3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2023; 14(1):9-23. doi:10.7150/jca.76809 This issue Cite
Research Paper
Six Cell Cycle-related Genes Serve as Potential Prognostic Biomarkers and Correlated with Immune Infiltrates in Hepatocellular Carcinoma
1. State Key Laboratory of Biocontrol, Guangdong Province Key Laboratory of Pharmaceutical Functional Genes, College of Life Sciences, Sun Yat-Sen University, Guangzhou, China.
2. School of Life Sciences, Beijing University of Chinese Medicine, Beijing, China.
3. Department of Central Laboratory, Shenzhen Hospital, Beijing University of Chinese Medicine, Shenzhen, China.
Abstract
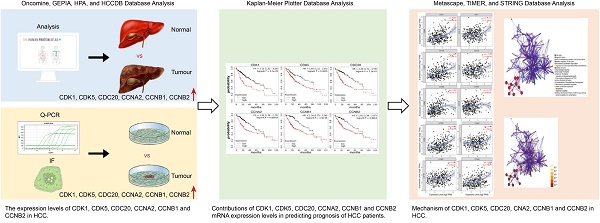
Background: Cell cycle-related genes (CDK1, CDK5, CDC20, CCNA2, CCNB1, and CCNB2) play important roles in the regulation of mitotic cell cycle in eukaryotes. However, the correlation between cell cycle-related genes and tumor-infiltrating and prognosis of hepatocellular carcinoma (HCC) needs further investigation.
Methods: Two public websites, Tumor Immune Estimate Resource (TIMER) and Oncomine, were used to assess the expression levels of cycle-related genes. We also analyzed the protein expression levels of six cell cycle-related genes using the HPA database. In addition, Kaplan-Meier plotter and Gene Expression Profiling Interactive Analysis (GEPIA) database were used to investigate the impact of cell cycle-related gene expression levels on the clinical prognosis of HCC. The correlation between cell cycle-related genes and cancer immune infiltrates was analyzed through TIMER site. Subsequently, GEPIA and TIMER database were used to assess the correlation between the expression of six cell cycle-related genes and polygenic markers in monocytes and macrophages, respectively. The cell cycle-related genes were also analyzed to find the associated genes with the highest alteration frequency, by the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) approaches of Metascape and String database, respectively.
Results: The expression levels of cell cycle-related genes were up-regulated in tumor tissues compared with normal tissues. Subsequently, the expression of high cell cycle-related genes was positively correlated with poor overall survival (OS) and progression-free survival (PFS) in HCC, for CDK1 (OS: HR = 2.15, P = 1.1E-05 PFS: HR = 2.03, P = 2.3E-06), CDK5 (OS: HR = 1.85, P = 0.0035 PFS: HR = 1.26, P = 0.17), CDC20 (OS: HR = 2.49, P = 5.1E-07 PFS: HR = 1.77, P = 0.00012), CCNA2 (OS: HR = 1.92, P = 0.00018 PFS: HR = 1.96, P = 5.2E-06), CCNB1 (OS: HR = 2.34, P = 3.4E-05 PFS: HR = 1.97, P = 5.3E-06), and CCNB2 (OS: HR = 1.91, P = 0.0013 PFS: HR = 1.63, P = 0.0011), respectively. Furthermore, the transcription level of cell cycle-related genes was significantly correlated with immune infiltrating levels of CD4+ T and CD8+ T cells, neutrophils, macrophages, and dendritic cells (DCs) in HCC, respectively. Amongst them, the expression levels of CDK1, CDC20, CCNA2, CCNB1 and CCNB2 manifest strongly correlated with diverse immune marker sets in HCC.
Conclusions: Our results demonstrated that cell cycle-related genes played key roles in the prognosis of HCC. Meanwhile, they were significantly correlated with immune infiltrating levels of CD4+ T cells, CD8+ T cells, neutrophils, macrophages and DCs in HCC, respectively. In addition, CDK1, CDC20, CCNA2, CCNB1 and CCNB2 expressions may be involved in the regulation of monocytes and tumor-associated macrophages (TAMs) in HCC, respectively. These findings strongly suggested that cell cycle-related genes could be used as novel biomarkers for exploring the prognosis and immune cells infiltration of HCC.
Keywords: cell cycle-related genes, hepatocellular carcinoma, tumor-infiltrating, prognostic biomarker, immune

 Global reach, higher impact
Global reach, higher impact