Impact Factor
ISSN: 1837-9664
J Cancer 2025; 16(5):1747-1753. doi:10.7150/jca.106426 This issue Cite
Research Paper
The Impact of MET Variants in Oral Cancer Progression and Clinicopathological Characteristics
1. Department of Dentistry, Changhua Christian Hospital, Changhua, Taiwan.
2. Department of Post-Baccalaureate Medicine, College of Medicine, National Chung Hsing University, Taichung, Taiwan.
3. Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan.
4. School of Medicine, Chung Shan Medical University, Taichung, Taiwan.
5. Department of Otolaryngology, Chung Shan Medical University Hospital, Taichung, Taiwan.
6. Department of Otolaryngology, St. Martin De Porres Hospital, Chiayi, Taiwan.
7. Department of Medical Research, Chung Shan Medical University Hospital, Taichung, Taiwan.
8. Department of Mathematics and Statistics, Florida Atlantic University, Boca Raton, Florida, USA.
9. Institute of Oral Sciences, Chung Shan Medical University, Taichung, Taiwan.
10. Department of Dentistry, Chung Shan Medical University Hospital, Taichung, Taiwan.
Received 2024-11-5; Accepted 2025-1-26; Published 2025-2-11
Abstract
Epigenetic, genetic predisposition and epidemiological risk factors were suggested to be involved in the carcinogenesis of oral cancer. In this study, we focused on the associations of MET single-nucleotide polymorphisms (SNPs) to oral cancer susceptibility and clinicopathological characteristics. The MET SNPs rs41736, rs41739, rs1621, and rs33917957 in 1198 controls and 1318 male patients with oral cancer were analyzed with real-time polymerase chain reaction. Our results revealed that the cigarette smokers among the oral cancer patients who carried the MET rs1621 polymorphic variant “G” were significantly associated with lower risk to develop oral cancer [OR (95% CI) = 0.463 (0.226-0.948)]. The male oral cancer patients who with the genotypic variant “G” of MET rs33917957 were associated with lower risk of cell differentiated grade (p = 0.041). In the TCGA database, the MET expressions were upregulated in oral cancer tissues compared to normal tissues, and were correlated with poor cell differentiated and poorer prognoses in smoker groups. In conclusion, these novel findings underscore the role of MET genetic variants in oral cancer susceptibility, particularly in smokers, and highlight the potential of these variants for prognosis and disease prediction.
Keywords: oral cancer, MET, polymorphism, susceptibility, SNP, prognosis
Introduction
Oral cancer is the sixth leading cause of cancer mortality worldwide [1]. About 90% of the oral cancer was originated from squamous cells, which is classified as oral squamous cell carcinoma (OSCC) [2, 3]. Risk factors such as cigarette smoking, alcohol consumption, and betel nut chewing were suggested as major contributors to oral cancer [4-6]. In Taiwan, the prevalence of these carcinogenic substances use in males was larger than in females, and the incidence rate of oral cancer was obviously higher in men than in women [7, 8].
MET is a proto-oncogene located on chromosome 7q31.2 which encodes the cellular-mesenchymal epithelial transition factor (c-Met) transmembrane tyrosine kinase receptor for hepatocyte growth factor (HGF) [9-11]. The c-Met was suggested to play a crucial role in regulating cell proliferation, differentiation, metastasis, and apoptosis through various signaling pathways, and its aberrant expression has been implicated in many human cancers [12-14]. In oral cancer, it was suggested that the MET activation may represent an early driver in oral premalignancy and may be target for chemoprevention of oral cancer, and the c-met was identified as a potential prognostic marker of cancer risk in patients with oral leukoplakia [12, 15-17]. In gastric cancer, it was suggested that the MET amplification is often accompanied by human epidermal growth factor receptor 2 (HER2) overexpression, and co-expression of MET and HER2 can synergistically enhance tumor invasion, and metastasis, which is a crucial factor for poor prognosis [18].
Previous studies have associated the overexpression of MET with cancer progression and prognosis including oral cancer [15, 17, 19], and the MET polymorphisms were suggested to be associated with cancer development and prognosis in various cancers such as small cell lung cancer [20], gastric cancer [21, 22], hepatocellular carcinoma (HCC) [23], and papillary thyroid carcinoma [23]. However, the associations and influences of MET polymorphisms to oral cancer tumor progression and clinicopathologic characteristics remained unclear. In the current study, we focused on four SNPs of MET rs41736, rs41739, rs1621, and rs33917957, and try to elucidate the associations of MET SNPs to oral cancer susceptibility and clinicopathologic characteristics with environmental risk factors.
Materials and Methods
Study subjects
A total of 1318 male patients with oral cancer and 1198 cancer-free controls were enrolled in our study. These participants who enrolled in our study were recruited during 2016 to 2021 at Chung Shan Medical University Hospital in Taichung, Taiwan. For the TNM staging of the oral cancer patients who enrolled in our study, the TNM staging were clinically staged at the time of diagnosis according to the American Joint Committee on Cancer (AJCC) [24]. The rating of tumor differentiation was examined by pathologist according to the AJCC classification. This project was approved by the institutional review board of Chung Shan Medical University Hospital (IRB number: CS1-21151).
Sample preparation and DNA extraction
For genomic DNA extraction, we collected the peripheral blood specimens from oral cancer patients and normal controls who participated in our study [25]. All the samples of peripheral whole blood were preserved with EDTA containing tubes. Each sample of whole blood were centrifuged under the settings of 3000 rpm, 10 minutes. The buffy coats were collected from centrifuged whole blood specimens and further used for DNA extraction [26]. Following the manufacturer's manual of QIAamp DNA blood mini kits, the genomic DNA extraction assay was performed to acquire the DNA. The Tris-EDTA (TE) buffer was applied to complete the final step of DNA elution. The extracted DNA was used as the DNA template in real-time polymerase chain reactions (PCRs).
MET SNPs genotyping
Assessment of allelic discrimination for the MET rs41736, rs41739, rs1621, and rs33917957 SNP was performed with an ABI StepOne Software v2.3 Real-Time PCR System. The TaqMan assay was adopted for the analysis of genotyping. The final data of genotyping was analyzed and calculated with the SDS 7000 series software (Applied Biosystems, Foster City, CA, USA).
Statistical analysis
The student's t test or Chi-squared test was performed between the patients with oral cancer and the controls to compare the age (years), betel quid chewing, cigarette smoking, alcohol drinking, tumor stage, tumor T status, lymph node status, metastasis, and cell differentiation. A p < 0.05 was suggested to present statistically significant. To compare the associations of the odds ratio (OR) with their 95% confidence intervals (CIs), and the clinical pathological statuses between the oral cancer risk and genotypic frequencies, the data was analyzed and assessed with multiple logistic regression models. All of the data in our study was analyzed and evaluated with SAS statistical software (Version 9.1, 2005; SAS Institute, Cary, NC).
Results
The distribution of demographical characteristics in 1198 controls and 1318 male patients with oral cancer was listed in Table 1. In the current study, we observed that the distributions of age (years) < 55 was 565 (47.2%) in controls and 619 (47.0%) in oral cancer patients, and the age ≧ 55 in controls and oral cancer patients was 633 (52.8%) and 699 (53.0%), respectively. The distributions of environmental risk factors exposure between the controls and oral cancer patients were 199 (16.6%) and 985 (74.7%) in betel quid chewing (p < 0.001), 636 (53.1%) and 1110 (84.2%) in cigarette smoking (p < 0.001), and 237 (19.8%) and 626 (47.5%) in alcohol drinking (p < 0.001), respectively.
The genotype distributions of MET gene polymorphisms in 1198 controls and 1318 male patients with oral cancer were listed in Table 2. The highest distribution frequencies in oral cancer patients of MET genetic polymorphisms rs41736, rs41739, rs1621, and rs33917957 were polymorphic variant C, polymorphic variant A, polymorphic variant A, and polymorphic variant A, respectively. The logistic regression models were applied to estimate the odds ratios (ORs) with their 95% confidence intervals (CIs). Our findings revealed no significant association between MET SNPs and the incidence of oral cancer (Table 2). Additionally, we applied propensity score matching (PSM), a statistical technique, to evaluate the intrinsic impact of MET variants on oral cancer progression. After matching for age, betel quid chewing, cigarette smoking, and alcohol consumption, no significant associations were observed between MET variants and oral cancer incidence (Table 3).
The distributions of demographical characteristics in 1198 controls and 1318 patients with oral cancer.
| Variable | Controls (N=1198) | Patients (N=1318) | p value |
|---|---|---|---|
| Age (yrs) | p=0.921 | ||
| <55 | 565 (47.2%) | 619 (47.0%) | |
| ≥55 | 633 (52.8%) | 699 (53.0%) | |
| Betel quid chewing | p < 0.001* | ||
| No | 999 (83.4%) | 333 (25.3%) | |
| Yes | 199 (16.6%) | 985 (74.7%) | |
| Cigarette smoking | p < 0.001* | ||
| No | 562 (46.9%) | 208 (15.8%) | |
| Yes | 636 (53.1%) | 1110 (84.2%) | |
| Alcohol drinking | p< 0.001* | ||
| No | 961 (80.2%) | 692 (52.5%) | |
| Yes | 237 (19.8%) | 626 (47.5%) | |
| Stage | |||
| I+II | 618 (46.9%) | ||
| III+IV | 700 (53.1%) | ||
| Tumor T status | |||
| T1+T2 | 662 (50.2%) | ||
| T3+T4 | 656 (49.8%) | ||
| Lymph node status | |||
| N0 | 866 (65.7%) | ||
| N1+N2+N3 | 452 (34.3%) | ||
| Metastasis | |||
| M0 | 1308 (99.2%) | ||
| M1 | 10 (0.8%) | ||
| Cell differentiation | |||
| Well differentiated | 183 (13.9%) | ||
| Moderately or poorly differentiated | 1135 (86.1%) |
Mann-Whitney U test or Chi-square test was used between healthy controls and patients with oral cancer. * p value < 0.05 as statistically significant.
We further analyzed the ORs and 95% CIs of oral cancer patients associated with MET genotyping and allele frequency among cigarette smokers. A significant association was found in those individuals who carried the MET rs1621 polymorphic GG genotype, with a lower risk of oral cancer susceptibility [The odds ratio (OR) (95% CI):0.463 (0.226-0.948); p = 0.035] (Table 4). Moreover, after we analyzed the odds ratio (OR) and 95% CI of clinical statuses associated with genotypic frequencies of MET rs33917957 in male oral cancer patients, we found that in 1110 cigarette smokers among the total 1318 male oral cancer patients, carriers who with the rs33917957 polymorphic “G” variant have a lower risk to develop poorer cell differentiated grade (p = 0.041) (Table 5).
Genotyping and allele frequency of MET single nucleotide polymorphism (SNP) in oral cancer and normal controls.
| Variable | Controls (N=1198) n (%) | Patients (N=1318) n (%) | OR (95% CI) |
|---|---|---|---|
| rs41736 | |||
| CC | 371 (31.0%) | 406 (30.8%) | 1.000 (reference) |
| CT | 584 (48.8%) | 658 (49.9%) | 1.030 (0.860-1.232) |
| TT | 243 (20.2%) | 254 (19.3%) | 0.955 (0.762-1.196) |
| CT+TT | 827 (69.0%) | 912 (69.2%) | 1.008 (0.851-1.194) |
| rs41739 | |||
| AA | 369 (30.8%) | 405 (30.7%) | 1.000 (reference) |
| AG | 584 (48.8%) | 657 (49.9%) | 1.025 (0.856-1.227) |
| GG | 245 (20.4%) | 256 (19.4%) | 0.952 (0.760-1.192) |
| AG+GG | 829 (69.2%) | 913 (69.3%) | 1.003 (0.847-1.189) |
| rs1621 | |||
| AA | 902 (75.3%) | 996 (75.6%) | 1.000 (reference) |
| AG | 273 (22.8%) | 305 (23.1%) | 1.012 (0.840-1.219) |
| GG | 23 (1.9%) | 17 (1.3%) | 0.669 (0.355-1.261) |
| AG+GG | 296 (24.7%) | 322 (24.4%) | 0.985 (0.821-1.181) |
| rs33917957 | |||
| AA | 1036 (86.5%) | 1139 (86.4%) | 1.000 (reference) |
| AG | 158 (13.2%) | 171 (13.0%) | 0.984 (0.781-1.242) |
| GG | 4 (0.3%) | 8 (0.6%) | 1.819 (0.546-6.058) |
| AG+GG | 162 (13.5%) | 179 (13.6%) | 1.005 (0.800-1.263) |
The ORs with analyzed by their 95% CIs were estimated by logistic regression models.
Genotyping and allele frequency of MET single nucleotide polymorphism (SNP) in oral cancer and normal controls after propensity score matchinga.
| Variable | Controls (N=530) n (%) | Patients (N=530) n (%) | OR (95% CI)b |
|---|---|---|---|
| rs41736 | |||
| CC | 173 (32.6%) | 174 (32.8%) | 1.000 (reference) |
| CT | 251 (47.4%) | 265 (50.0%) | 1.050 (0.800-1.378) |
| TT | 106 (20.0%) | 91 (17.2%) | 0.854 (0.601-1.212) |
| CT+TT | 357 (67.4%) | 356 (67.2%) | 0.991 (0.767-1.281) |
| rs41739 | |||
| AA | 172 (32.5%) | 174 (32.8%) | 1.000 (reference) |
| AG | 251 (47.4%) | 264 (49.8%) | 1.040 (0.792-1.365) |
| GG | 107 (20.1%) | 92 (17.4%) | 0.850 (0.599-1.205) |
| AG+GG | 358 (67.5%) | 356 (67.2%) | 0.983 (0.760-1.271) |
| rs1621 | |||
| AA | 402 (75.8%) | 385 (72.6%) | 1.000 (reference) |
| AG | 116 (21.9%) | 137 (25.8%) | 1.233 (0.928-1.638) |
| GG | 12 (2.3%) | 8 (1.6%) | 0.696 (0.281-1.721) |
| AG+GG | 128 (24.2%) | 145 (27.4%) | 1.183 (0.898-1.558) |
| rs33917957 | |||
| AA | 457 (86.2%) | 455 (85.8%) | 1.000 (reference) |
| AG | 72 (13.6%) | 75 (14.2%) | 1.046 (0.738-1.482) |
| GG | 1 (0.2%) | 0 (0.0%) | --- |
| AG+GG | 73 (13.8%) | 75 (14.2%) | 1.032 (0.729-1.461) |
a Propensity score matching for age, betel quid chewing, cigarette smoking, and alcohol consumption,
b The ORs with analyzed by their 95% CIs were estimated by logistic regression models.
Genotyping and allele frequency of MET single nucleotide polymorphism (SNP) in oral cancer among cigarette smokers.
| Variable | Controls (N=636) n (%) | Patients (N=1110) n (%) | OR (95% CI) |
|---|---|---|---|
| rs41736 | |||
| CC | 208 (32.7%) | 336 (30.3%) | 1.000 (reference) |
| CT | 298 (46.9%) | 558 (50.3%) | 1.159 (0.928-1.449) |
| TT | 130 (20.4%) | 216 (19.4%) | 1.029 (0.779-1.358) |
| CT+TT | 428 (67.3%) | 774 (69.7%) | 1.119 (0.908-1.380) |
| rs41739 | |||
| AA | 207 (32.5%) | 335 (30.2%) | 1.000 (reference) |
| AG | 298 (46.9%) | 557 (50.2%) | 1.155 (0.924-1.444) |
| GG | 131 (20.6%) | 218 (19.6%) | 1.028 (0.779-1.357) |
| AG+GG | 429 (67.5%) | 775 (69.8%) | 1.116 (0.905-1.377) |
| rs1621 | |||
| AA | 476 (74.8%) | 846 (76.2%) | 1.000 (reference) |
| AG | 143 (22.5%) | 250 (22.5%) | 0.984 (0.778-1.243) |
| GG | 17 (2.7%) | 14 (1.3%) | 0.463 (0.226-0.948)a |
| AG+GG | 160 (25.2%) | 264 (23.8%) | 0.928 (0.740-1.164) |
| rs33917957 | |||
| AA | 552 (86.8%) | 962 (86.7%) | 1.000 (reference) |
| AG | 81 (12.7%) | 140 (12.6%) | 0.992 (0.740-1.329) |
| GG | 3 (0.5%) | 8 (0.7%) | 1.530 (0.404-5.790) |
| AG+GG | 84 (13.2%) | 148 (13.3%) | 1.011 (0.758-1.348) |
The ORs with analyzed by their 95% CIs were estimated by logistic regression models.
ap = 0.035.
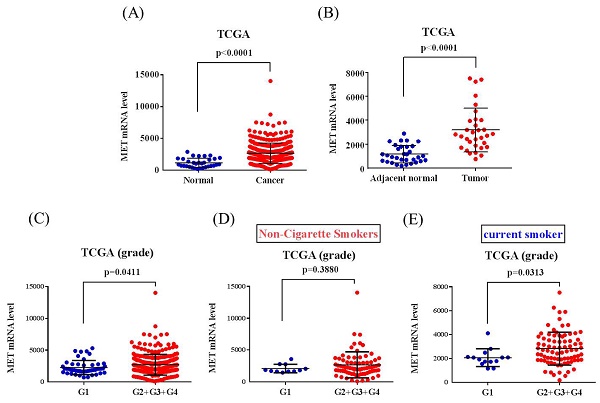
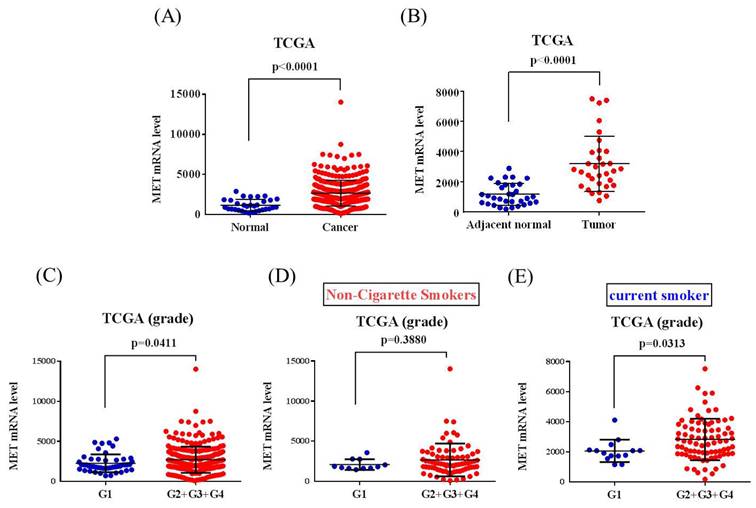
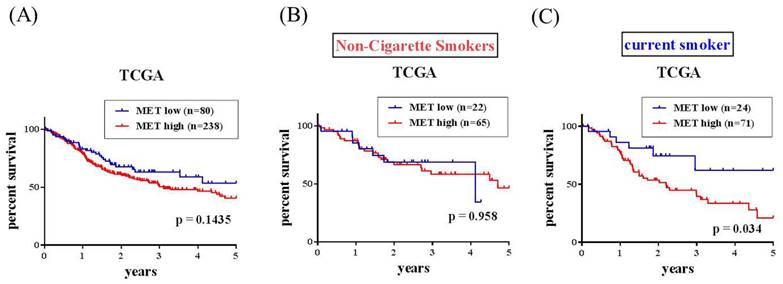
The correlations of MET expression levels with clinical significance and survival rates in head and neck squamous cell carcinoma (HNSCC) patients were further analyzed from the TCGA dataset. We observed that MET expression was prone to be upregulated in HNSCC carcinoma tissue compared with normal tissues (Figure 1A-1B). Furthermore, patients with moderately differentiated (G2) and poorly differentiated (G3) showed significantly higher MET expression in tumors compared to patients at well-differentiated (G1) both in all the HNSCC patients (p=0.0411) and the smoker group (p=0.0313) (Figures 1C-E). Most importantly, HNSCC patients who had METhigh tumors had shorter disease-specific survival times compared with those who had METlow tumors in the smoker group (p=0.034) (Figures 2A-2C). Taken together, the above clinical data indicated that upregulation of MET is a critical event in promoting HNSCC progression.
Discussion
In this study, we demonstrated the associations between the MET SNPs and oral cancer. Alcohol consumption, betel quid chewing, and cigarette smoking are the three major and well-known risk factors for head and neck cancer [27-30]. In Taiwan, most of the oral cancer patients were male who with the habits of cigarette smoking and/or betel nut chewing, and over 90% of oral cancer was OSCC [31-33]. Consistent with these results, in our current study, statistically significant associations of these risk factors including betel quid chewing, cigarette smoking, and alcohol drinking were found between the 1198 controls and 1318 male patients with oral cancer, respectively (p < 0.001, table 1). We further analyzed the associations of genotyping and allele frequency of MET SNP in oral cancer patients and normal controls.
Odds ratio (OR) and 95% confidence intervals (CI) of clinical statuses associated with genotypic frequencies of MET rs33917957in male oral cancer patients.
| Total (N=1318) | Cigarette Smokers (N=1110) | Non-Cigarette Smokers (N=208) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Variable | AA (N=1139) | AG+GG (N=179) | p value | AA (N=962) | AG+GG (N=148) | p value | AA (N=177) | AG+GG (N=31) | p value |
| Clinical Stage | |||||||||
| Stage I+II | 527 (46.3%) | 91 (50.8%) | 0.255 | 446 (46.4%) | 75 (50.7%) | 0.327 | 81 (45.8%) | 16 (51.6%) | 0.548 |
| StageIII+IV | 612 (53.7%) | 88 (49.2%) | 516 (53.6%) | 73 (49.3%) | 96 (54.2%) | 15 (48.4%) | |||
| Tumor size | |||||||||
| ≦ T2 | 576 (50.6%) | 86 (48.0%) | 0.530 | 502 (52.2%) | 72 (48.6%) | 0.423 | 74 (41.8%) | 14 (45.2%) | 0.728 |
| > T2 | 563 (49.4%) | 93 (52.0%) | 460 (47.8%) | 76 (51.4%) | 103 (58.2%) | 17 (54.8%) | |||
| Lymph node metastasis | |||||||||
| No | 742 (65.1%) | 124 (69.3%) | 0.280 | 627 (65.2%) | 104 (70.3%) | 0.225 | 115 (65.0%) | 20 (64.5%) | 0.961 |
| Yes | 397 (34.9%) | 55 (30.7%) | 335 (34.8%) | 44 (29.7%) | 62 (35.0%) | 11 (35.5%) | |||
| Metastasis | |||||||||
| M0 | 1129 (99.1%) | 179 (100.0%) | - | 953 (99.1%) | 148 (100.0%) | - | 176 (99.4%) | 31 (100.0%) | - |
| M1 | 10 (0.9%) | 0 (0.6%) | 9 (0.9%) | 0 (0.0%) | 1 (0.6%) | 0 (2.2%) | |||
| Cell differentiated grade | |||||||||
| Well | 151 (13.3%) | 32 (17.9%) | 0.098 | 133 (13.8%) | 30 (20.3%) | 0.041 | 18 (10.2%) | 2 (6.4%) | 0.521 |
| Moderate or poor | 988 (86.7%) | 147 (82.1%) | 829 (86.2%) | 118 (79.7%) | 159 (89.8%) | 29 (93.6%) | |||
MET levels in the head and neck squamous cell carcinoma (HNSCC) patients from TCGA database. (A) MET levels were compared between the HNSCC tumor tissues and normal tissue. (B) MET levels were compared between the HNSCC tumor tissues and adjacent noncancerous normal tissue. (C-E) MET levels were compared between grade I (G1, well-differentiated), grade II (G2, moderately differentiated), grade III (G3, poorly differentiated), and grade IV (G4, undifferentiated or anaplastic) in (C) all patients, (D) non-cigarette smokers and (E) smokers.
MET expression and overall survival in the head and neck squamous cell carcinoma (HNSCC) patients from TCGA database. MET expression and overall survival in (A) All HNSCC, (B) non-cigarette smokers (C) cigarette smoker population.
However, no significant association was found between the oral cancer patients and normal controls, suggesting a limited effect of these MET SNPs to oral cancer carcinogenesis. Intriguingly, after we analyzed the genotyping and allele frequency of MET SNP in oral cancer among cigarette smokers, a significant association was found in MET rs1621 polymorphisms between the oral cancer patients and controls, suggesting a lower risk to develop oral cancer carcinogenesis of these individuals who carried the MET rs1621 “GG” genotype.
A previous study has suggested that the SNP rs1621 in the seed-matching sequence of MET was related to affect the activity of miR-199a, which mediates the downregulation of the MET gene through targeting the 3'-UTR [34]. In a study of HCC, the variant GG genotype of MET rs1621 was suggested to be associated with a decreased risk for HCC, and the MET rs1621 polymorphism may influence susceptibility to HCC, alone and combined with miR-199a rs74723057 [23]. Consistent with this result, our data exhibited the same result in oral cancer among cigarette smokers. Of note, in a study of micropapillary-predominant subtype pulmonary adenocarcinoma (MPPAC), it was suggested that the c-MET protein overexpression was significantly associated with smoking status, lymphatic and venous invasion, and tumor-node-metastasis stage, but c-MET gene amplification showed no relation with any of these characteristics [35]. Moreover, cigarette smoking was suggested to induce overexpression of c-Met receptor in microvessels of oral lichen planus [36], and the influence of cigarette smoking to induce overexpression of HGF in type II pneumocytes and lung cancer cells was also observed [37]. These studies have indicated that the smoking status was highly linked to c-MET protein overexpression, overexpression of c-Met receptor, and overexpression of HGF in various cancers. Taken together, although the expression of c-MET protein, c-Met receptor, and HGF was not detected in our study, it can be proposed that these oral cancer patients involved in our study who smokes may have higher level of these key regulators of MET pathway. Even under the circumstances, the oral cancer patients who carried the MET rs1621 “GG” genotype still represented to be associated with decreased risk of oral cancer. The MET rs1621 to regulate the activity of miR-199a which mediates the downregulation of the MET gene through targeting the 3'-UTR might provide a possible mechanism to explain this phenomenon [23].
For MET rs33917957, although the information for rs33917957 is limited, a previous study focused on gastric cancer has suggested that the MET N375S variant genotypes (NS/SS) were associated with a significantly decreased risk of gastric cancer [22]. The MET N375S (rs33917957 A>G) was revealed as a germline missense variant in exon 2 of the MET gene which corresponds to the semaphorin domain of the MET protein [22], and the MET N375 forms two potential hydrogen bonds, whereas S375 modeling structure retains only one hydrogen bond, thereby has weaker ligand binding affinity compared with the N375 [22, 38]. Another study which focused on the effect of c-Met expression on survival in head and neck squamous cell has suggested the possible role of c-Met as an early marker of poor prognosis and a hallmark of aggressive biological behavior in oral cancer [39]. Compared with these results, our data showed consistency with the rs33917957 expressed in gastric cancer that the cigarette smokers of male oral cancer patients who carried the MET rs33917957 “AG + GG” polymorphic variants were associated with lower risk to develop moderate or poorer cell differentiated grade, and the ligand binding affinity from MET N375 to S375 may play an essential role in oral cancer. Besides, it was suggested that high expression of c-Met was associated with the primary location of head and neck carcinomas [40]. The squamous cell carcinoma expressed c-Met was found to be more frequently than undifferentiated carcinoma, and positive c-Met expression was suggested to correlated with high probability of lymph node metastasis [40]. Therefore, although the oral cancer patients who carried the MET rs33917957 polymorphisms showed no statistically significant association of clinical variables such as the clinical stage and TNM staging in our study, and most of the oral cancer patients enrolled in our study are individuals without lymph node metastasis, the MET rs33917957 may hence be interpreted as an early marker to evaluate cancer progression and prognosis oral cancer. However, the exact expressions of MET/HGF pathway correspond to MET polymorphisms and oral cancer progression and prognosis require future well-designed study to elucidate it.
In conclusion, our study first demonstrated the associations of MET polymorphisms to oral cancer disease susceptibility and clinical statuses. The study's findings have practical applications in both early detection and personalized treatment for oral cancer. By identifying individuals at risk based on their MET SNPs and considering factors such as smoking status, clinicians could offer more individualized approaches to prevention, screening, and treatment. Moreover, further research on MET expression and its role in cancer prognosis could lead to the development of new therapeutic targets.
Acknowledgements
We thank the Human Biobank of Chung Shan Medical University Hospital, Taichung, Taiwan for specimen preparation. This study was supported by grants from Chung Shan Medical University and Changhua Christian Hospital (CSMU-CCH-112-05).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ghelichli M, Mohtasham N, Mohajertehran F, Farshbaf A, Anvari K, Taghipour A. et al. Associations between RORgammat and T-bet Expressions, clinicopathological indices and survival rate in oral Squamous cell carcinoma patients. Cytokine. 2023;163:156116
2. Ghaderi H, Roshan-Zamir M, Jafarinia M, Kruger E. Oral Squamous Cell Carcinoma: Focus on Biomarkers for Screening. J Dent (Shiraz). 2024;25:1-16
3. Su CW, Lin CW, Yang WE, Yang SF. TIMP-3 as a therapeutic target for cancer. Ther Adv Med Oncol. 2019;11:1758835919864247
4. Rich BJ, Samuels SE, Azzam GA, Kubicek G, Freedman L. Oral Cavity Squamous Cell Carcinoma: Review of Pathology, Diagnosis, and Management. Crit Rev Oncog. 2024;29:5-24
5. Lin CW, Yang WE, Su CW, Lu HJ, Su SC, Yang SF. IGF2BP2 promotes cell invasion and epithelial-mesenchymal transition through Src-mediated upregulation of EREG in oral cancer. Int J Biol Sci. 2024;20:818-30
6. Yang SF, Lin CW, Chuang CY, Lee YC, Chung WH, Lai HC. et al. Host Genetic Associations with Salivary Microbiome in Oral Cancer. J Dent Res. 2022;101:590-8
7. Yeh JC, Chen YT, Chou YE, Su SC, Chang LC, Chen YL. et al. Interactive effects of CDKN2B-AS1 gene polymorphism and habitual risk factors on oral cancer. J Cell Mol Med. 2023;27:3395-403
8. Su SC, Chang LC, Huang HD, Peng CY, Chuang CY, Chen YT. et al. Oral microbial dysbiosis and its performance in predicting oral cancer. Carcinogenesis. 2021;42:127-35
9. Ma PC, Jagadeeswaran R, Jagadeesh S, Tretiakova MS, Nallasura V, Fox EA. et al. Functional expression and mutations of c-Met and its therapeutic inhibition with SU11274 and small interfering RNA in non-small cell lung cancer. Cancer Res. 2005;65:1479-88
10. Raj S, Kesari KK, Kumar A, Rathi B, Sharma A, Gupta PK. et al. Molecular mechanism(s) of regulation(s) of c-MET/HGF signaling in head and neck cancer. Mol Cancer. 2022;21:31
11. Mo HN, Liu P. Targeting MET in cancer therapy. Chronic Dis Transl Med. 2017;3:148-53
12. Saintigny P, William WN Jr, Foy JP, Papadimitrakopoulou V, Lang W, Zhang L. et al. Met Receptor Tyrosine Kinase and Chemoprevention of Oral Cancer. J Natl Cancer Inst. 2018;110:250-7
13. Crepaldi T, Gallo S, Comoglio PM. The MET Oncogene: Thirty Years of Insights into Molecular Mechanisms Driving Malignancy. Pharmaceuticals (Basel). 2024;17:448
14. Sivakumar M, Jayakumar M, Seedevi P, Sivasankar P, Ravikumar M, Surendar S. et al. Meta-analysis of functional expression and mutational analysis of c-Met in various cancers. Curr Probl Cancer. 2020;44:100515
15. Yokokawa M, Morita KI, Oikawa Y, Kayamori K, Sakamoto K, Ikeda T. et al. Co-expression of EGFR and MET has a synergistic effect on the prognosis of patients with oral squamous cell carcinoma. J Oral Pathol Med. 2020;49:235-42
16. Li L, Sun Z, Huang X, Li X, Sun L, Zhang L. et al. Role of c-Met expression on prognosis of head and neck cancer: A literature review and meta-analysis. Head Neck. 2019;41:1999-2006
17. Sun Z, Liu Q, Ye D, Ye K, Yang Z, Li D. Role of c-Met in the progression of human oral squamous cell carcinoma and its potential as a therapeutic target. Oncol Rep. 2018;39:209-16
18. Zhang C, Dong HK, Gao JM, Zeng QQ, Qiu JT, Wang JJ. Advances in the diagnosis and treatment of MET-variant digestive tract tumors. World J Gastrointest Oncol. 2024;16:4338-53
19. Gumustekin M, Kargi A, Bulut G, Gozukizil A, Ulukus C, Oztop I. et al. HGF/c-Met overexpressions, but not met mutation, correlates with progression of non-small cell lung cancer. Pathol Oncol Res. 2012;18:209-18
20. Cao X, Hong X, Jia X, Zhang L, Chen G. Single-nucleotide polymorphism rs41736 located in MET was significantly associated with prognosis of small cell lung cancer patients. Med Oncol. 2014;31:333
21. Yang JJ, Cho LY, Ko KP, Shin A, Ma SH, Choi BY. et al. Genetic susceptibility on CagA-interacting molecules and gene-environment interaction with phytoestrogens: a putative risk factor for gastric cancer. PLoS One. 2012;7:e31020
22. Liu Y, Zhang Q, Ren C, Ding Y, Jin G, Hu Z. et al. A germline variant N375S in MET and gastric cancer susceptibility in a Chinese population. J Biomed Res. 2012;26:315-8
23. Wang Q, Yu X, Li Q, Qin L, Tan S, Zeng X. et al. Association between miR-199a rs74723057 and MET rs1621 polymorphisms and the risk of hepatocellular carcinoma. Oncotarget. 2016;7:79365-71
24. Huang SH, O'Sullivan B. Overview of the 8th Edition TNM Classification for Head and Neck Cancer. Curr Treat Options Oncol. 2017;18:40
25. Su SC, Hsieh MJ, Lin CW, Chuang CY, Liu YF, Yeh CM. et al. Impact of HOTAIR Gene Polymorphism and Environmental Risk on Oral Cancer. J Dent Res. 2018;97:717-24
26. Chen YT, Lin CW, Chou YE, Su SC, Chang LC, Lee CY. et al. Potential impact of ADAM-10 genetic variants with the clinical features of oral squamous cell carcinoma. J Cell Mol Med. 2023;27:1144-52
27. Tsai YS, Chen YC, Chen TI, Lee YK, Chiang CJ, You SL. et al. Incidence trends of oral cavity, oropharyngeal, hypopharyngeal and laryngeal cancers among males in Taiwan, 1980-2019: a population-based cancer registry study. BMC Cancer. 2023;23:213
28. Hashibe M, Brennan P, Benhamou S, Castellsague X, Chen C, Curado MP. et al. Alcohol drinking in never users of tobacco, cigarette smoking in never drinkers, and the risk of head and neck cancer: pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. J Natl Cancer Inst. 2007;99:777-89
29. Wyss A, Hashibe M, Chuang SC, Lee YC, Zhang ZF, Yu GP. et al. Cigarette, cigar, and pipe smoking and the risk of head and neck cancers: pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. Am J Epidemiol. 2013;178:679-90
30. Lee YA, Li S, Chen Y, Li Q, Chen CJ, Hsu WL. et al. Tobacco smoking, alcohol drinking, betel quid chewing, and the risk of head and neck cancer in an East Asian population. Head Neck. 2019;41:92-102
31. Su SC, Chang LC, Lin CW, Chen MK, Yu CP, Chung WH. et al. Mutational signatures and mutagenic impacts associated with betel quid chewing in oral squamous cell carcinoma. Hum Genet. 2019;138:1379-89
32. Su SC, Yeh CM, Lin CW, Hsieh YH, Chuang CY, Tang CH. et al. A novel melatonin-regulated lncRNA suppresses TPA-induced oral cancer cell motility through replenishing PRUNE2 expression. J Pineal Res. 2021;71:e12760
33. Lu HJ, Su CW, Su SC, Chang LC, Wu MF, Lin CW. et al. Prognostic impact of caspase-8 mutation in oral cavity squamous cell carcinoma. Oral Dis. 2024 Sep 17. doi: 10.1111/odi.15124. Online ahead of print
34. Ning L, Yu Y, Liu X, Ai L, Zhang X, Rao W. et al. Association Analysis of MET Gene Polymorphism with Papillary Thyroid Carcinoma in a Chinese Population. Int J Endocrinol. 2015;2015:405217
35. Zhang J, Sun J, Zhang Z, Liang X, Luo Y, Wu S. et al. Protein overexpression and gene amplification of cellular mesenchymal-epithelial transition factor is associated with poor prognosis in micropapillary-predominant subtype pulmonary adenocarcinoma. Hum Pathol. 2018;72:59-65
36. Klosek SK, Sporny S, Stasikowska-Kanicka O, Kurnatowska AJ. Cigarette smoking induces overexpression of c-Met receptor in microvessels of oral lichen planus. Arch Med Sci. 2011;7:706-12
37. Chen JT, Lin TS, Chow KC, Huang HH, Chiou SH, Chiang SF. et al. Cigarette smoking induces overexpression of hepatocyte growth factor in type II pneumocytes and lung cancer cells. Am J Respir Cell Mol Biol. 2006;34:264-73
38. Krishnaswamy S, Kanteti R, Duke-Cohan JS, Loganathan S, Liu W, Ma PC. et al. Ethnic differences and functional analysis of MET mutations in lung cancer. Clin Cancer Res. 2009;15:5714-23
39. Lo Muzio L, Farina A, Rubini C, Coccia E, Capogreco M, Colella G. et al. Effect of c-Met expression on survival in head and neck squamous cell carcinoma. Tumour Biol. 2006;27:115-21
40. Choe JY, Yun JY, Nam SJ, Kim JE. Expression of c-Met Is Different along the Location and Associated with Lymph Node Metastasis of Head and Neck Carcinoma. Korean J Pathol. 2012;46:515-22
Author contact
![]() Corresponding authors: Ying-Erh Chou, PhD. or Chiao-Wen Lin, PhD. School of Medicine, Chung Shan Medical University, 110, Section 1, Chien-Kuo N. Road, Taichung, Taiwan, ROC. Fax: 886-4-24723229. E-mail: intointo814com (Ying-Erh Chou); cwlinedu.tw (Chiao-Wen Lin).
Corresponding authors: Ying-Erh Chou, PhD. or Chiao-Wen Lin, PhD. School of Medicine, Chung Shan Medical University, 110, Section 1, Chien-Kuo N. Road, Taichung, Taiwan, ROC. Fax: 886-4-24723229. E-mail: intointo814com (Ying-Erh Chou); cwlinedu.tw (Chiao-Wen Lin).

 Global reach, higher impact
Global reach, higher impact