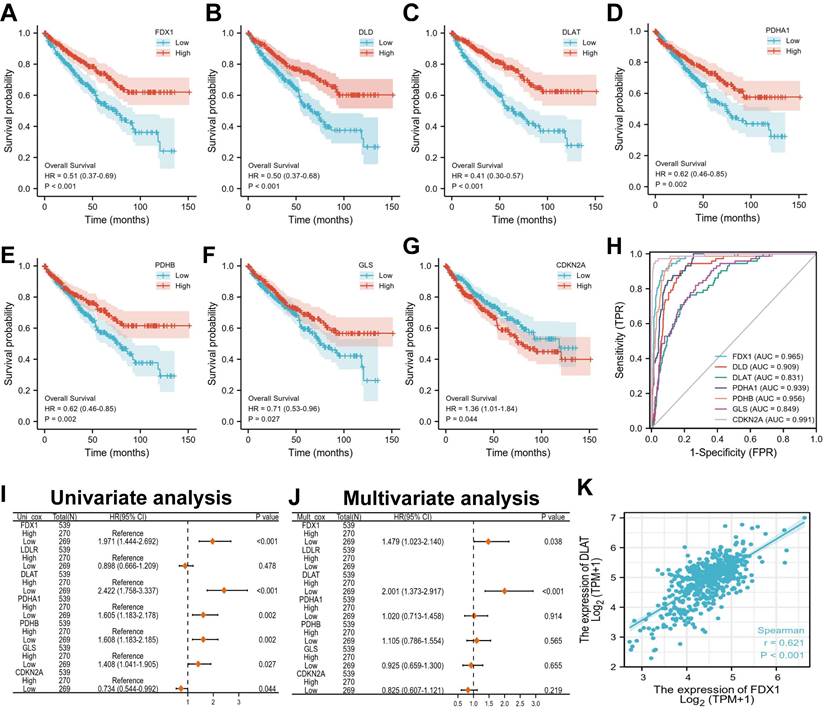
Next, we evaluated the correlation between the expression of cuproptosis differential genes and the prognosis in ccRCC patients. The results indicated that ccRCC patients with low expression of FDX1, DLD, DLAT, PDHA1, GLS and PDHB had poor survival probability (HR < 1, P < 0.05), but not CDKN2A (Fig. 3A-G). Furthermore, the evaluation of univariate cox regression model revealed that low expression of FDX1 (HR 1.971; 95% CI 1.444-2.692, P < 0.001), DLAT (HR 2.422; 95% CI 1.758-3.337, P < 0.001), PDHA1 (HR 1.605; 95% CI 1.183-2.178, P = 0.002), PDHB (HR 1.608; 95% CI 1.183-2.185, P = 0.002) and GLS (HR 1.408; 95% CI 1.041-1.905, P = 0.027) were poor predictors for OS in ccRCC patients (Fig. 3I). Conversely, low CDKN2A expression (HR 0.734; 95% CI 0.544-0.992, P = 0.044) predicted a favorable prognosis in patients suffering from ccRCC. The multivariate cox results suggested that only FDX (HR 1.479; 95% CI 1.023-2.140, P = 0.038) and DLAT (HR 2.001; 95% CI 1.373-2.917, P < 0.001) were identified as independent prognostic factors affecting OS in patients with ccRCC (Fig. 3J). For the diagnostic value of cuproptosis differential genes in ccRCC patients, the AUCs of FDX1, DLD, DLAT, PDHA1, GLS, PDHB and CDKN2A were 0.965, 0.909, 0.813, 0.939, 0.956, 0.849 and 0.991, respectively (Fig. 3H). These findings suggest that ccRCC patients with strong (CDKN2A) or weak (FDX1, DLAT, etc.) CDGs expressions possess high diagnostic accuracy. Overall, FDX and DLAT act as independent prognostic factors and have a highly accurate diagnostic value for ccRCC patients. Thus, they may have important implications for the treatment of ccRCC patients. The correlation between FDX1 and DLAT in ccRCC was investigated by the “corrplot” R package. Notably, the results found that FDX1 and DLAT exhibited a strong positive correlation (r = 0.621, P < 0.01, Fig. 3K).
Co-expression network selection and gene functional enrichment analysis of FDX1 and DLAT genes
We further identified differentially expressed genes related to FDX1 and DLAT in ccRCC using the LinkedOmics database. The differentially expressed genes associated with FDX1 and DLAT were found under the Pearson test (Fig. 4A, 4D), and their top 50 positively (r > 0) and negatively (r < 0) correlated genes were shown in the heatmap (Fig. 4B, 4C, 4E, 4F). The positive and negative correlation genes of FDX1 and DLAT were selected based on the following criteria: |r| > 0.5, P < 0.05. Eventually, 101 genes were positively and 83 genes were negatively correlated with FDX1 and 625 genes were positively and 642 were genes negatively correlated with DLAT, respectively. Among them, a total of 43 positively and 30 negatively associated genes with FDX1 and DLAT were identified for further analysis (Fig. 4G, 4H). These genes were used for GO and KEGG enrichment analysis. For genes positively associated with FDX1 and DLAT, the functional analysis demonstrated that acetyl-CoA biosynthetic process, respiratory electron transport chain, lipid oxidation, mitochondrial respiratory chain, active ion transmembrane transporter activity and acetyl-CoA metabolic process were severely affected. The KEGG results indicated that these genes were primarily involved in TCA cycle, oxidative phosphorylation and secretion, and carbon metabolism (Fig. 4I). For genes negatively correlated with FDX1 and DLAT, the functional analysis indicated that the regulation of signal transduction by p53 class mediator, mitotic cytokinetic process, host intracellular domains, iron-sulfur cluster binding, and metal cluster binding were impacted on the biological processes, molecular functions, and cellular component terms. The KEGG results revealed that these genes were mainly involved in the process of endocytosis, base excision repair and ether lipid metabolism (Fig. 4J).
The identification of cuproptosis differential genes and enrichment analysis of function and pathway. (A) The volcano plot reveals differential genes in ccRCC. (B) Heatmap displays the top 20 significantly up- and down-regulated differentially expressed genes. (C) Venn diagram of cuproptosis differentially expressed genes. (D) GO functional enrichment analysis (BP, MF and CC). (E) KEGG pathway enrichment analysis of cuproptosis differential genes.
Expression and mapping analysis of cuproptosis differential genes. (A-C) The mRNA and protein expression patterns of cuproptosis differential genes in ccRCC by TCGA and HPA databases. (D) Western blot analysis of DLAT and FDX1 in five paired clinical ccRCC and paracancerous tissue samples. “ns” represents P ≥ 0.05, *P < 0.05, **P < 0.01, ***P < 0.001.
Genomic alterations and methylation analysis
The mutation frequencies of FDX1 and DLAT in ccRCC patients were explored through the cBioPortal database. The dataset including 392 patients (TCGA-ccRCC, Nature 2013, RNA Seq V2 RSEM) was selected for analysis. The somatic mutation frequency of DLAT in ccRCC was 0.3%, consisting mainly of missense mutations (Fig. S1B), which were comparatively rare, with only 1 in 392 samples. Surprisingly, FDX1 did not exhibit any somatic mutations (Fig. S1A). Therefore, FDX1 and DLAT mutations were not found to affect the overall survival of patients (Fig. S1C, S1D). Furthermore, we found that the total FDX1 and DLAT methylation levels were reduced in ccRCC tissues compared with normal tissues (Fig. 5C, 5D), and their methylation status was both dramatically correlated with OS and CpG sites in ccRCC patients (Fig. 5A, 5B). Thus, the methylation sites of FDX1 and DLAT genes were examined, along with the prognostic value of each CpG, based on the TCGA database. The data showed that cg02239377, cg06674932 and cg26061355 of FDX1 and cg08065721 of DLAT were the most methylated sites (Fig. 5E, 5F). Nevertheless, the methylation sites of the FDX1 gene included cg05485370 (HR:0.518, 95%CI: 0.311-0.863, P = 0.012), cg13258606 (HR:0.42, 95%CI: 0.245-0.718, P = 0.0015), cg23587050 (HR:0.411, 95%CI: 0.269-0.639, P < 0.001) and cg26061355 (HR:0.584, 95%CI: 0.351-0.974, P = 0.039), suggesting a good prognosis for patients suffering from ccRCC. In contrast, patients with cg05741490 (HR:2.054, 95%CI: 1.198-3.548, P = 0.0099), cg06674932 (HR:1.996, 95%CI: 1.172-3.4, P = 0.011), cg09762563 (HR:1.557, 95%CI: 1.061-2.286, P = 0.024) and cg26763524 (HR:1.655, 95%CI: 1.007-2.721, P = 0.047) conferred a poor prognosis (Fig. 5G). For DLAT gene, cg00327185 (HR:0.555, 95%CI: 0.34-0.905, P = 0.018) and cg13372927 (HR:0.498, 95%CI: 0.328-0.756, P = 0.0011) revealed a good prognosis in ccRCC patients, but cg10616121 (HR:2.442, 95%CI: 1.647-3.623, P < 0.001) and cg27191019 (HR:3.243, 95%CI: 1.774-5.929, P < 0.001) were associated with adverse patient outcomes (Fig. 5H).
Overall survival and diagnostic value of different cuproptosis differential genes in ccRCC patients. (A-G) Prognostic analysis of cuproptosis differential gene expression in ccRCC patients. (H) The diagnostic value of different cuproptosis differential genes for ccRCC. (I, J) Univariate and multivariate regression model of differential genes for different cuproptosis. (K) Correlation analysis of FDX1 and DLAT genes.
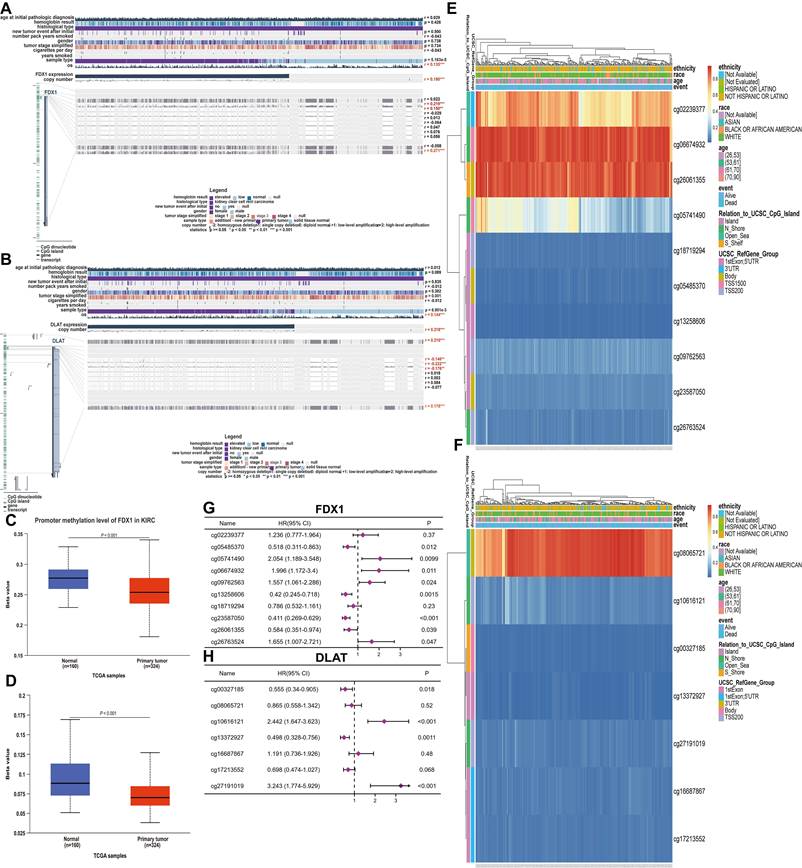
Correlation analysis of FDX1 and DLAT expression with immune infiltration level in ccRCC
To understand the relationship between FDX1 and DLAT in cellular immunity, the potential correlation of their expression with 24 types of immune cells was analyzed by ssGSEA from the R package with a Spearman test. The findings showed that FDX1 expression was significantly related to neutrophils, mast cells, eosinophils, TReg, aDC and cytotoxic cells (Fig. 6A). However, DLAT was significantly associated with eosinophils, neutrophils, T helper cells, cytotoxic cells, B cells, NK CD56bright cells and TReg (Fig. 6B). Further studies demonstrated that FDX1 expression was positively correlated with infiltration levels of eosinophils and mast cells (r = 0.168, P < 0.001), neutrophils (r = 0.172, P < 0.001), but negatively correlated with cytotoxic cells (r = -0.239, P < 0.001) and TReg (r = -0.314, P < 0.001), while there was no significant correlation with B cells (r = -0.073, P < 0.090). DLAT expression was positively correlated with infiltration levels of eosinophils (r = 0.310, P < 0.001), neutrophils (r = 0.224, P < 0.001), T helper cells (r = 0.205, P < 0.001), but negatively correlated with CD8 T cells (r = -0.266, P < 0.001), cytotoxic cells (r = -0.393, P < 0.001) and B cells (r = -0.135, P < 0.002) (Fig. S2). Subsequently, the expression of FDX1 and DLAT were classified into high and low groups according to their expression levels. Significant differences were found in the levels of infiltrating immune cells, such as neutrophils, mast cells, eosinophils, Treg, DC and cytotoxic cells (P < 0.05), while FDX1 was not significant different in B cells. But for DLAT, there were obvious differences in the levels of infiltrating immune cells, including eosinophils, neutrophils, T helper cells, cytotoxic cells, B cells, NK CD56bright cells and TReg (P < 0.05) (Fig. 6C). Finally, we found that high levels of mast cells and Treg were remarkably associated with survival in ccRCC patients with low FDX1 expression (Fig. 6E, 6G), and high levels of eosinophils, neutrophils and NK cells were significantly correlated with survival in patients with high DLAT expression.
Differentially co-expressed genes that correlated with FDX1 or DLAT and functional analysis in ccRCC patients. (A) Correlations between FDX1 and differently expressed genes (Pearson correlation analysis). (B, C) Heatmaps show the genes positively or negatively associated with FDX1 (Top 50 genes). (D) Correlations between DLAT and differently expressed genes (Pearson correlation analysis). (E, F) Heatmaps show the genes that are positively or negatively correlated with DLAT (Top 50 genes). (G) The venn diagram results show that 43 genes are positively correlated with both FDX1 and DLAT. (H) The venn diagram results suggest that 30 genes are negatively correlation with both FDX1 and DLAT. (I, J) GO analysis and KEGG enrichment of the 43 and 30 genes, respectively.
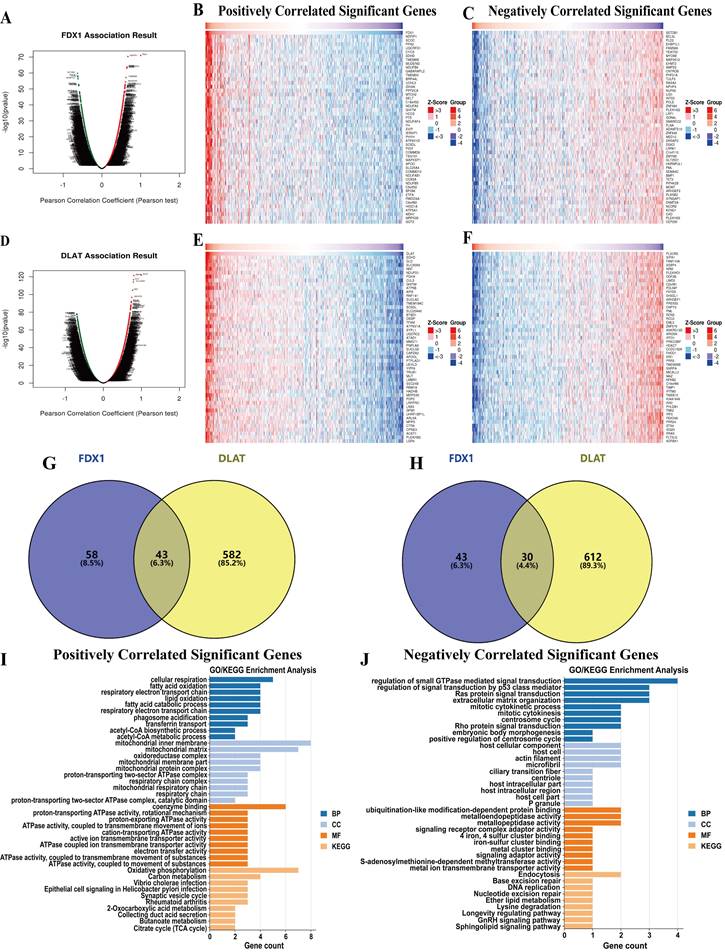
Methylation analysis of FDX1 and DLAT genes in ccRCC patients. (A, B) The methylation of FDX1 and DLAT was markedly correlated with OS and CpG islands in ccRCC patients. (C, D) The methylation levels of FDX1 and DLAT genes. The CpG methylation levels and forest plots of FDX1 (E, G) and DLAT (F, H).
Correlation analysis of FDX1 and DLAT expression with immune checkpoints
The correlation of FDX1 and DLAT genes with immunosuppressant checkpoints in pan-cancer were shown in Fig. 7A, 7B. Specially, FDX1 and DLAT genes were closely associated with EDNRB, CD274, HAVCR2, IL10, TGFB1, LAG3, PDCD1, IL4 and VTCN1 in ccRCC patients. Further studies revealed that FDX1 was positively correlated with CD274 (r = 0.309, P <0.001) and EDNRB (r = 0.421, P <0.001) (Fig. S3A, B) according to the following criteria: |r| > 0.3 and P <0.05. DLAT was also related to CD274 (r = 0.415, P <0.001) and EDNRB (r = 0.418, P <0.001) (Fig. S3C, D). Additionally, the correlations between immunostimulants and FDX1 and DLAT genes was investigated by Spearman correlation. The results showed that FDX1 and DLAT expression were markedly correlated with HMGB1, TLR4, CX3CL1, CD27, CCL5, CXCL10 and TNFRSF18 (Fig. 7C, 7D). We found that FDX1 expression was positively correlated with CX3CL1 (r = 0.395, P <0.001), TLR4 (r = 0.353, P < 0.001) and HMGB1 (r = 0.466, P <0.001), but negatively correlated with TNFRSP18 (r = -0.342, P <0.001) (Fig. S3E-H). As of interest, DLAT expression was also positively correlated with CX3CL1 (r = 0.331, P <0.001), TLR4 (r = 0.551, P < 0.001) and HMGB1 (r = 0.448, P <0.001), but negatively correlated with TNFRSP18 (r = -0.461, P <0.001) in patients with ccRCC (Fig. S3I-L). These results suggest that FDX1 and DLAT genes are highly relevant to immune checkpoints, which may lead to a favorable response from patients to immune checkpoint therapy.
Exploring the sensitivity of ccRCC to cuproptosis
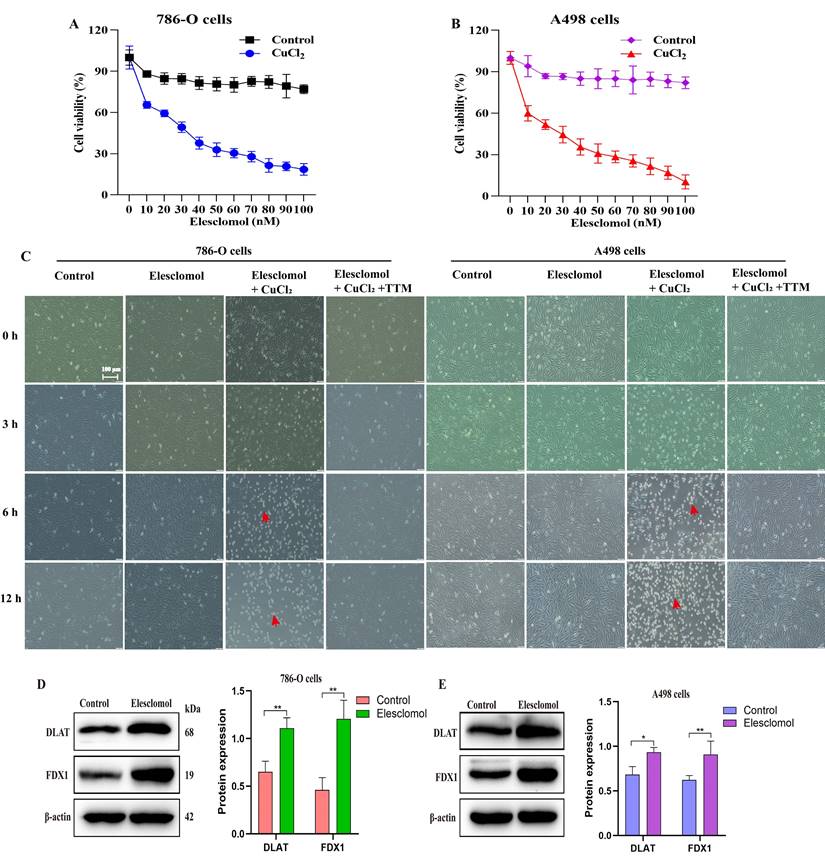
To explore the sensitivity of ccRCC to cuproptosis, we analyzed the cytotoxic effect of the cuproptosis inducer elesclomol in the presence of CuCl2 on 786-O and A498 cells. The results profiled that different elesclomol (0-100 nM) induced cell death in a concentration-dependent manner. Contrarily, cells grown in the absence of CuCl2 were resistant to elesclomol (Fig. 8A, 8B). The brightfield images of cell growth showed that co-treatment with elesclomol (100 nM) and CuCl2 (1 µM) resulted in the death of 786-O and A498 cells after 6 and 12 hours, while chelating copper with 20 µM tetra-thiomolybdate (TTM) prevented cell death (Fig. 8C). Of note, the expression of both FDX1 and DLAT proteins was up-regulated when 786-O and A498 cells were co-treated with 30 nM elesclomol and 1 µM CuCl2 (Fig. 8D, 8E).
Correlation analysis of FDX1 and DLAT with the levels of immune cell infiltration. (A, B) The association of FDX1 and DLAT with 24 types of immune cells. (C) Comparison of immune cells between high- and low-FDX1 and DLAT expression groups. (D-G) Effects of neutrophils, mast cells, eosinophils and Treg levels on survival in ccRCC patients with high and low FDX1 expression. (H-L) Effects of eosinophils, neutrophils, NK cells and Treg levels on survival of ccRCC patients with high and low DLAT expression.
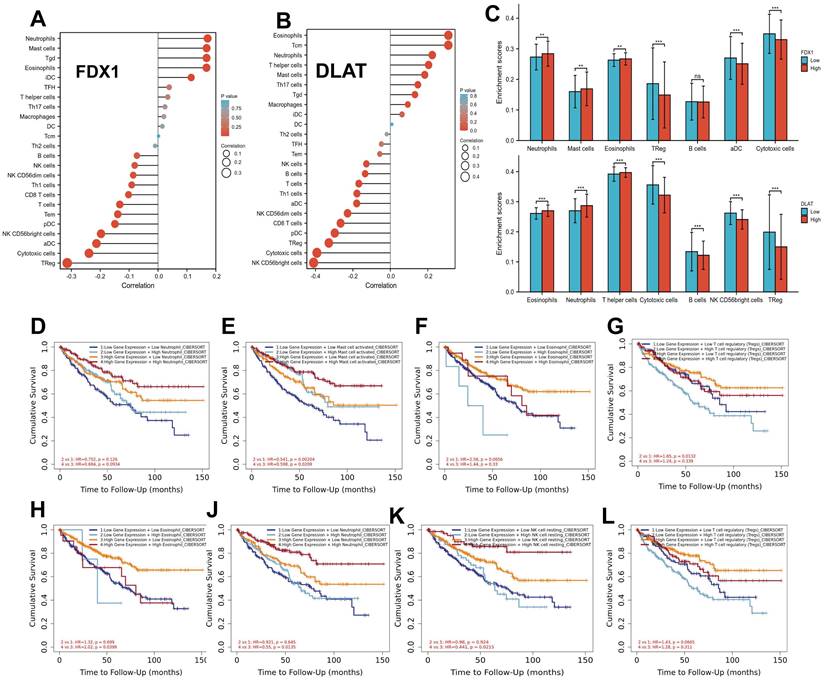
Correlation evaluation of immune checkpoint with the expression of FDX1 and DLAT. (A, B) The heatmaps displayed the correlation between immunoinhibitors with FDX1 and DLAT expression. (C, D) The heatmaps represented the correlation between immunostimulators with FDX1 and DLAT expression.
Screening and validation of potential drug targets for FDX1 and DLAT
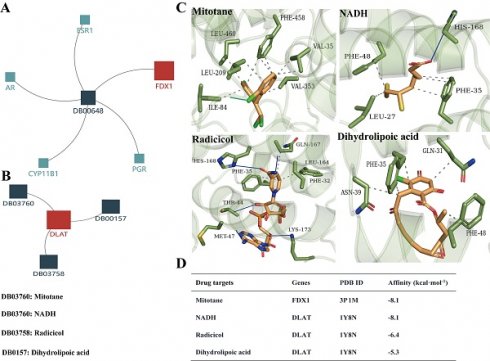
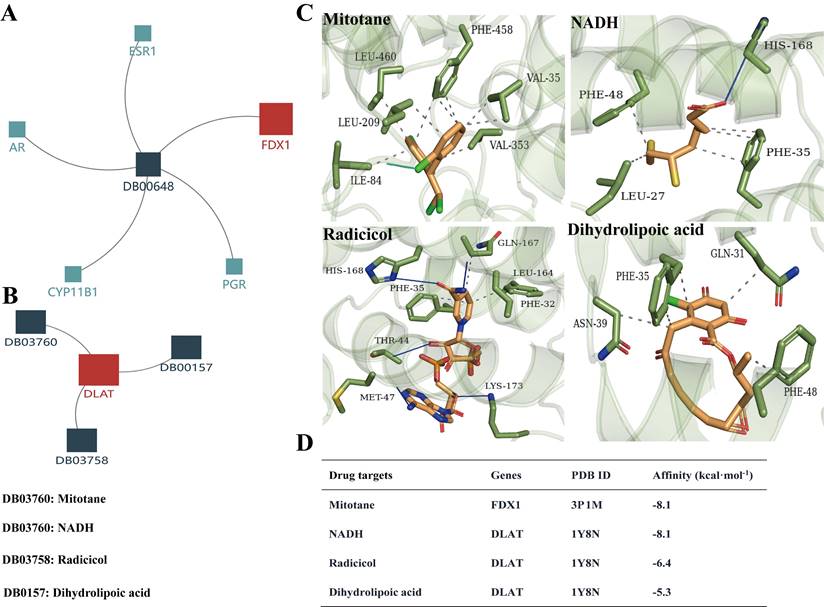
Based on the above analysis results showing that FDX1 and DLAT are promising targets for ccRCC therapy, we predicted the corresponding drug targets through the DrugBank database, a drug target information source. The data indicated that FDX1 was assigned only one agent named mitotane, whereas NADH, radicicol and dihydrolipoic acid are potential drug targets for DLAT (Fig. 9A, 9B). Molecular docking results revealed that Leu460/209, Phe458, Val35/353 and Ile84 of FDX1 were key sites for mitotane binding, its binding affinity up to -8.1 kcal. mol-1. NADH, radicicol and dihydrolipoic acid docked to several critical sites of DLAT, including Phe48/35/32, His168, Met47, Asn39, Gln31, etc., and the binding affinities were -8.1 kcal. mol-1, -6,4 kcal. mol-1 and -5,3 kcal. mol-1, respectively (Fig. 9C, 9D).
FDX1 and DLAT are pivotal targets of elesclomol-induced cuproptosis in ccRCC. (A, B) The cell viability of 786-O and A498 cells treated with elesclomol or elesclomol with CuCl2 (1 µM) for 12 h. (C) Representative brightfield images of 786-O and A498 cells incubated with either vehicle, 1µM CuCl2 or 1µM CuCl2 with 20 µM tetra-thiomolybdate (TTM), and then treated with 100 nM of elesclomol for 3, 6 and 12 h. (D, E) The expression of FDX1 and DLAT proteins in 786-O and A498 cells co-treated with elesclomol (30 nM) and CuCl2 (1 µM) for 12 h. Scale bar: 100 μm. *P < 0.05, **P < 0.01, ***p < 0.001 vs. normal or control. All experiments were repeated independently three times.
The drug targets of FDX1 and DLAT proteins. (A, B) Illustrations represent the drug targets of FDX1 and DLAT. (C) Three-dimensional illustrations displayed interactions of mitotane, NADH, adicicol and dihydrolipoic acid in the amino acid sites of FDX1 and DLAT proteins. (D) The affinity and related information of drug targets for FDX1 and DLAT. Blue solid line: hydrogen bonds; Grey dotted line: hydrophobic interaction. NADH: beta-Nicotinamide adenine dinucleotide.
Discussion
Copper is one of the most important trace elements in the human body, integral to various essential proteins, and involved in extremely relevant physiological and biochemical processes[23, 24]. The metabolic abnormalities of copper can cause related diseases, such as immune dysfunction, anaemia and endocrine disorders[25]. Intracellular copper overload leads to cell death, termed cuproptosis, and is considered a state-of-the-art strategy for the treatment of cancers[15]. However, few reports on the therapeutic strategies and mechanisms behind cuproptosis in a variety of tumors. Therefore, extensive works are required to further investigate the underlying mechanism and potential targets.
In the present study, we found that ccRCC was highly sensitive to cuproptosis and identified cuproptosis-related genes FDX1 and DLAT as the most prognostic and diagnostic candidates for ccRCC. FDX1, one of the ferredoxin families, contains soluble iron-sulfur (FeS) proteins[26]. Importantly, FDX1 encodes FeS protein that reduces Cu2+ to Cu1+ and is engaged in protein lipoylation in the TCA cycle, thereby regulating cuproptosis[15, 27]. DLAT is a mitochondrial protein that is an integral component of the pyruvate dehydrogenase complex (PDH)[28] and oligomerizes because the integration of copper and lipid acylated proteins in the TCA cycle plays an important role in cuproptosis[15]. Noteworthy, we found that the expressions of FDX1 and DLAT was lower in patients with ccRCC than in normal tissues. The high expression of both FDX1 and DLAT underscores the great clinical significance and diagnostic value, correlating with the favorable survival in ccRCC patients. These results highlight that they may serve as possible therapeutic targets against ccRCC. Emphatically, the relationship between FDX1 and DLAT exhibited a strong positive correlation. Previous evidence indicated that FDX1 leads to the accumulation of the toxic lipoylated DLAT, resulting cuproptosis[15, 29]. Protein lipoylation is a highly conserved post-translational modification of lysine observed on only four enzymes: dihydrolipoamide branched-chain transacylase E2 (DBT), glycine cleavage system protein H (GCSH), dihydrolipoamide S-succinyltransferase (DLST), and these enzymes participate in regulating the TCA cycle[30, 31]. The lipoylation of these proteins is essential for the proper function of these enzymes. Furthermore, FDX1 is an upstream regulator of protein lipoylation. This might explain the positive correlation between FDX1 and DLAT in ccRCC. Next, focusing on the functional analysis of the differentially expressed genes that are positively and negatively associated with both FDX1 and DLAT genes in ccRCC. It is revealed that these genes were mainly involved in the process of acetyl-CoA metabolism, acetyl-CoA biosynthesis, lipid oxidation, mitochondrial respiratory chain, acetyl-CoA metabolism and iron-sulfur cluster binding.
Gene mutations across tumor progression and treatment failure, such as drug resistance and immunotherapy resistance. The common tumor suppressor gene TP53 is frequently deleted or mutated in human cancers and is involved in the progression of drug resistance in carcinomas[32, 33]. Mutations in JAK1, JAK2 and STAT1 gene induce resistance to immunotherapy in patients with metastatic melanoma[34]. Most studies focus on the survival prognosis, clinical features, methylation, and biological functions of the cuprotosis-related gene FDX. However, there is little research on the significance of DLAT methylation and mutation in cuprotosis in ccRCC. Accumulating evidence suggests that FDX1 mutations result in permanent neonatal diabetes with subclinical exocrine deficiency[35], while DLAT mutations cause atypical Pantothenate Kinase Associated Neurodegeneration (PKAN)[36]. Our data indicated that the likelihood of mutations in FDX1 (with no mutations observed) and DLAT (0.3%) is very low in ccRCC. This reason could be that many important genes are not mutated or deleted, and aberrant gene expression is achieved through DNA methylation in cancer[37, 38]. The correlation between methylation and gene expression levels is complex and can be positive or negative, and the causality of this relationship remains unclear[39]. Our results revealed that the promoter methylation levels of FDX1 and DLAT were substantially reduced in ccRCC compared to normal tissues, suggesting that the changes in FDX1 and DLAT expression are due to this epigenetic modification. A natural follow-up issue is whether methylation of FDX1 and DLAT can be useful as prognostic predictors in patients with ccRCC. Earlier work has reported that the hereditary genetic polymorphisms of ATG5 serves as a prognostic predictor in patients with esophageal squamous cell carcinoma[40]. The study indicated the methylated CpG sites of FDX1 were beneficial for OS, and DLAT gene has only two methylated CpG sites beneficial to OS. The relationship between DNA methylation and gene expression largely depends on the genomic location of DNA methylation, which also influences the clinicopathological features of cancers[41, 42]. This may explain the different prognostic significance of FDX1 and DLAT methylation sites in ccRCC patients.
ccRCC is an immunogenic cancer typically infiltrated by immune cells, including macrophages and T lymphocytes[43]. In particular, T cells are the most abundant immune infiltrating cells in ccRCC[44]. Higher levels of tumor-infiltrating lymphocytes and CD8+T cells have been proven to be associated with poorer survival in ccRCC patients[45-47]. In this study, we found that FDX1 and DLAT were inversely correlated with the infiltration of most immune cells, particularly Tregs and CD8+ T cells. The expressions of FDX1 and DLAT are tightly associated with immune checkpoints, including PD-1, LAG3, PD-L1 and TLR4. Previous studies consistently reported that the expressions of LAG-3, PD-1, PD-L1, CD28, CD80, CD27 and CTLA-4 were relevant to the immunosuppression in the tumor microenvironment[48-50]. LAG3, CD28, CTLA4, PD-L1 have been identified as adverse prognostic factors in ccRCC patients[48, 51]. PD-1 is highly expressed in ccRCC tissues, which increases the infiltration of CD8+ T cells and Tregs, leading to immunosuppression within the immune microenvironment[52, 53]. Consistent with the above literature, our study explains that high levels of immune infiltration checkpoints lead to suppression of the tumor immune microenvironment. However, FDX1 and DLAT genes are highly correlated with immune checkpoints, which result in favorable patient responses to immune checkpoint therapy. More importantly, we confirmed the low expression of FDX1 and DLAT in clinical ccRCC samples and the sensitivity of ccRCC to cuproptosis in vitro. Furthermore, we analyzed the cytotoxic effects of cuproptosis inducer elesclomol in the presence of CuCl2 on ccRCC cells (786-O and A498) in the present study. The results showed that elesclomol reduced the cell viability in a concentration-dependent manner, whereas Cu chelator TTM prevented cell death of ccRCC. FDX1 as a potential target for mitotane and DLAT as a potential target for NADH, radiaticol, and dihydrolipoic acid were predicted by DrugBank database. Existing literatures have reported the antitumor activity of mitotane, adicicol and dihydrolipoic acid[54-56], but its molecular mechanisms still remains to be elucidated. The molecular docking results indicated that FDX1 and DLAT might be potential drug targets for mitotane, adicicol and dihydrolipoic acid against ccRCC by inducing cuproptosis. Despite these intriguing results, our study inevitably has some shortcomings. These results need to be further validated by biological experiments.
Conclusion
In the present study, we investigated the sensitivity of ccRCC to cuproptosis and applied comprehensive bioinformatics analysis to reveal the roles of cuproptosis-related genes FDX1 and DLAT in ccRCC. The findings demonstrated that both FDX1 and DALT were independent prognostic markers for poor survival in ccRCC patients, showing a strong positive correlation as well as the fact that they were correlated significantly with immune infiltration and immune checkpoints. Taken together, FDX1 and DLAT are promising targets for the prevention of ccRCC, and cuproptosis may be a novel and attractive strategy for therapy of ccRCC. The combination of targeting FDX1 and DLAT may be a novel insight into the induction of cuproptosis in ccRCC.
Abbreviations
ccRCC: clear cell renal cell carcinoma; CDGs: cuproptosis differential genes; OS: overall survival; PCD: programmed cell death; TCA: tricarboxylic acid; ROC: receiver operating characteristic; GO: gene ontology; KEGG: kyoto Encyclopedia of Genes and Genomes; FDX1: ferredoxin 1; DLAT: dihydrolipoyl transacetylase; ssGSEA: single-sample GSEA; TIMER: Tumor Immune Estimation Resource; SDS-PAGE: SDS-polyacrylamide gel electrophoresis; TCA cycle: the citrate cycle; FeS: iron-sulfur; PDH: pyruvate dehydrogenase complex; DBT: branched-chain transacylase E2; GCSH: glycine cleavage system protein H; DLST: dihydrolipoamide S-succinyltransferase.
Supplementary Material
Supplementary figures and table.
Acknowledgements
This work was supported by the National Natural Sciences Foundation of China (82474133, 82274159), the National Science Fund of Hunan Province (2022JJ80088), Key Project of Hunan Provincial Health Commission (202213055529), the Youth Project of Hunan Provincial Natural Science Foundation (2023JJ40485), the Scientific Research Project of Hunan Provincial Health Commission (R2023007), the Outstanding Youth Project of Educational Department of Hunan Province (23B0387, 22B0355), the Outstanding Youth Project of Hunan University of Chinese Medicine (2024XJZB002), National Natural Science Foundation Pre research Project of Hunan University of Traditional Chinese Medicine (2024XJYY08), Academician Liu Liang Workstation Guidance Project (24YS002), Open Fund Project of National Key Laboratory Cultivation Base of Chinese Medicinal Powder & Innovative Medicinal Jointly Established by Province and Ministry (23PTKF1016), Hunan Province College Students Innovation Training Program Project (S202410541062), the First-Class Discipline of Pharmaceutical Science of Hunan.
Author contributions
All the authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Hongfang Li, Chanjuan Zhang, Neng Zhu and Yaning Shi. The first draft of the manuscript was written by Hongfang Li. Li Qin conceived and supervised the research. All the authors have read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Teng R, Liu Z, Tang H, Zhang W, Chen Y, Xu R. et al. HSP60 silencing promotes Warburg-like phenotypes and switches the mitochondrial function from ATP production to biosynthesis in ccRCC cells. Redox biology. 2019;24:101218
2. He Y, Luo Y, Huang L, Zhang D, Wang X, Ji J. et al. New frontiers against sorafenib resistance in renal cell carcinoma: From molecular mechanisms to predictive biomarkers. Pharmacological research. 2021;170:105732
3. Hong Q, Wang S, Liu S, Chen X, Cai G. LRG1 May Accelerate the Progression of ccRCC via the TGF-β Pathway. BioMed research international. 2020;2020:1285068
4. Liu G, Ye Z, Zhao X, Ji Z. SP1-induced up-regulation of lncRNA SNHG14 as a ceRNA promotes migration and invasion of clear cell renal cell carcinoma by regulating N-WASP. American journal of cancer research. 2017;7(12):2515-25
5. Ni L, Yuan C, Zhang C, Xiang Y, Wu J, Wang X. et al. Co-Expression Network Analysis Identified LTF in Association with Metastasis Risk and Prognosis in Clear Cell Renal Cell Carcinoma. Onco targets and therapy. 2020;13:6975-86
6. Kesavardhana S, Malireddi RKS, Kanneganti TD. Caspases in Cell Death, Inflammation, and Pyroptosis. Annual review of immunology. 2020;38:567-95
7. Liu HZ, Shan TD, Han Y, Liu XS. Silencing long non-coding RNA CASC9 inhibits colorectal cancer cell proliferation by acting as a competing endogenous RNA of miR-576-5p to regulate AKT3. Cell death discovery. 2020;6(1):115
8. Yoshida M, Minagawa S, Araya J, Sakamoto T, Hara H, Tsubouchi K. et al. Involvement of cigarette smoke-induced epithelial cell ferroptosis in COPD pathogenesis. Nature Communications. 2019;10(1):3145
9. Zeh HJ, Bahary N, Boone BA, Singhi AD, Miller-Ocuin JL, Normolle DP. et al. A Randomized Phase II Preoperative Study of Autophagy Inhibition with High-Dose Hydroxychloroquine and Gemcitabine/Nab-Paclitaxel in Pancreatic Cancer Patients. Clinical cancer research. 2020;26(13):3126-34
10. Marsh T, Kenific CM, Suresh D, Gonzalez H, Shamir ER, Mei W. et al. Autophagic Degradation of NBR1 Restricts Metastatic Outgrowth during Mammary Tumor Progression. Development cell. 2020;52(5):591-604.e596
11. Iniguez AB, Alexe G, Wang EJ, Roti G, Patel S, Chen L. et al. Resistance to Epigenetic-Targeted Therapy Engenders Tumor Cell Vulnerabilities Associated with Enhancer Remodeling. Cancer cell. 2018;34(6):922-38.e927
12. Li M, Rui Y, Peng W, Hu J, Jiang A, Yang Z. et al. FIGNL1 promotes non-small cell lung cancer cell proliferation. International journal of oncology research. 2021;58(1):83-99
13. Giuliano S, Dufies M, Ndiaye PD, Viotti J, Borchiellini D, Parola J. et al. Resistance to lysosomotropic drugs used to treat kidney and breast cancers involves autophagy and inflammation and converges in inducing CXCL5. Theranostics. 2019;9(4):1181-99
14. Lv H, Liu X, Zeng X, Liu Y, Zhang C, Zhang Q. et al. Comprehensive Analysis of Cuproptosis-Related Genes in Immune Infiltration and Prognosis in Melanoma. Frontiers in pharmacology. 2022;13:930041
15. Tsvetkov P, Coy S, Petrova B, Dreishpoon M, Verma A, Abdusamad M. et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science. 2022;375(6586):1254-61
16. Hakimi AA, Reznik E, Lee CH, Creighton CJ, Brannon AR, Luna A. et al. An Integrated Metabolic Atlas of Clear Cell Renal Cell Carcinoma. Cancer cell. 2016;29(1):104-116
17. Wettersten HI, Aboud OA, Lara PN Jr. and Weiss RH. Metabolic reprogramming in clear cell renal cell carcinoma. Nature reviews nephrology. 2017;13(7):410-9
18. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43-9
19. Bian Z, Fan R, Xie L. A Novel Cuproptosis-Related Prognostic Gene Signature and Validation of Differential Expression in Clear Cell Renal Cell Carcinoma. Genes (Basel). 2022;13(5):851
20. Koch A, Jeschke J, Van Criekinge W, van Engeland M, De Meyer T. MEXPRESS update 2019. Nucleic acids research. 2019;47(W1):W561-5
21. Chaikuad A, Johansson C, Krojer T, Yue W, Phillips C, Bray J. et al. Crystal structure of human ferredoxin-1 (FDX1) in complex with iron-sulfur cluster. Protein Data Bank 3P1M Released. 2010:11-03
22. Kato M, Chuang JL, Tso SC, Wynn RM, Chuang DT. Crystal structure of pyruvate dehydrogenase kinase 3 bound to lipoyl domain 2 of human pyruvate dehydrogenase complex. EMBO journal. 2005;24(10):1763-74
23. Kong C, Zhao X, Li Y, Yang S, Chen YM, Yang Z. Ion-Induced Synthesis of Alginate Fibroid Hydrogel for Heavy Metal Ions Removal. Frontiers in chemistry. 2019;7:905
24. Waters BM, Chu HH, Didonato RJ, Roberts LA, Eisley RB, Lahner B. et al. Mutations in Arabidopsis yellow stripe-like1 and yellow stripe-like3 reveal their roles in metal ion homeostasis and loading of metal ions in seeds. Plant Physiology. 2006;141(4):1446-58
25. Huang XY, Deng F, Yamaji N, Pinson SR, Fujii-Kashino M, Danku J. et al. A heavy metal P-type ATPase OsHMA4 prevents copper accumulation in rice grain. Nature communications. 2016;7:12138
26. Sawyer A, Winkler M. Evolution of Chlamydomonas reinhardtii ferredoxins and their interactions with [FeFe]-hydrogenases. Photosynthesis research. 2017;134(3):307-16
27. Dörsam B, Fahrer J. The disulfide compound α-lipoic acid and its derivatives: A novel class of anticancer agents targeting mitochondria. Cancer letters. 2016;371(1):12-9
28. Goh WQ, Ow GS, Kuznetsov VA, Chong S, Lim YP. DLAT subunit of the pyruvate dehydrogenase complex is upregulated in gastric cancer-implications in cancer therapy. American journal of translational research. 2015;7(6):1140-51
29. Chen Y, Tang L, Huang W, Zhang Y, Abisola FH, Li L. Identification and validation of a novel cuproptosis-related signature as a prognostic model for lung adenocarcinoma. Frontiers in endocrinology. 2022;13:963220
30. Solmonson A, DeBerardinis RJ. Lipoic acid metabolism and mitochondrial redox regulation. Journal of biological chemistry. 2018;293(20):7522-30
31. Rowland EA, Snowden CK, Cristea IM. Protein lipoylation: an evolutionarily conserved metabolic regulator of health and disease. Current opinion in chemical biology. 2018;42:76-85
32. Van Roosbroeck K, Fanini F, Setoyama T, Ivan C, Rodriguez-Aguayo C, Fuentes-Mattei E. et al. Combining Anti-Mir-155 with Chemotherapy for the Treatment of Lung Cancers. Clinical cancer research. 2017;23(11):2891-904
33. Shen S, Faouzi S, Bastide A, Martineau S, Malka-Mahieu H, Fu Y. et al. An epitranscriptomic mechanism underlies selective mRNA translation remodelling in melanoma persister cells. Nature communications. 2019;10(1):5713
34. Liu XD, Kong W, Peterson CB, McGrail DJ, Hoang A, Zhang X. et al. PBRM1 loss defines a nonimmunogenic tumor phenotype associated with checkpoint inhibitor resistance in renal carcinoma. Nature communications. 2020;11(1):2135
35. Nicolino M, Claiborn KC, Senée V, Boland A, Stoffers DA, Julier C. A novel hypomorphic PDX1 mutation responsible for permanent neonatal diabetes with subclinical exocrine deficiency. Diabetes. 2010;59(3):733-40
36. McWilliam CA, Ridout CK, Brown RM, McWilliam RC, Tolmie J, Brown GK. Pyruvate dehydrogenase E2 deficiency: a potentially treatable cause of episodic dystonia. European journal of paediatric neurology. 2010;14(4):349-53
37. Kumari A, Iwasaki T, Pyndiah S, Cassimere EK, Palani CD, Sakamuro D. Regulation of E2F1-induced apoptosis by poly(ADP-ribosyl)ation. Cell death differentiation. 2015;22(2):311-22
38. Xiao J, Liu Y, Wu F, Liu R, Xie Y, Yang Q. et al. miR-639 Expression Is Silenced by DNMT3A-Mediated Hypermethylation and Functions as a Tumor Suppressor in Liver Cancer Cells. Molecular therapy. 2020;28(2):587-98
39. Husquin LT, Rotival M, Fagny M, Quach H, Zidane N, McEwen LM. et al. Exploring the genetic basis of human population differences in DNA methylation and their causal impact on immune gene regulation. Genome biology. 2018;19(1):222
40. Yang PW, Hsieh MS, Chang YH, Huang PM, Lee JM. Genetic polymorphisms of ATG5 predict survival and recurrence in patients with early-stage esophageal squamous cell carcinoma. Oncotarget. 2017;8(53):91494-504
41. Fang F, Wang X, Song T. Five-CpG-based prognostic signature for predicting survival in hepatocellular carcinoma patients. Cancer biology & medicine. 2018;15(4):425-33
42. Maruyama R, Toyooka S, Toyooka KO, Harada K, Virmani AK, Zöchbauer-Müller S. et al. Aberrant promoter methylation profile of bladder cancer and its relationship to clinicopathological features. Cancer research. 2001;61(24):8659-63
43. Liu XD, Hoang A, Zhou L, Kalra S, Yetil A, Sun M. et al. Resistance to Antiangiogenic Therapy Is Associated with an Immunosuppressive Tumor Microenvironment in Metastatic Renal Cell Carcinoma. Cancer immunology research. 2015;3(9):1017-29
44. Şenbabaoğlu Y, Gejman RS, Winer AG, Liu M, Van Allen EM, de Velasco G. et al. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome biology. 2016;17(1):231
45. Nakano O, Sato M, Naito Y, Suzuki K, Orikasa S, Aizawa M. et al. Proliferative activity of intratumoral CD8(+) T-lymphocytes as a prognostic factor in human renal cell carcinoma: clinicopathologic demonstration of antitumor immunity. Cancer research. 2001;61(13):5132-6
46. Patel HD, Puligandla M, Shuch BM, Leibovich BC, Kapoor A, Master VA. et al. The future of perioperative therapy in advanced renal cell carcinoma: how can we PROSPER? Future oncology. 2019;15(15):1683-95
47. Braun DA, Hou Y, Bakouny Z, Ficial M, Sant' Angelo M, Forman J. et al. Interplay of somatic alterations and immune infiltration modulates response to PD-1 blockade in advanced clear cell renal cell carcinoma. Nature Medicine. 2020;26(6):909-18
48. Wang Q, Zhang J, Tu H, Liang D, Chang DW, Ye Y. et al. Soluble immune checkpoint-related proteins as predictors of tumor recurrence, survival, and T cell phenotypes in clear cell renal cell carcinoma patients. Journal for immunotherapy of cancer. 2019;7(1):334
49. Gao Y, Nihira NT, Bu X, Chu C, Zhang J, Kolodziejczyk A. et al. Acetylation-dependent regulation of PD-L1 nuclear translocation dictates the efficacy of anti-PD-1 immunotherapy. Nature cell biology. 2020;22(9):1064-75
50. Baumann D, Hägele T, Mochayedi J, Drebant J, Vent C, Blobner S. et al. Proimmunogenic impact of MEK inhibition synergizes with agonist anti-CD40 immunostimulatory antibodies in tumor therapy. Nat communications. 2020;11(1):2176
51. Montemagno C, Hagege A, Borchiellini D, Thamphya B, Rastoin O, Ambrosetti D. et al. Soluble forms of PD-L1 and PD-1 as prognostic and predictive markers of sunitinib efficacy in patients with metastatic clear cell renal cell carcinoma. Oncoimmunology. 2020;9(1):1846901
52. Liu S, Wang F, Tan W, Zhang L, Dai F, Wang Y. et al. CTLA4 has a profound impact on the landscape of tumor-infiltrating lymphocytes with a high prognosis value in clear cell renal cell carcinoma (ccRCC). Cancer cell international. 2020;20:519
53. Herrmann A, Priceman SJ, Swiderski P, Kujawski M, Xin H, Cherryholmes GA. et al. CTLA4 aptamer delivers STAT3 siRNA to tumor-associated and malignant T cells. Journal of clinical investigation. 2014;124(7):2977-87
54. Li X, Kim W, Juszczak K, Arif M, Sato Y, Kume H. et al. Stratification of patients with clear cell renal cell carcinoma to facilitate drug repositioning. iScience. 2021;24(7):102722
55. Ho YS, Lai CS, Liu HI, Ho SY, Tai C, Pan MH. et al. Dihydrolipoic acid inhibits skin tumor promotion through anti-inflammation and anti-oxidation. Biochemical pharmacology. 2007;73(11):1786-95
56. Kim YJ, Lee SA, Myung SC, Kim W, Lee CS. Radicicol, an inhibitor of Hsp90, enhances TRAIL-induced apoptosis in human epithelial ovarian carcinoma cells by promoting activation of apoptosis-related proteins. Molecular and cellular biochemistry. 2012;359(1-2):33-43
Author contact
![]() Corresponding author: Li Qin, E-mail: lqinedu.cn; ORCID: 0000-0002-9797-2636.
Corresponding author: Li Qin, E-mail: lqinedu.cn; ORCID: 0000-0002-9797-2636.

 Global reach, higher impact
Global reach, higher impact