Impact Factor
ISSN: 1837-9664
J Cancer 2025; 16(7):2353-2359. doi:10.7150/jca.103209 This issue Cite
Review
SF3a1: A Novel Potential Tumor Biomarker or Therapeutic Target
1. Department of Obstetrics and Gynecology, Shanghai Tenth People's Hospital, Tongji University School of Medicine, Shanghai, 200040, China.
2. Shanghai Fourth People's Hospital, Tongji University School of Medicine, Shanghai, 200434, China.
3. Gynecological Minimally Invasive Surgery Institute, Tongji University School of Medicine, Shanghai, 200331, China.
Received 2024-12-1; Accepted 2025-3-21; Published 2025-4-9
Abstract
Alternative splicing is an evolutionarily conserved and essential cellular process that is catalyzed by a multi-complex spliceosome. Dysregulation of this process has been implicated in various tumors over the recent years. SF3a1 is a critical subunit of U2 small nuclear ribonucleoprotein (snRNP) in the spliceosome, which has been found to be aberrant in several human diseases. Recent reports suggest that SF3a1 might be a novel therapeutic target. However, a comprehensive description of SF3a1 is lacking. In this review, we present the findings of SF3a1 from protein structure, biological function to strong associations with human diseases including cancer. Studies have reported that SF3a1 dysregulation and associated alternative splicing events mediate tumorigenesis and other immune-related disorders. However, further functional and mechanistic studies are needed to fully understand the regulatory network of SF3a1 in human diseases. In conclusion, SF3a1 could serve as a promising prognostic biomarker and therapeutic target for specific cancer types, including prostate cancer, colorectal cancer and hepatocellular carcinoma.
Keywords: alternative splicing, prognostic biomarker, SF3a1, therapeutic target, tumorigenesis.
Introduction
Pre-mRNA alternative splicing (AS) is an essential process that ensures proteome diversity from a limited number of protein-coding genes in eukaryotic genomes. This process is carried out by a dynamic ribonucleoprotein complex, also known as the spliceosome. Five snRNPs and over 100 non-snRNP proteins assemble sequentially to form the spliceosome during the process [1, 2]. Abnormal status of these proteins cause aberrant AS and lead to disease including tumor [3]. U2 snRNP is one of the snRNPs that assembles the E and A complexes at the early stage of AS [4]. It recognizes branchpoint sequence (BPS) of the intron through SF3a and SF3b complexes [5, 6]. Binding of SF3a and SF3b complexes to a 20-nucleotide region upstream of the BPS anchors U2 snRNP to pre-mRNA and formed A complex in an ATP-dependent manner [4-6]. As one of the main components of U2 snRNP, the SF3a complex contains three subunits: SF3a1/SF3a120, SF3a2/SF3a66 and SF3a3/SF3a60 [7, 8]. Each subunit of SF3a is essential for human cell viability and pre-mRNA splicing [9, 10]. Aberrant expression of these proteins has been found in several malignant tumors, suggesting their involvement in tumor development and progression [11-13].
Given the key functions of SF3a1 in spliceosome and the extensive regulatory effects of other U2 snRNP components on tumor biology [12], we present a comprehensive overview of SF3a1's structure and biological function, particularly in the context of tumorigenesis. The correlation between SF3a1 and cancers, as well as other immune-related diseases, including autoimmune disorders and infections, suggests that it could serve as a promising prognostic marker and therapeutic target. However, there remains a significant gap in functional and mechanistic research that needs to be addressed.
The structure of SF3a1 and its role in spliceosome
The structure of the SF3a1 gene and protein
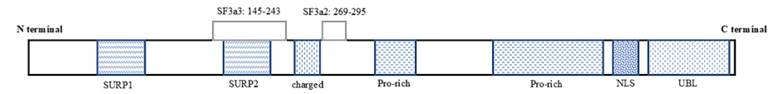
Human SF3a1 gene is located on chromosome 22q12.2, which containing 16 exons and 15 introns [14]. It is transcribed into three transcript variants with 3.2, 3.8 and 5.7 kilobases respectively, and produces a 793-amino acid protein rich in prolines and charged amino acids. SF3a1 has been identified to be homologous to splicing factor PRP21p in Saccharomyces cerevisiae [15]. The N-terminal of SF3a1 contains two suppressor-of-white-apricot and prp21/spp91 (SURP) domains, and the C-terminal contains a nuclear localization signal (NLS) and a ubiquitin-like (UBL) domain [15, 16] (Fig. 1). The SURP2 domain mediates the interaction of SF3a1 with SF3a3, while the 130-amino acid region between SURP2 and Pro-rich domains mediates the interaction of SF3a1 with SF3a2 [15]. Nuclear magnetic resonance (NMR) spectroscopy revealed that Leu169 in SURP2 domain of SF3a1 was indispensable for the SF3a complex formation (Fig. 1). It was proposed that SF3a1 functioned as the scaffold and nuclear import determinant of SF3a complex, because no direct interaction between SF3a2 and SF3a3 was found [10, 17, 18].
The interplay between SF3a1 and various molecules
As one of the core components of spliceosome, SF3a1 participates in multiple physiological activities by interacting with various molecules including proteins and RNAs.
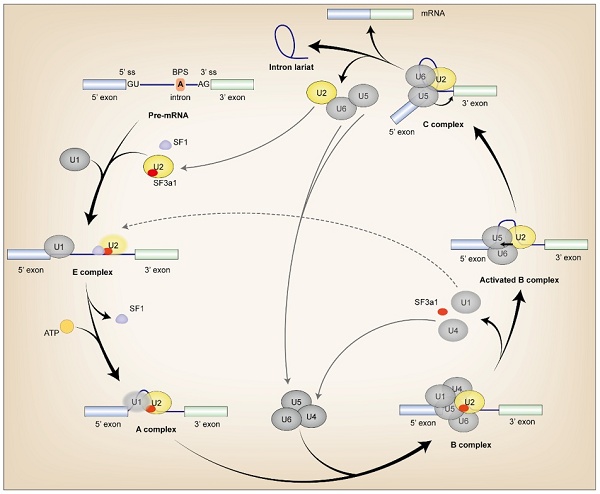
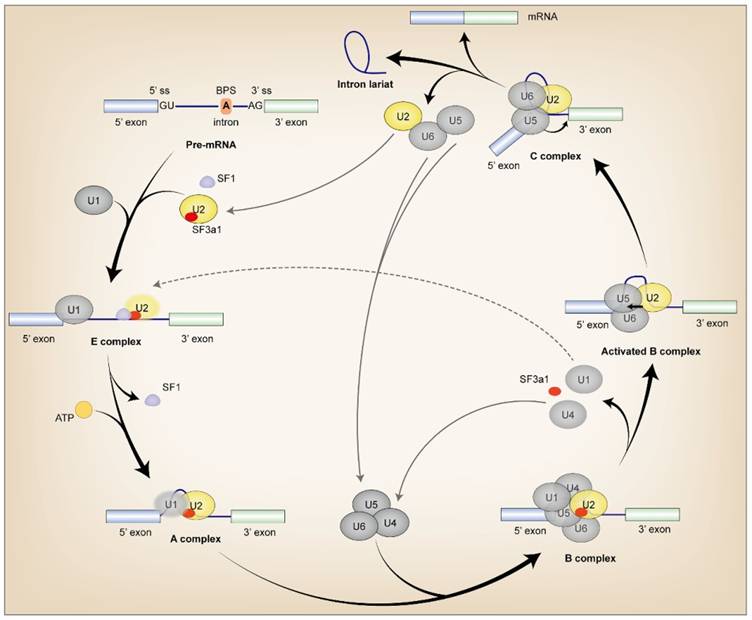
During pre-mRNA AS, SF3a1 interacts dynamically with various proteins and RNAs throughout spliceosome assembly. The SF3a heterotrimer is imported into the nucleus independently and targeted to Cajal bodies (CBs), where it is incorporated into the U2 snRNP [18-20]. Although SF3a was found to be related to the 3' portion of the U2 snRNA and SF3b, the precise interaction between SF3a1 and U2 snRNA has not yet been elucidated, although SF3a3 contacts both the bases of stem-loop I and a bulge in stem-loop III of U2 snRNA [21, 22]. Then SF3a1 binds to splicing factor 1 (SF1) via SURP1 domain to mediate recruitment of the U2 snRNP for efficient complex E assembly [1]. During transition from the E to A complex, SF3a1 interacts with the GCG/CGC RNA stem and the apical UUCG tetraloop of U1-SL4 in a sequence-specific manner. This interaction mediates the connection between U1 and U2 snRNPs, which identifies the 5' and 3' splice site of pre-mRNA respectively for functional spliceosome assembly and intron lariat formation. The RGG motif in the UBL domain at the carboxyl terminal of SF3a1 plays a pivotal role in the interaction, while mutations at Y772 and Y773 in the UBL domain of SF3a1 perturb the interplay [23-26]. In the B complex, SF3a1 binds to prp3 which is indispensable for U4/U6•U5 tri-snRNP formation [27] (Fig. 2).
In addition to spliceosome components, SF3a1 also interacts with other proteins such as chromodomain helicase DNA-binding protein 1 (CHD1) to regulate pre-mRNA splicing. SF3a1 bridged with CHD1 to enhance chromatin association and pre-mRNA splicing on specific genes by recognizing tri-methylation of histone H3 on lysine 4 (H3K4me3) [28]. SF3a1 activation facilitated proper splicing and accumulation of cohesion factor Sororin during S phase to maintain functional sister chromatid cohesion and subsequent cell cycle progression [29]. SF3a1 could also bind the N-terminal region of transcription factor SP1 with the function of this interaction remained unclear [30].
Thus, SF3a1 exhibits complex interactions with various proteins and RNA motifs to ensure efficient and proper splicing of pre-mRNAs. Further exploration is needed to determine the precise role of SF3a1 in the spliceosome and its upstream and downstream regulators.
The role of SF3a1 in tumors
Human tumors are characterized by splicing vulnerabilities [31]. Like other core components of spliceosome, SF3a1 has been identified as a risk factor in both hematologic and solid tumors (Table 1).
Schematic representation of SF3a1 protein. The numeric range referred to the amino acids spanning on SF3a1 that interacted with SF3a2 or SF3a3. SURP, suppressor-of-white-apricot and prp21/spp91; NLS, nuclear localization signal; UBL, ubiquitin-like.
The clinical association between SF3a1 mutation and various hematologic and solid malignancies
| Tumor type | Sample sizea | SNP | Genotypesb | P value/ OR (95% CI) | Mutation frequencyc | Clinical correlation | Ref. |
|---|---|---|---|---|---|---|---|
| BPDCN | 50 | N | N | N | 0.02 | no prognostic relation | [32] |
| Pancreatic cancer | 298 | rs2074733 | (TT+CT)/CC | P=1.4E-02 /0.65(0.48-0.88) | 0.60/0.70 | lower risk | [33] |
| Pancreatic cancer | 413 | rs2074733 | (TT+CT)/CC | P=3.0E-04 /0.54(0.41-0.73) | 0.66/0.78 | lower risk | [33] |
| Familial gastric and rectal cancer | 8 | N | T/C | N | 0.88 | possibly contributing to cancer | [34] |
| Familial gastric and rectal cancer | 98 | N | G/T | N | 0.01 | possibly contributing to cancer | [34] |
| CRC | 1 | rs1370165248 | A/G | N | 1 | major clonal mutation | [35] |
| CRC | 801 | rs5753073 | (AG+GG)/AA | P>0.05 | 0.25/0.28 | no risk correlation | [36] |
| CRC | 801 | rs2839998 | (GA+AA)/GG | P>0.05 | 0.55/0.53 | no risk correlation | [36] |
| CRC | 801 | rs10376 | (AC+AA)/CC | P>0.05 | 0.17/0.20 | no risk correlation | [36] |
| CRC | 801 | rs2074733 | (TC+CC)/TT | P>0.05 | 0.72/0.73 | no risk correlation | [36] |
| HCC | 378 | rs5994293 | (TG+GG)/TT | P=5.0E-04 /0.70(0.58-0.84) | 0.65/0.49 | higher risk | [37] |
| HCC | 428 | rs5994293 | (TG+GG)/TT | P=5.0E-04 /0.70(0.58-0.84) | 0.58/0.55 | higher risk | [37] |
N: not present.
a: The size referred to patient number.
b: The last genotype was used as the reference for OR calculations.
c: The last number indicated mutation frequency of control group in case-control study.
BPDCN, blastic plasmacytoid dendritic cell neoplasm; CRC, colorectal cancer; HCC, hepatocellular carcinoma; SNPs, single nucleotide polymorphisms.
The incorporation of SF3a1 in spliceosome assembly. Firstly, SF1 interacted with SF3a1 to recruit U2 snRNP during complex E assembly, and SF3a1 was necessary for anchoring U2 snRNP to pre-mRNA. Then, the interaction between SF3a1 and U1 snRNA facilitated intron lariat formation in an ATP-dependent manner in complex A. Finally, before or during the first catalytic step of splicing, SF3a1 was dissociated from the spliceosome. Except for the labels in the Figure, black arrow in the complex: transesterification; ss, splice site; gray lines referred to the splicing cycle; oval without black line edge indicated unstable binding.
The role of SF3a1 in hematologic malignancies
Authoritative studies have shown SF3a1's involvement in hematologic malignancies. A whole-exome sequencing analysis revealed that SF3a1 was mutated in myelodysplasia (MDS) patients with a predisposition to acute myeloid leukemia, which was further confirmed in a large series of myeloid neoplasms exhibiting features of MDS [38]. Patients with blastic plasmacytoid dendritic cell neoplasm (BPDCN) were found to harbor somatic mutations in SF3a1 and other RNA splicing components, however, SF3a1 mutation status did not show statistically significant correlation with overall survival in this malignancy [32]. In addition to tumorigenesis, SF3a1 might also play a role in drug resistance. Recurrent SF3a1 mutations were only present in non-responders to azacitidine (Aza) and erythropoietin (Epo) in transfusion-dependent, growth factor-resistant, low- and Int-1-risk MDS patients, although they were not able to predict therapeutic response [39]. Dysregulated epigenetic modulation of SF3a1 has also been found. DNA methylation analysis revealed that SF3a1 was not downregulated by promoter hypermethylation in leukemia cells [40]. The above studies demonstrated that SF3a1 mutations occurred in specific types and subsets of hematologic malignancies. Although less frequently studied compared to SF3b1, SF3a1 could be used as a complementary molecule to predict prognosis and treatment response in specific patients.
The role of SF3a1 in solid cancers
Research on SF3a1 has been more abundant and in-depth in cancers compared to hematologic malignancies. A two-stage case-control study demonstrated a consistent association between two C alleles at rs2074733 in the SF3a1 gene and an increased risk of pancreatic cancer. In addition, smoking or drinking cooperated with the harmful genotype of SF3a1 to increase pancreatic cancer risk [33]. SF3a1 was also among the 12 mutated genes with novel but not penetrant non-synonymous SNPs in a family with gastric and rectal cancer [34]. However, another hospital-based case-control study in a Chinese population indicated that four selected SNPs (rs10376, rs5753073, rs2839998, and rs2074733) of SF3a1 were not significantly associated with colorectal cancer (CRC) risk, even after normalizing for smoking and alcohol use status [36]. Limitations, including a small number of SNPs, samples, and environmental factors in the latter study, probably led to the contradictory results, which require further research for validation. Blood plasma cell-free DNA sequencing revealed major clonal mutations of SF3a1 and others were found in all samples from one advanced CRC patient who showed rapid but not sustained response to chemotherapy, implying SF3a1 might contribute to chemotherapy resistance and the pathogenesis of CRC [35]. In a two-stage case-control study in China, TT alleles at rs5994293 in SF3a1 gene also exhibited a significant correlation with higher hepatocellular carcinoma (HCC) risk, which additively interacted with smoking and alcohol consumption to increase HCC risk in HBsAg-negative participants in both the screened and combined cohorts [37]. After applying multiple bioinformatic methods on the TCGA cohort, SF3a1 was identified as one of the hub genes in the gene network of the top 800 OS-related AS events in HCC patients [41]. Furthermore, SF3a1 was among the 20 lactylation-related genes with prognostic grouping value in HCC. The low-risk group exhibited a more active immune response [42]. In endometrial cancer, SF3a was among the up-regulated proteins in the human cell line Ishikawa after megestrol acetate treatment, implying a potential function of SF3a in predicting the response of gynecologic malignancy to hormonotherapy [43]. In conclusion, SF3a1 mutations are significantly correlated with the risk and prognosis of various cancers, particularly those of the digestive system, although some discordance exists, and further functional experiments are needed.
Several aberrantly expressed proteins in various cancers have been found to be regulated by SF3a1. Different isoforms of muscleblind-like 1 (MBNL1) were found to exert opposing functions in prostate cancer (PC). MBNL1 lacking exon 7, which tumor preferentially abandoned, led to DNA damage and subsequent inhibition of cell viability and migration [44]. While the overall expression of MBNL1 was downregulated, exon 7 in the MBNL1 transcript was the most differentially included exon in several cancers including PC, and was essential for the homodimerization of the MBNL1 protein. SF3a1 was necessary for the retention of exon 7 to exert pro-tumor effects [44]. When PC3 cells were treated with novel and specific PolyPurine Reverse Hoogsteen (PPRH) hairpins for gene silencing against anti-apoptotic Survivin, SF3a1 was identified among the important interactive gene nodes in the STRING network analysis and gene sets in GSEA of differentially expressed genes, indicating that SF3a1 participated in the apoptosis-related pathway of PC [45]. The axis of SF3a1/ MBNL1 or Survivin isoforms might provide a promising subset of therapeutic targets and prognostic markers for PC. Furthermore, SF3a1 was found to promote splicing landscape reprogramming and progression of metastatic castration-resistant PC through SOX6- GH22I030351 axis [46]. And an integrative lactylome and proteome analysis based on liquid chromatography-tandem mass spectrometry (LC-MS/MS) revealed SF3A1 was lactylated, a novel type of post-translational modification, under both normoxia and hypoxia in oral squamous cell carcinoma (OSCC), which might contribute to the altered pre-mRNA splicing pattern and pathogenesis of OSCC [47].
Collectively, SF3a1 presented as variant mutated forms in different hematologic and solid malignancies, and specific mutations were associated with patient outcomes and treatment responses, despite a few discordant findings, indicating SF3a1 and related AS events could be potential and promising therapeutic targets. However, most studies have focused only on prognostic correlations, while functional and mechanistic research remains largely underexplored, particularly in terms of how mutation patterns of SF3a1 influence U2 snRNP functionality, the subsequent mis-splicing of specific genes, and resulting tumorigenesis.
The role of SF3a1 in other immune-related diseases
SF3a1 has also been implicated in non-tumor immune-related disorders, including autoimmune and infectious diseases. A systematic meta-analysis revealed that SF3a1 SNPs were involved in genomic susceptibility repertoire of the most prevalent juvenile idiopathic arthritis subtype [48]. Comprehensive functional genomic screens showed that SF3a promoted osteoarthritis progression by inducing collagen IIA expression [49]. The expression level of SF3a, defined as an autoantigen gene of systemic lupus erythematosus (SLE), was upregulated in T lymphocytes of glomerulonephritis patients after immunosuppressive therapy [50]. These findings indicated that SF3a1 might act as an etiological factor and therapeutic target in specific auto-immune diseases.
SF3a was downregulated in THP-1 macrophage cells following mycobacterium tuberculosis (M. tb) H37Rv infection, implying the importance of SF3a in innate immune cells for pathogen defense [51]. In addition to promoting inflammatory response, SF3a1 also limited excessive and persistent inflammation by regulating the expression of mRNA isoforms in the toll-like receptor (TLR) signaling pathway. Negative regulators including sTLR4, Rab7b, and possibly IKKβb were upregulated through various AS events, while positive factors such as IRAK1, IKKβ, and CD14 were downregulated via intron retention regulated by SF3a1. MyD88 was another gene regulated by SF3a1 via AS in the innate immune response [52, 53]. Decreased expression of SF3a1 by liver X receptor agonist T0901317 had been shown to inhibit inflammation via up-regulating the alternative splice short form of MyD88 mRNA [54].
These studies revealed that SF3a1 dysregulation led to immune homeostasis disruption and the onset of immune-related disorders. Furthermore, SF3a1 could serve as a target to prevent excessive inflammatory responses and the development of subsequent diseases.
Conclusion
The idea that altered splicing underlay oncogenesis has gained attention and expanded the therapeutic scope of multiple tumors over the years. SF3a1 appears to perform key functions within the SF3a complex in U2 snRNP through its dynamical interactions with various molecules. Currently, SF3a1 mutations have been shown to be significantly correlated with hematologic and solid tumors. Several functional and mechanistic studies have elucidated the potential contribution of SF3a1 to the pathogenesis of cancers including CRC, PC and HCC. Additionally, SF3a1 also participates in autoimmune disorders and infectious diseases by regulating expression of isoforms of inflammatory factors. These findings suggest that SF3a1 is involved in both tumorigenic and immune-related diseases, although further investigations are needed. It also suggests that SF3a1 could be a novel therapeutic target for tumor and autoimmune disease treatment. The induction of immunogenic tumor-specific neoepitopes by SF3b1 mutations further implies its potential in tumor immunotherapy [31, 55].
Collectively, SF3a1 is a component of the SF3a complex within the spliceosome, functions as a core regulatory factor in pre-mRNA AS, and plays pivotal roles in multiple biological processes, including tumor development. Aberrant expression of SF3a1 and related AS events may serve as biomarkers or therapeutic targets for related diseases, especially cancers such as PC, CRC and HCC.
Abbreviations
snRNPs: small nuclear ribonucleoproteins; AS: alternative splicing; BPS: branchpoint sequence; SURP2: the second suppressor-of-white-apricot and prp21/spp91; NLS: nuclear localization signal; UBL: ubiquitin-like; NMR: nuclear magnetic resonance; CBs: Cajal bodies; SF1: splicing factor 1; CHD1: chromodomain helicase DNA binding protein 1; H3K4me3: histone H3 on lysine 4; MDS: myelodysplasia; BPDCN: blastic plasmacytoid dendritic cell neoplasm; Aza: azacitidine; Epo: erythropoietin; CRC: colorectal cancer; HCC: hepatocellular carcinoma; MBNL1: muscleblind-like 1; PC: prostate cancer; PPRH: PolyPurine Reverse Hoogsteen; LC-MS/MS: liquid chromatography-tandem mass spectrometry; OSCC: oral squamous cell carcinoma; SLE: systemic lupus erythematosus; M. tb: mycobacterium tuberculosis; TLR: toll-like receptor.
Acknowledgements
Funding
This review was supported by Shanghai Tenth People's Hospital (Climbing talent program, 2021SYPDRC006), the Fundamental Research Funds for the Central Universities (22120220601); and the Emerging Frontier Technology Research Program of Shanghai Shen-Kang Hospital Development Center (No. SHDC12021113). We apologize to colleagues whose work was not cited due to space limitations.
Author contributions
XQ S: Investigation, Writing-Original Draft, Visualization. YQ S: Writing-Review & Editing, Visualization. SP L: Conceptualization, Resources, Writing-Review & Editing, Supervision, Project administration, Funding acquisition. ZP C: Conceptualization, Resources, Supervision, Project administration, Funding acquisition.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Crisci A, Raleff F, Bagdiul I, Raabe M, Urlaub H, Rain JC. et al. Mammalian splicing factor SF1 interacts with SURP domains of U2 snRNP-associated proteins. Nucleic acids research. 2015;43:10456-73
2. Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Molecular cell. 2003;12:5-14
3. Taylor J, Xiao W, Abdel-Wahab O. Diagnosis and classification of hematologic malignancies on the basis of genetics. Blood. 2017;130:410-23
4. Das R, Zhou Z, Reed R. Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Molecular cell. 2000;5:779-87
5. Gozani O, Feld R, Reed R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes & development. 1996;10:233-43
6. Bessonov S, Anokhina M, Will CL, Urlaub H, Lührmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846-50
7. Zuo Y, Feng F, Qi W, Song R. Dek42 encodes an RNA-binding protein that affects alternative pre-mRNA splicing and maize kernel development. Journal of integrative plant biology. 2019;61:728-48
8. Brosi R, Gröning K, Behrens SE, Lührmann R, Krämer A. Interaction of mammalian splicing factor SF3a with U2 snRNP and relation of its 60-kD subunit to yeast PRP9. Science (New York, NY). 1993;262:102-5
9. Tanackovic G, Krämer A. Human splicing factor SF3a, but not SF1, is essential for pre-mRNA splicing in vivo. Molecular biology of the cell. 2005;16:1366-77
10. Nesic D, Krämer A. Domains in human splicing factors SF3a60 and SF3a66 required for binding to SF3a120, assembly of the 17S U2 snRNP, and prespliceosome formation. Molecular and cellular biology. 2001;21:6406-17
11. Liu KL, Yin YW, Lu BS, Niu YL, Wang DD, Shi B. et al. E2F6/KDM5C promotes SF3A3 expression and bladder cancer progression through a specific hypomethylated DNA promoter. Cancer cell international. 2022;22:109
12. Cieśla M, Ngoc PCT, Cordero E, Martinez Á S, Morsing M, Muthukumar S. et al. Oncogenic translation directs spliceosome dynamics revealing an integral role for SF3A3 in breast cancer. Molecular cell. 2021;81:1453-68.e12
13. Hakobyan S, Loeffler-Wirth H, Arakelyan A, Binder H, Kunz M. A Transcriptome-Wide Isoform Landscape of Melanocytic Nevi and Primary Melanomas Identifies Gene Isoforms Associated with Malignancy. International journal of molecular sciences. 2021;22(13):7165
14. Hu Z, Wu C, Shi Y, Guo H, Zhao X, Yin Z. et al. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nature genetics. 2011;43:792-6
15. Krämer A, Mulhauser F, Wersig C, Gröning K, Bilbe G. Mammalian splicing factor SF3a120 represents a new member of the SURP family of proteins and is homologous to the essential splicing factor PRP21p of Saccharomyces cerevisiae. RNA (New York, NY). 1995;1:260-72
16. Online Mendelian lnheritance in Man, OMIM@. McKusick-Nathans lnstitute of Genetic Medicine, JohnsHopkins University (Baltimore, MD), {date). World Wide Web URL: https://omim.org/.
17. Huang CJ, Ferfoglia F, Raleff F, Krämer A. Interaction domains and nuclear targeting signals in subunits of the U2 small nuclear ribonucleoprotein particle-associated splicing factor SF3a. The Journal of biological chemistry. 2011;286:13106-14
18. Kuwasako K, He F, Inoue M, Tanaka A, Sugano S, Güntert P. et al. Solution structures of the SURP domains and the subunit-assembly mechanism within the splicing factor SF3a complex in 17S U2 snRNP. Structure (London, England: 1993). 2006;14:1677-89
19. Krämer A, Ferfoglia F, Huang CJ, Mulhaupt F, Nesic D, Tanackovic G. Structure-function analysis of the U2 snRNP-associated splicing factor SF3a. Biochemical Society transactions. 2005;33:439-42
20. Nesic D, Tanackovic G, Krämer A. A role for Cajal bodies in the final steps of U2 snRNP biogenesis. Journal of cell science. 2004;117:4423-33
21. Dybkov O, Will CL, Deckert J, Behzadnia N, Hartmuth K, Lührmann R. U2 snRNA-protein contacts in purified human 17S U2 snRNPs and in spliceosomal A and B complexes. Molecular and cellular biology. 2006;26:2803-16
22. Krämer A, Grüter P, Gröning K, Kastner B. Combined biochemical and electron microscopic analyses reveal the architecture of the mammalian U2 snRNP. The Journal of cell biology. 1999;145:1355-68
23. de Vries T, Martelly W, Campagne S, Sabath K, Sarnowski CP, Wong J. et al. Sequence-specific RNA recognition by an RGG motif connects U1 and U2 snRNP for spliceosome assembly. Proceedings of the National Academy of Sciences of the United States of America. 2022;119(6):e2114092119
24. Martelly W, Fellows B, Senior K, Marlowe T, Sharma S. Identification of a noncanonical RNA binding domain in the U2 snRNP protein SF3A1. RNA (New York, NY). 2019;25:1509-21
25. Martelly W, Fellows B, Kang P, Vashisht A, Wohlschlegel JA, Sharma S. Synergistic roles for human U1 snRNA stem-loops in pre-mRNA splicing. RNA biology. 2021;18:2576-93
26. Sharma S, Wongpalee SP, Vashisht A, Wohlschlegel JA, Black DL. Stem-loop 4 of U1 snRNA is essential for splicing and interacts with the U2 snRNP-specific SF3A1 protein during spliceosome assembly. Genes & development. 2014;28:2518-31
27. Pozzi B, Bragado L, Will CL, Mammi P, Risso G, Urlaub H. et al. SUMO conjugation to spliceosomal proteins is required for efficient pre-mRNA splicing. Nucleic acids research. 2017;45:6729-45
28. Sims RJ 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P. et al. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Molecular cell. 2007;28:665-76
29. Watrin E, Demidova M, Watrin T, Hu Z, Prigent C. Sororin pre-mRNA splicing is required for proper sister chromatid cohesion in human cells. EMBO reports. 2014;15:948-55
30. Gunther M, Laithier M, Brison O. A set of proteins interacting with transcription factor Sp1 identified in a two-hybrid screening. Molecular and cellular biochemistry. 2000;210:131-42
31. Kahles A, Lehmann KV, Toussaint NC, Hüser M, Stark SG, Sachsenberg T. et al. Comprehensive Analysis of Alternative Splicing Across Tumors from 8,705 Patients. Cancer cell. 2018;34:211-24.e6
32. Yin CC, Pemmaraju N, You MJ, Li S, Xu J, Wang W. et al. Integrated Clinical Genotype-Phenotype Characteristics of Blastic Plasmacytoid Dendritic Cell Neoplasm. Cancers. 2021;13(23):5888
33. Tian J, Liu Y, Zhu B, Tian Y, Zhong R, Chen W. et al. SF3A1 and pancreatic cancer: new evidence for the association of the spliceosome and cancer. Oncotarget. 2015;6:37750-7
34. Thutkawkorapin J, Picelli S, Kontham V, Liu T, Nilsson D, Lindblom A. Exome sequencing in one family with gastric- and rectal cancer. BMC genetics. 2016;17:41
35. Hendricks A, Rosenstiel P, Hinz S, Burmeister G, Röcken C, Boersch K. et al. Rapid response of stage IV colorectal cancer with APC/TP53/KRAS mutations to FOLFIRI and Bevacizumab combination chemotherapy: a case report of use of liquid biopsy. BMC medical genetics. 2020;21:3
36. Chen X, Du H, Liu B, Zou L, Chen W, Yang Y. et al. The Associations between RNA Splicing Complex Gene SF3A1 Polymorphisms and Colorectal Cancer Risk in a Chinese Population. PloS one. 2015;10:e0130377
37. Song W, Zhu B, Tian Y, Zhong R, Tian J, Miao X. et al. [Research on the association between U2-dependent spliceosome gene and hepatocellular cancer]. Zhonghua liu xing bing xue za zhi = Zhonghua liuxingbingxue zazhi. 2015;36:634-8
38. Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R. et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64-9
39. Tobiasson M, Dybedahl I, Holm MS, Karimi M, Brandefors L, Garelius H. et al. Limited clinical efficacy of azacitidine in transfusion-dependent, growth factor-resistant, low- and Int-1-risk MDS: Results from the nordic NMDSG08A phase II trial. Blood cancer journal. 2014;4:e189
40. Wong JJ, Lau KA, Pinello N, Rasko JE. Epigenetic modifications of splicing factor genes in myelodysplastic syndromes and acute myeloid leukemia. Cancer science. 2014;105:1457-63
41. Yang L, He Y, Zhang Z, Wang W. Systematic analysis and prediction model construction of alternative splicing events in hepatocellular carcinoma: a study on the basis of large-scale spliceseq data from The Cancer Genome Atlas. PeerJ. 2019;7:e8245
42. Chen Y, Chang L, Hu L, Yan C, Dai L, Shelat VG. et al. Identification of a lactylation-related gene signature to characterize subtypes of hepatocellular carcinoma using bulk sequencing data. Journal of gastrointestinal oncology. 2024;15:1636-46
43. Zhang Y, Zhang Y, Lin QH. [Progesterone-modulated proteins in human endometrial cancer cell line Ishikawa]. Nan fang yi ke da xue xue bao = Journal of Southern Medical University. 2006;26:1110-3
44. Tabaglio T, Low DH, Teo WKL, Goy PA, Cywoniuk P, Wollmann H. et al. MBNL1 alternative splicing isoforms play opposing roles in cancer. Life science alliance. 2018;1:e201800157
45. Félix AJ, Ciudad CJ, Noé V. Functional pharmacogenomics and toxicity of PolyPurine Reverse Hoogsteen hairpins directed against survivin in human cells. Biochemical pharmacology. 2018;155:8-20
46. Woo BJ, Moussavi-Baygi R, Karner H, Karimzadeh M, Yousefi H, Lee S. et al. Integrative identification of non-coding regulatory regions driving metastatic prostate cancer. Cell reports. 2024;43:114764
47. Song F, Hou C, Huang Y, Liang J, Cai H, Tian G. et al. Lactylome analyses suggest systematic lysine-lactylated substrates in oral squamous cell carcinoma under normoxia and hypoxia. Cellular signalling. 2024;120:111228
48. McIntosh LA, Marion MC, Sudman M, Comeau ME, Becker ML, Bohnsack JF. et al. Genome-Wide Association Meta-Analysis Reveals Novel Juvenile Idiopathic Arthritis Susceptibility Loci. Arthritis & rheumatology (Hoboken, NJ). 2017;69:2222-32
49. Daouti S, Latario B, Nagulapalli S, Buxton F, Uziel-Fusi S, Chirn GW. et al. Development of comprehensive functional genomic screens to identify novel mediators of osteoarthritis. Osteoarthritis and cartilage. 2005;13:508-18
50. Pereira E, Tamia-Ferreira MC, Cardoso RS, Mello SS, Sakamoto-Hojo ET, Passos GA. et al. Immunosuppressive therapy modulates T lymphocyte gene expression in patients with systemic lupus erythematosus. Immunology. 2004;113:99-105
51. Zhang W, Niu C, Fu RY, Peng ZY. Mycobacterium tuberculosis H37Rv infection regulates alternative splicing in Macrophages. Bioengineered. 2018;9:203-8
52. O'Connor BP, Danhorn T, De Arras L, Flatley BR, Marcus RA, Farias-Hesson E. et al. Regulation of toll-like receptor signaling by the SF3a mRNA splicing complex. PLoS genetics. 2015;11:e1004932
53. De Arras L, Alper S. Limiting of the innate immune response by SF3A-dependent control of MyD88 alternative mRNA splicing. PLoS genetics. 2013;9:e1003855
54. Li N, Li Y, Han X, Zhang J, Han J, Jiang X. et al. LXR agonist inhibits inflammation through regulating MyD88 mRNA alternative splicing. Frontiers in pharmacology. 2022;13:973612
55. Bigot J, Lalanne AI, Lucibello F, Gueguen P, Houy A, Dayot S. et al. Splicing Patterns in SF3B1-Mutated Uveal Melanoma Generate Shared Immunogenic Tumor-Specific Neoepitopes. Cancer discovery. 2021;11:1938-51
Author contact
![]() Corresponding authors: lshupedu.cn; mdcheng18edu.cn.
Corresponding authors: lshupedu.cn; mdcheng18edu.cn.

 Global reach, higher impact
Global reach, higher impact