Impact Factor
ISSN: 1837-9664
J Cancer 2025; 16(3):812-820. doi:10.7150/jca.104214 This issue Cite
Review
Research Progress of DNA Methylation Markers for Endometrial Carcinoma Diagnosis
1. Clinical Pathology Department, Shandong Second Medical University, Shandong Province, Weifang, Shandong 261042, P.R. China.
2. Department of Pathology, The seventh Medical Center, Chinese PLA General Hospital, Beijing, 100700, P.R. China.
3. Department of Gynecology and Obstetrics of Qingdao West Coast New Area People's Hospital, Shandong Province, Qingdao, Shandong 266000, P.R. China.
# These authors contributed equally to this work.
Received 2024-9-25; Accepted 2024-11-25; Published 2025-1-1
Abstract
Endometrial carcinoma (EC) is the most common malignancies of the female reproductive system in developed countries and areas. Ultrasound-guided and hysteroscopic samplings are commonly used to diagnose EC. However, clinicians question their diagnostic efficacy and the associated patient discomfort. DNA methylation is the widely studied epigenetic alteration in human tumors, and tumor screening and diagnosis. This review summarized common methods for collecting clinical samples for methylation testing. Furthermore, we analyzed the diagnostic evaluation indices of different methylation marker assays in clinical diagnosis and discussed the challenges of methylation testing in the future application of EC diagnosis.
Keywords: endometrial carcinoma, DNA methylation, epigenetics, diagnostic biomarkers
1. Introduction
Endometrial carcinoma (EC) is the most common malignancy in the female reproductive system in developed countries. According to Global Cancer Statistics 2020, EC is the sixth most common cancer in women, with a global incidence of 417,000 new cases, mostly prevalent between the ages of 65 and 75[1, 2]. Over the past three decades, EC diagnoses have increased by 132% globally, and deaths have nearly doubled[3].
In 1983, Oncol proposed the traditional classification of EC; estrogen-dependent EC (Type I EC) is associated with obesity, hyperlipidemia, and excessive estrogen Estrogen-independent EC (Type II EC) occurs without the aforementioned etiological factors[4]. Type I EC is represented by endometrioid carcinoma, which is characterized by a high degree of differentiation, generally favorable prognosis, slow tumor growth, and low grade and invasiveness. Type II EC encompasses various histological types, including serous, clear cell, and mixed carcinomas, and other rare types. Type II ECs typically exhibit poor prognosis, rapid tumor growth, and high invasiveness, often presenting with metastasis at the time of diagnosis[5]. In 2013, The Cancer Genome Atlas project conducted a large-scale molecular characterization of and proposed a new classification method based on tumor genetic mutations, copy number variations, mRNA expression, methylation profiles, and protein expression. There are four molecular subtypes. The POLE ultramutated type is characterized by a high mutation rate due to mutations in the POLE gene. Microsatellite instability (MSI) hypermutated type exhibits a high level of MSI and a moderate mutation rate. The copy number low (endometrioid) type displays low copy number alterations and is often associated with Type I EC[6]. The copy number high (serous-like) type is characterized by high copy number alterations and is typically correlated with Type II EC and a poorer prognosis. The molecular classification of EC allows clinicians to select appropriate treatments improving patient prognosis[5, 7].
Early diagnosis of EC is associated with better prognostic outcomes; therefore, accurate diagnosis and timely treatment are crucial for its management[8]. Transvaginal ultrasound (TVU) is the preferred early screening and diagnosis option. A study of over 1,000 patients showed that an endometrial thickness of ≥ 5 mm in postmenopausal women had a sensitivity of 96.2% and a negative predictive value of 99.3% for detecting EC. However, TVU specificity was only 51.5%, indicating the need for additional screening to rule out other malignancies[9]. Hysteroscopy allows the direct sampling of suspicious lesions and can be used in TVU-positive and recurrently symptomatic women. Information from endometrial biopsy is often used for preoperative disease staging, is essential for surgical management, and guides the scope of the procedure. However, hysteroscopy also increases the risk of cancer spread, and physical discomfort and false-negative results are common complications[10, 11].
In recent years, AI has shown extensive potential for application in the diagnosis of EC, particularly in areas of pathologic analysis, molecular diagnosis, predictive model construction, and so on. For example, Li et al. developed an artificial intelligence system for screening and diagnosing EC, successfully classifying malignant and benign EC cells[12]. AI could also provide more precise assessments in the molecular classification of EC[13]. Consequently, safer, more effective, and reliable means of screening and early diagnosis in high-risk populations is needed to reduce lethality, improving the clinical prognosis of EC.
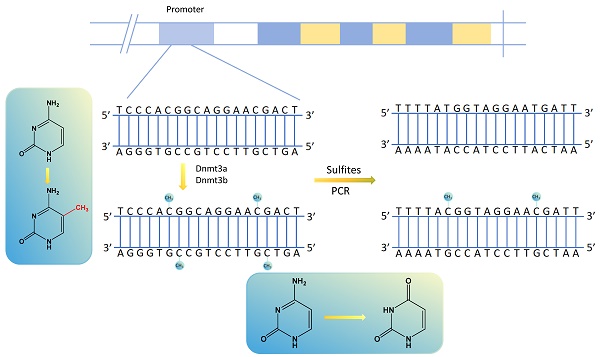
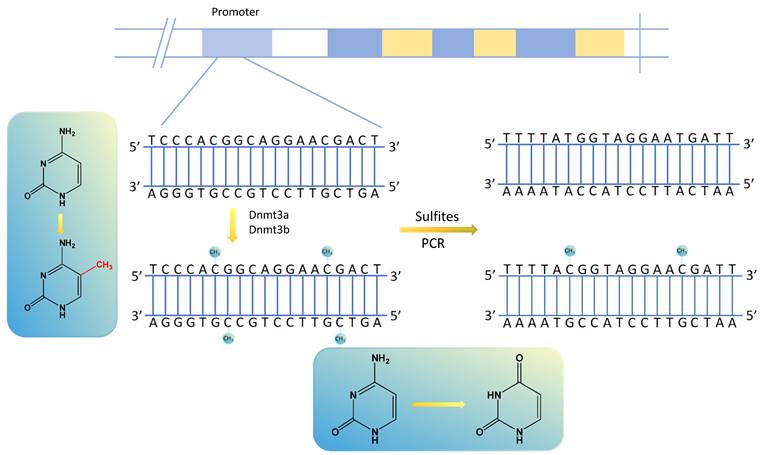
Methylation transfers methyl groups from active methyl compounds to other compounds and occurs in specific proteins or nucleic acids chemically modified to form methylated products[14, 15]. DNA methylation, in which methyltransferases catalyze the transfer of active methyl groups to target chemicals without altering DNA sequence composition, is an important type of epigenetic regulation[16]. It usually occurs in the promoter region of the DNA sequence, and its aberrant expression can result in the aberrant expression of tumor-associated genes in various human tumors[17, 18]. Among the DNA methylation sites, the most prominent manifestation is using DNA methyltransferases to transfer methyl groups to cytosine 5 carbon atom of cytosine-phosphate-guanine (CpG) dinucleotides to form 5-methylcytosine (5mC), interfering with promoter recognition and gene regulation[19]. The regions of aggregated CpG dinucleotides are called CpG islands, and the promoter regions of genes usually contain many CpG islands. Methylated cytosine is unrecognized by sulfite, determining whether and to what extent the DNA is methylated[18] (Fig. 1). There are many ways to detect DNA methylation, including genome-wide methylation detection technologies (WGBS, RRBS, Illumina EPIC BeadChip Microarray, MeDIP-seq and MethylRAD) and site-specific methylation detection technologies (pyrophosphate sequencing, Massarray, BSP and MSP)[20-22].
EC is diagnosed by detecting promoter methylation levels of single or multiple genes in the specimen of endometrium[23]. Genes with aberrant DNA methylation interfere with various biological pathways, such as cell adhesion and proliferation, cell cycle regulation, and apoptosis, contributing to the development and progression of EC[24]. Big data and bioinformatics have revealed that methylation markers can help predicting less aggressive tumors and are suitable for fertility preservation therapy[25]. This review summarized the commonly used clinical tests for methylation sample collection, genes or genomes that play a key role in the organism, and the diagnostic evaluation of methods for detecting methylation in clinical EC diagnostic applications. We also explored the prospects for applying DNA methylation in EC screening and diagnosis.
2. DNA methylation markers for EC
The development and progression of EC involve multiple biological processes and numerous genes. Gene methylation plays a crucial role in regulating the expression of related genes or controlling other genes. By focusing on the CpG islands within gene promoter regions and utilizing next-generation sequencing technology supported by a large number of clinical samples, the methylation status of some specific genes can be observed. From the perspective of EC etiology, we collected data on methylation-related genes in EC and clinical diagnostic information for certain genes to explore their feasibility as early diagnostic markers for EC (Table 1).
2.1 Genes involved in regulating cell proliferation and differentiation
Ras-binding domain family 1 isoform A (RASSF1A) regulates cellular processes such as cell cycle arrest, migration, microtubule stabilization, and pro-apoptosis in response to various stimuli[26]. Hypermethylation of the RASSF1A promoter is frequently associated with poor prognostic characters, including advanced clinical stage, lymph node and/or distant metastasis, and drug resistance[27]. RASSF1A may play a role in cellular proliferation and apoptosis via regulating the RAS-MAPK signaling pathway[28]. Pijnenborg et al. reported that 85% of patients with EC exhibited RASSF1A methylation, and RASSF1A promoter methylation was present in 70% of cases in premenopausal EC[29]. Fiolka et al. found a significant correlation between RASSF1A and higher tumor grade, deeper infiltration of the uterine myometrium, and metastases in the pelvic lymph nodes[30]. As a therapeutic target, 5-aza-2'-deoxycytidine (5-Aza-CdR) may reverse the methylation status of the RASSF1A gene, restored its mRNA and protein expression, and control the growth of EC cell lines by inducing apoptosis[31].
The calcineurin 13 (CDH13) gene is a novel member of the calcineurin superfamily. It is primarily expressed on transmembrane glycoproteins on the surface of epithelial cells and mediates intercellular Ca2+dependent adhesion to maintain normal tissue structure[32]. CDH13 expression in many tumor cell lines inhibits cell proliferation and invasiveness, increases susceptibility to apoptosis, and reduces tumor growth in vivo models[33]. CDH13 hypermethylation is an independent prognostic factor in EC. Early reports have identified significant changes in the methylation of the CDH13 promoter during the development and progression of EC[34]. Yan Sheng and colleagues demonstrated that treatment with 5-Aza-CdR or trichostatin A partially reversed mRNA levels, proving that the methylation of CDH13 is one of the main reasons for its decreased expression in cancer cells[35]. Clinical analysis of EC samples revealed that the methylation level of the CDH13 promoter ranged from 80% to 81.36%, while in atypical hyperplasia samples, it ranged from 50% to 51.7%[36, 37]. Krasnyi's team found that cysteine dioxygenase type 1 (CDO1) and CDH 13 gene methylation levels in EC tissue samples (stage IA) predicted the outcome of drug treatment[38].
Sensitivity, specificity, and AUC values for using gene methylation to diagnose EC
| Gene | AUC | Sensitivity | Specificity | Reference |
|---|---|---|---|---|
| Single-gene | ||||
| CDO1 | 0.842-0.968 | 82.0% | 93.8% | [66, 50, 116] |
| BHLHE22 | 0.95 | 83.7% | 93.7% | [50] |
| CELF4 | 0.94 | 96.0% | 78.7% | [50] |
| ZNF662 | 0.89 | 92.0% | 80% | [50] |
| ZNF454 | 0.938 | 79.55% | 93.42% | [50] |
| CDH13 | 0.67-0.88 | 81.36% | / | [37, 117, 118] |
| RASSF1A | 0.75 | 85.4% | 70% | [37, 117, 119] |
| CELF4 | 0.96 | 96.0% | 78.7% | [31] |
| HAND2 | 0.91 | / | / | [28] |
| ROR2 | 0.665 | / | / | [66] |
| EDNRB | 0.845 | / | / | [66] |
| NDN | 0.985 | / | / | [66] |
| DCAF12L1 | 0.704 | / | / | [120] |
| MSX1 | 0.73 | / | / | [120] |
| Multi-gene | ||||
| CDO1+CELF4 | / | 87.5% | 90.8% | [50] |
| CDO1+BHLHE22 | 0.86 | 92.9% | 77.7% | [121] |
| BHLHE22+CDO1+HAND2 | / | 87% | 86% | [46] |
| BHLHE22+CDO1+TBX5 | / | 89.1% | 88% | [46] |
| CDO1+CELF4+ BHLHE22 | / | 91.8% | 95.5% | [50] |
| CDO1+ ZNF454 | 0.931 | 90.91% | 86.84% | [116] |
| EMX2OS+NBPF8+ SFMBT2 | 0.98 | 97% | 97% | [99] |
| DCAF12L1+MSX1 | 0.867 | / | / | [120] |
Altered gene methylation sites and gene changes after PCR amplification by sulfite treatment.
Heart and neural crest derivatives expressed 2 (HAND2) gene encode a transcription factor that belongs to the basic Helix-Loop-Helix (bHLH) family. It is primarily expressed in cardiac development and neural crest derivatives[39]. HAND2 is crucial in regulating embryonic development, cell proliferation, and differentiation, particularly in the development of the right ventricle of the heart, neural crest cells, and limb buds[40]. HAND2 is present in endometrial stromal cells, where it inhibits ligand-dependent transcriptional activation of estrogen receptor alpha (ERα) and activates interleukin-15 transcription[41]. Reportedly, increased methylation of HAND2 is a hallmark of endometrial precancer, typically correlated with decreased RNA and protein levels. Women with high levels of HAND2 methylation respond less effectively to progesterone therapy for endometrial precancer[42].
The bHLH Family Member e22 (BHLHE22) is a transcriptional repressor and regulates cell differentiation during neuronal development[43]. The expression of BHLHE22 protein was significantly lower in EC than in normal endometrium. High expression of BHLHE22 correlated with the MSI subtype, tumor grade, and patient age, and significantly improved survival outcomes[44, 45]. Furthermore, BHLHE22 overexpression inhibited the proliferation and migration of EC cells[45]. Phui-Ly Liew discovered that the highly methylated panels BHLHE22/CDO1/HAND2 (87.0% sensitivity and 86.0% specificity) and BHLHE22/CDO1/TBX5 (89.1% sensitivity and 80.0% specificity) exhibited significant differences, effectively distinguishing between benign and malignant endometrial lesions[46]. Rui-Lan Huang used methylation-specific PCR (QMSP) on 146 cervical scrapings and found that a panel consisting of any two of the three highly methylated genes BHLHE22, CDO1, and CELF4 demonstrated a sensitivity of 91.8% and a specificity of 95.5%[31]. In a recent study, the team led by Kuo-Chang Wen utilized MPap detection technology and observed that the sensitivity and specificity of the genes CDO1 / BHLHE22 in EC were above 90% and 70%, respectively[47].
As an oncogene, the tumor suppressor activity of phosphatase and tensin homolog (PTEN) primarily depends on its lipid phosphatase function, inhibiting PI3K/AKT activation. Consequently, PTEN regulates various cellular processes, including proliferation, survival, energy metabolism, cell structure, and motility[48]. Khatami's team observed PTEN promoter methylation in 52.0% of EC tumor tissues, compared to 13.6% in non-tumor tissues[49]. Gotoh et al. employed DNA methylome microarray sequencing and found PTEN mutations and clonal expansion of tumor cells in atypical hyperplasia samples[50]. Liew et al. also demonstrated that PTEN mutations were seldomly present in cervical scrapings of normal endometrium (25%) and benign uterine lesions (10%), but adding PTEN mutation testing to the BHLHE22/CDO1-based methylation assay did not enhance the detection efficiency of EC[46].
Adenomatous polyposis coli (APC) is a gene that disrupts the Wnt/β-catenin signaling pathway, preventing it from taking part in organ development, cell proliferation, survival, differentiation, and migration[51]. Zysman's team studied 114 endometrial adenocarcinoma specimens, and illustrated that the frequency of APC hypermethylation was increased in MSI+ endometrial tumors[52]. Ignatov et al. reported that DNA methylation frequency of the APC gene increased from atypical hyperplasia (23.5%) to early-stage EC (77.4%) and then gradually decreased in advanced carcinoma (24.2%)[53]. However, in a recent study, Lou et al. observed hypomethylation in the promoter of APC and upregulation of gene expression in mutant EC samples[54].
The P16 gene belongs to the inhibitor of cyclin-dependent kinase 4 (INK4) gene family. It consists of four members, p16 INK4A, p15 INK4B, p18 INK4C, and p19 INK4D, and possesses biological properties of cell growth inhibition and tumor suppression[55]. Moreover, p16 inhibits cell cycle protein-dependent kinases, leading to G1 cell cycle arrest, whereas methylated p16 leads to tumor development[56, 57]. In a meta-analysis including 264 cases of EC patients, hypermethylation of the p16 gene promoter was associated with an increased risk of EC[58]. Multi-institutional studies have demonstrated that p16 methylation is rare in precancerous lesions, but predominant in advanced EC, and therefore is not indicated for early screening. However, it can be used as a potential prognostic marker[59].
2.2 Genes related to hormone and metabolism
The imbalance of estrogen and progesterone is a significant cause of endometrial carcinogenesis, particularly in Type I EC. Hyperlipidemia and slow fat metabolism are also high-risk factors for EC.
CDO1 is a critical enzyme in cysteine catabolism and vital in physiological processes, including lipid metabolism, organismal growth, and development[60]. CDO1 enhances the production of reactive oxygen species to induce apoptosis. It interacts with peroxisome proliferator-activated receptor γ, activating the key tumor suppressor transcription factor CCAAT/enhancer-binding protein (C/EBP) α, thereby inhibiting tumor progression[61, 62]. CDO1 is a potential tumor suppressor gene. In cancers such as renal cell, breast, and colorectal cancer, the promoter region of CDO1 frequently undergoes hypermethylation, leading to reduced or absent expression[63, 64]. This methylation-induced silencing may allow cancer cells to evade cell death by oxidative stress, promoting tumor survival and progression[65]. Clinical data suggest that CDO1 is significantly hypermethylated in Types I and II of EC, and it can serve as a diagnostic marker to distinguish between cancerous and normal tissues[66, 67].
Estrogen receptor 1 (ESR1) is the gene that encodes ERα. This nuclear receptor predominantly regulates gene expression by binding to estrogens, such as 17β-estradiol. It plays a vital role in the development of the reproductive system, bone health, cardiovascular function, and the growth and differentiation of breast tissue[68, 69]. SNAI2 promotes ESR1 methylation by recruiting DNA methyltransferase 3B rather than DNA methyltransferase 1 in ERα-positive breast cancer cells and may contribute to cell adhesion and junctions[70]. However, the role of ESR1 methylation in the development of EC remains controversial. Carla Bartosch and colleagues analyzed ERα and PRA promoter methylation in 45 cases of EC and concluded that the methylation of these genes plays a limited role in the etiology of the disease[71]. Vanessa Todorow demonstrated that in three out of five EC cell lines, the promoter region of ESR1 was methylated, suggesting that ESR1 methylation may influence EC development[72]. Results of bioinformatics analysis also support this viewpoint[73, 74]. PIWIL1 mediates the ERα signaling pathway involved in E2-stimulated carcinoma cell proliferation, which may be one of the mechanisms by which ESR1 methylation contributes to EC progression[75].
2.3 Genes involved in expression regulation and epigenetics
The impairment of gene repair function and the body's immune system affect cancer progression in terms of prevention and elimination. Meanwhile, focusing on the methylation changes of the genes themselves, it is important to consider that alteration of genes that promote methylation can also impact EC development and progression. These genes have broader effects and can assist in the early diagnosis and prognostic assessment when used alongside methylation biomarkers.
The human mutL homolog 1 (hMLH1) gene undergoes DNA mismatch repair (MMR) gene mutations in Lynch syndrome, commonly used in the pathologic diagnosing of rectal carcinoma and ECs[6, 76]. Two-thirds of EC exhibit dMMR, mainly caused by methylation of the MLH1 promoter[77]. Three-quarters of EC patients aged 36-59 exhibited methylation of the MLH1 gene. Annukka Pasanen et al. found that 76% of 244 dMMR cases were associated with methylation[78, 79]. Kahn et al. analyzed that 86.3% (1016/1159) patients' loss of MLH1 staining were due to MLH1 methylation[80]. Clinical studies have found that tumor size is significantly associated with MLH1 methylation[81, 82]. Thus, hMLH1 methylation testing may be used as an early clinical screening and prognostic indicator for patients with EC[34].
Methyl-CpG-binding protein 2 (MeCP2) is a protein essential to regulating gene expression and DNA methylation[83]. It was initially studied extensively for its involvement in Rett syndrome, a neurodevelopmental disorder that affects brain development[84]. MeCP2 binds to methylated CpG dinucleotides and interacts with other proteins to either repress or activate the expression of specific genes[85]. In EC research, MeCP2 is associated with decreased promoter methylation, leading to higher expression levels and promoting methylation of other genes. Gene mutations in MeCP2 are associated with a favorable prognosis[86, 87]. Yuning Xiong et al. discovered that MeCP2 specifically binds to and methylates the hMLH1 promoter[88]. Yongli Chu's team found that MeCP2 plays a key role in the silencing of the progesterone receptor-B (PR-B) gene, suggesting that epigenetic reactivation of PR-B could be explored as a potential strategy to sensitize PR-B-negative EC to progestin therapy[89].
CUGBP Elav-like family member 4 (CELF4) is a member of the CELF protein family, which is involved in regulating processes such as alternative RNA splicing, post-transcriptional regulation, translation, and mRNA degradation[90]. Similar to CDO1 and BHLHE22, CELF4 was also first identified and reported by Huang et al. to exhibit abnormally elevated methylation in EC. Statistical analysis showed that CELF4 had a sensitivity of 96.0%, a specificity of 78.7%, and an AUC of 0.94[31]. In recent studies, researchers have shown a preference for the combined detection of CDO1 and CELF4 methylation, with sensitivity ranging from 84.9% to 87.5% and specificity from 86.6% to 95.9%[67, 91, 92]. Additionally, Zhao et al. and Kong et al. both suggested that combining BMI index and the joint use of TVU could improve the prediction of EC screening[67, 93]. However, a recent study also indicated that the combined use of CDO1 and CELF4 did not lead to better screening outcomes, with AUC values of only 0.6000 and 0.5286, respectively[94].
3. Sample collection for DNA methylation detection
In populations with a low incidence of EC, TVU followed by endometrial biopsy is the most cost-effective strategy[95]. Surgical anesthesia can greatly reduce the pain associated with endometrial sampling. Nonetheless, it is difficult to ensure that patients can be painlessly sampled in outpatients for large-scale screening[96]. With the advancement in molecular methylation technology, the method of collecting vaginal fluid, urine, and blood samples to detect tumor methylation markers and understand their prognosis is gradually being applied in clinical diagnosis (Table 2).
Changes in AUC values across genes for different clinical modes of endometrial sampling.
| Gene | Vaginal fluid (Tao brush) | Vaginal fluid (Pap brush) | Vaginal fluid (tampons) | Urine | Blood |
|---|---|---|---|---|---|
| BHLHE22 | 0.878 | ||||
| CDO1 | 0.842 | ||||
| TBX5 | 0.703 | ||||
| HAND2 | 0.767 | ||||
| MME | 0.7 | ||||
| PCDHGB 7 | 0.83 | 0.86 | |||
| HIST1H4F | 0.951 | ||||
| RASSF1 | 0.938 | 0.75 | |||
| HTR1B | 0.82 | ||||
| HOXA9 | 0.74 | ||||
| GHSR | 0.95 | ||||
| SST | 0.92 | ||||
| ZIC1 | 0.86 | ||||
| ZSCAN12/OXT | 0.99 | ||||
| MLH1 | 0.96 | ||||
| CDH13 | 0.67 | ||||
| ADCYAP1 | 0.86 | 0.67 | |||
| ASCL2 | 0.86 | 0.68 | |||
| HS3HT2 | 0.81 | 0.67 | |||
| HTR1B | 0.8 | 0.82 | |||
| MME | 0.68 | 0.7 | |||
| NYP | 0.86 | 0.66 | |||
| GTF2A1 | 0.76 | 0.43 | |||
| HAAO | 0.68 | ||||
| HSP2A | 0.54 | ||||
| Reference | [46, 122-124] | [122, 123] | [37] | [111] | [114, 125] |
3.1 Vaginal fluid collection
The collection of vaginal secretions can capture exfoliated endometrial cells, allowing for pathological and molecular diagnostics. Though this approach is non-invasive and relatively simple to perform, its sensitivity remains relatively low, which may result in false-negative outcomes, thus limiting its accuracy in detecting early-stage EC. Commonly used method to collect vaginal fluid is tampon, which significantly reduces the pain caused by endometrial sampling. A clinical trial used a visceral analog scale to compare pain associated with tampon collection, tampon brushing, and endometrial biopsy. Concludingly, pain linked to tampon collection was easy to tolerate by the patients while still achieving good diagnostic efficiency[97-99]. Moreover, Bakkum-Gamez et al. reported that methylation of vaginal fluid collected from tampons for detecting EC exhibited high sensitivity and strong specificity, and adding EDTA buffer to the PBS-based tampon buffer improved sensitivity and specificity[100].
3.2 Endometrial cell collection with special collectors
Endometrial cell collection is the method for direct collection of endometrial cells. The widely used devices to collect endometrial samples include ceramic and pasteurized brushes. The ceramic brush method is one of the most widely used endometrial sampling devices[101]. In a meta-analysis of more than 700 individuals, the ceramic brush method of endometrial cytology sampling was demonstrated to be less invasive, less expensive, and more suitable than endometrial biopsy for screening and diagnosing precancer and malignancy[102, 103]. Another study illustrated that the sensitivity of endometrial cells obtained by ceramic brushing was comparable to that of biopsy tissue for detecting atypical hyperplasia and EC[104] (Table 2). Compared to collecting vaginal fluid, the use of an endometrial brush allows for the direct collection of abnormal cells, offering higher sensitivity and specificity. This approach aids in detecting early cancerous changes and reduces the rate of false-negative results.
3.3 The urine collection
Urine testing enables comprehensive screening, improves patient autonomy, and ensures high diagnostic efficiency[105, 106]. Urine testing can obtain DNA from locally shed cellular tumors and tumor-free cells, such as cells excreted through the kidneys Second-generation sequencing, microRNA detection, and quantitative SWATH analysis have recently been used to collect patients' urine to diagnose EC[107-109]. Wever et al. set up a value of urine, cervicovaginal self-sampling, and the clinician's cervical sampling of the three groups of sampling methods to carry out methylation detection[110]. The quantitative methylation-specific PCR was used to detect nine DNA methylation markers; significantly higher methylation levels were found in all groups compared with healthy controls[110]. Another study divided the urine samples into three fractions (whole urine, urine sediment, and urine supernatant) and analyzed DNA methylation markers. All DNA methylation markers exhibited increased methylation levels in all urine fractions of EC patients compared to healthy controls[111]. Whole urine samples demonstrated the highest ability to discriminate patients from controls[111]. However, using urine specimens for EC diagnostic studies is not widely used as compared to vaginal fluid specimens, possibly due to the inability to directly obtain endometrial cells, while its screening and diagnostic accuracy remain to be discussed.
3.4 Patient blood or plasma samples collection
Patient blood or plasma samples allow monitoring of small fragments of DNA shed into the bloodstream by the tumor cells, including circulating extracellular nucleic acid, circulating tumor DNA (ctDNA), and circulating tumor cells[112]. DNA from these samples is analyzed to detect point mutations, copy number alterations, gene fusions, and DNA methylation. This method demonstrated good applicability for cancer diagnosis, determining prognosis, targeting gene-specific therapies, monitoring/predicting disease recurrence, and treatment response[113]. Researchers have established the digital droplet PCR (ddPCR) method to detect hypermethylated ctDNA in plasma of EC patients with high analytical specificity and sensitivity in retrospective and cohort studies[114]. The ddPCR exhibited good clinical diagnostic performance in other cancer studies, such as ovarian and colorectal cancer[115]. However, diagnosis of EC by detecting DNA methylation in blood has not been studied on a large scale in clinical settings. Early detection of EC by using blood samples may not be suitable for screening of earlier stage of EC.
Despite various methods for EC screening via DNA methylation testing, a single test cannot fully replace cervical biopsy. Thus, a combined approach is needed to enhance screening accuracy. Herzog et al. focused on specific DNA regions within genes such as ZSCAN12 and GYPC using qMSP (quantitative methylation-specific PCR) in the WID quantitative EC (WID-qEC) assay, demonstrating high sensitivity and specificity in detecting different stages and types of EC[20]. Therefore, we should not be limited to DNA methylation alone. Diversifying biomarker detection and combining multiple methods can significantly improve screening accuracy.
4. Conclusion
Epigenetics and DNA methylation are novel and promising techniques for biomarker discovery and subsequent screening. Genome-wide and site-specific methylation assays play an important role in screening potential cancer gene methylation sites and in the targeted assessment of gene methylation levels. Clinical studies in EC have reported that detecting single or combined gene methylation markers is comparable to the diagnostic accuracy of endometrium biopsy. It is more acceptable to patients due to its convenience, speed, and painless sampling. However, there are many inconsistencies regarding the results of DNA methylation abnormalities of these tumor suppressor genes in EC. This is due to environment, lifestyle, and individual differences. Additionally, aside from the currently popular cancer gene methylation markers, most gene methylation studies are in the controversial stage. Large numbers of clinical samples are needed to validate and resolve controversies. In conclusion, research on DNA methylation could provide rich and complex information on epigenetic gene regulation in EC and its precursors and offer technical support for better and faster methylation assays in EC screening and diagnosis.
Funding
Project supported by Capital's Funds for Health Improvement and Research (No. 2024-2-5081).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Sung H, Ferlay J, Siegel RL. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mor tality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-49
2. Makker V, MacKay H, Ray-Coquard I. et al. Endometrial cancer. Nat Rev Dis Primers. 2021;7:88
3. Gu B, Shang X, Yan M. et al. Variations in incidence and mortality rates of endometrial cancer at t he global, regional, and national levels, 1990-2019. Gynecol Oncol. 2021;161:573-80
4. Bokhman JV. Two pathogenetic types of endometrial carcinoma. Gynecol Oncol. 1983;15:10-7
5. Berek JS, Matias-Guiu X, Creutzberg C. et al. FIGO staging of endometrial cancer: 2023. J Gynecol Oncol. 2023;34:e85
6. Cancer Genome Atlas Research N, Kandoth C, Schultz N. et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67-73
7. Murali R, Soslow RA, Weigelt B. Classification of endometrial carcinoma: more than two types. Lancet Oncol. 2014;15:e268-78
8. Expert Panel on GYN, Imaging OB, Robbins JB. et al. ACR Appropriateness Criteria® Abnormal Uterine Bleeding. J Am Coll Radiol. 2020;17:S336-S45
9. Long B, Clarke MA, Morillo ADM, Wentzensen N, Bakkum-Gamez JN. Ultrasound detection of endometrial cancer in women with postmenopausa l bleeding: Systematic review and meta-analysis. Gynecol Oncol. 2020;157:624-33
10. Polyzos NP, Mauri D, Tsioras S. et al. Intraperitoneal dissemination of endometrial cancer cells after hyster oscopy: a systematic review and meta-analysis. Int J Gynecol Cancer. 2010;20:261-7
11. Williams PM, Gaddey HL. Endometrial Biopsy: Tips and Pitfalls. Am Fam Physician. 2020;101:551-6
12. Li Q, Wang R, Xie Z. et al. Clinically Applicable Pathological Diagnosis System for Cell Clumps in Endometrial Cancer Screening via Deep Convolutional Neural Networks. Cancers. 2022;14:4109
13. Darbandsari A, Farahani H, Asadi M. et al. AI-based histopathology image analysis reveals a distinct subset of en dometrial cancers. Nature communications. 2024;15:4973
14. Huang E, Chen L. RNA N6-methyladenosine modification in female reproductive biology and pathophysiology. Cell Commun Signal. 2023;21:53
15. Chang S, Yim S, Park H. The cancer driver genes IDH1/2, JARID1C/ KDM5C, and UTX/ KDM6A: crosst alk between histone demethylation and hypoxic reprogramming in cancer metabolism. Exp Mol Med. 2019;51:1-17
16. Dai X, Ren T, Zhang Y, Nan N. Methylation multiplicity and its clinical values in cancer. Expert Rev Mol Med. 2021;23:e2
17. Deng J-Y, Wu X-Q, He W-J. et al. Targeting DNA methylation and demethylation in diabetic foot ulcers. J Adv Res. 2023;54:119-31
18. Jones MJ, Goodman SJ, Kobor MS. DNA methylation and healthy human aging. Aging Cell. 2015;14:924-32
19. Wang X, Kong X, Feng X, Jiang D-S. Effects of DNA, RNA, and Protein Methylation on the Regulation of Ferr optosis. Int J Biol Sci. 2023;19:3558-75
20. Herzog C, Marín F, Jones A. et al. A Simple Cervicovaginal Epigenetic Test for Screening and Rapid Triage of Women With Suspected Endometrial Cancer: Validation in Several Coh ort and Case/Control Sets. JCO. 2022;40:3828-38
21. Ma Y, Long C, Liu G. et al. WGBS combined with RNA-seq analysis revealed that Dnmt1 affects the me thylation modification and gene expression changes during mouse oocyte vitrification. Theriogenology. 2022;177:11-21
22. Gong T, Borgard H, Zhang Z. et al. Analysis and Performance Assessment of the Whole Genome Bisulfite Sequ encing Data Workflow: Currently Available Tools and a Practical Guide to Advance DNA Methylation Studies. Small Methods. 2022;6:e2101251
23. Zhao S, Chen L, Zang Y. et al. Endometrial cancer in Lynch syndrome. Int J Cancer. 2022;150:7-17
24. Bhootra S, Jill N, Shanmugam G, Rakshit S, Sarkar K. DNA methylation and cancer: transcriptional regulation, prognostic, an d therapeutic perspective. Med Oncol. 2023;40:71
25. Makabe T, Arai E, Hirano T. et al. Genome-wide DNA methylation profile of early-onset endometrial cancer: its correlation with genetic aberrations and comparison with late-ons et endometrial cancer. Carcinogenesis. 2019;40:611-23
26. Bin Y, Ding Y, Xiao W, Liao A. RASSF1A: A promising target for the diagnosis and treatment of cancer. Clin Chim Acta. 2020;504:98-108
27. Malpeli G, Innamorati G, Decimo I. et al. Methylation Dynamics of RASSF1A and Its Impact on Cancer. Cancers. 2019;11:959
28. Velasco A, Pallares J, Santacana M. et al. Promoter hypermethylation and expression of sprouty 2 in endometrial c arcinoma. Hum Pathol. 2011;42:185-93
29. Pijnenborg JMA, Dam-de Veen GC, Kisters N. et al. RASSF1A methylation and K-ras and B-raf mutations and recurrent endome trial cancer. Ann Oncol. 2007;18:491-7
30. Fiolka R, Zubor P, Janusicova V. et al. Promoter hypermethylation of the tumor-suppressor genes RASSF1A, GSTP1 and CDH1 in endometrial cancer. Oncology reports. 2013;30:2878-86
31. Huang R-L, Su P-H, Liao Y-P. et al. Integrated Epigenomics Analysis Reveals a DNA Methylation Panel for En dometrial Cancer Detection Using Cervical Scrapings. Clin Cancer Res. 2017;23:263-72
32. Yang J, Niu H, Huang Y, Yang K. A Systematic Analysis of the Relationship of CDH13 Promoter Methylatio n and Breast Cancer Risk and Prognosis. PLoS One. 2016;11:e0149185
33. Andreeva AV, Kutuzov MA. Cadherin 13 in cancer. Genes Chromosomes Cancer. 2010;49:775-90
34. Seeber LMS, Zweemer RP, Marchionni L. et al. Methylation profiles of endometrioid and serous endometrial cancers. Endocr Relat Cancer. 2010;17:663-73
35. Sheng Y, Wang H, Liu D. et al. Methylation of tumor suppressor gene CDH13 and SHP1 promoters and their epigenetic regulation by the UHRF1/PRMT5 complex in endometrial carc inoma. Gynecol Oncol. 2016;140:145-51
36. Dvorak O, Ndukwe M, Slavickova M, Laco J, Spacek J. DNA methylation of selected tumor suppressor genes in endometrial hype rplasia. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2024;168:68-73
37. Bakkum-Gamez JN, Sherman ME, Slettedahl SW. et al. Detection of endometrial cancer using tampon-based collection and meth ylated DNA markers. Gynecol Oncol. 2023;174:11-20
38. Krasnyi AM, Gadzhieva LT, Kokoeva DN. et al. Analysis of CDO1, PITX2, and CDH13 Gene Methylati on in Early Endometrial Cancer for Prediction of Medical Treatment Out comes. International journal of molecular sciences. 2024;25:4892
39. Tamura M, Amano T, Shiroishi T. The Hand2 Gene Dosage Effect in Developmental Defects and Human Congen ital Disorders. Curr Top Dev Biol. 2014;110:129-52
40. Xin M, Olson EN, Bassel-Duby R. Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat Rev Mol Cell Biol. 2013;14:529-41
41. Marinić M, Mika K, Chigurupati S, Lynch VJ. Evolutionary transcriptomics implicates HAND2 in the origins of implantation and regulation of gestation length. eLife. 2021;10:e61257
42. Yin XH, Shi M, Wu H, Zhou L, Xu B. Role of HAND2 gene and protein expression in endometrial carcinoma. Eur J Gynaecol Oncol. 2017;38:95-101
43. Ross SE, McCord AE, Jung C. et al. Bhlhb5 and Prdm8 form a repressor complex involved in neuronal circuit assembly. Neuron. 2012;73:292-303
44. Yin C, Wang M, Wang Y. et al. BHLHE22 drives the immunosuppressive bone tumor microenvironment and a ssociated bone metastasis in prostate cancer. J Immunother Cancer. 2023;11:e005532
45. Darmawi Chen L-Y, Su P-H et al. BHLHE22 Expression Is Associated with a Proinflammatory Immune Microen vironment and Confers a Favorable Prognosis in Endometrial Cancer. International journal of molecular sciences. 2022;23:7158
46. Liew P-L, Huang R-L, Wu T-I. et al. Combined genetic mutations and DNA-methylated genes as biomarkers for endometrial cancer detection from cervical scrapings. Clin Epigenetics. 2019;11:170
47. Wen K-C, Huang R-L, Chen L-Y. et al. Endometrial Cancer Detection Using a Cervical DNA Methylation Assay (M Pap) in Women with Abnormal Uterine Bleeding: A Multicenter Hospital-B ased Validation Study. Cancers. 2022;14:4343
48. Worby CA, Dixon JE. PTEN. Annu Rev Biochem. 2014;83:641-69
49. Khatami F, Shahriari S, Aminimoghaddam S. et al. PTEN promoter methylation and expression in endometrial cancer tissues. Epigenomics. 2023;15:507-16
50. Gotoh O, Sugiyama Y, Tonooka A. et al. Genetic and epigenetic alterations in precursor lesions of endometrial endometrioid carcinoma. J Pathol. 2024;263:275-287
51. Ghazanfari T, Asaadi Tehrani G, Maziri P. The Relationship between the Methylation of Promoter Regions of Tumor Suppressor Genes PTEN and APC with Endometrial Cancer. Asian Pac J Cancer Prev. 2019;20:2259-65
52. Zysman M, Saka A, Millar A. et al. Methylation of adenomatous polyposis coli in endometrial cancer occurs more frequently in tumors with microsatellite instability phenotype. Cancer Res. 2002;62:3663-6
53. Ignatov A, Bischoff J, Ignatov T. et al. APC promoter hypermethylation is an early event in endometrial tumorig enesis. Cancer Sci. 2010;101:321-7
54. Lou J, Chu X, Yang X, Jamil M, Zhu H. Deciphering DNA repair gene mutational landscape in uterine corpus end ometrial carcinoma patients using next generation sequencing. Am J Cancer Res. 2024;14:210-26
55. Serra S, Chetty R. p16. J Clin Pathol. 2018;71:853-8
56. Rayess H, Wang MB, Srivatsan ES. Cellular senescence and tumor suppressor gene p16. Int J Cancer. 2012;130:1715-25
57. Bihl MP, Foerster A, Lugli A, Zlobec I. Characterization of CDKN2A(p16) methylation and impact in colorectal c ancer: systematic analysis using pyrosequencing. J Transl Med. 2012;10:173
58. Hu Z-y, Tang L-d, Zhou Q, Xiao L, Cao Y. Aberrant promoter hypermethylation of p16 gene in endometrial carcinom a. Tumour Biol. 2015;36:1487-91
59. Kommoss FKF, Mar L-M, Howitt BE. et al. High-Grade Endometrial Stromal Sarcomas With YWHAE::NUTM2 Gene Fusion Exhibit Recurrent CDKN2A Alterations and Absence of p16 Staining is a Poor Prognostic Marker. Mod Pathol. 2023;36:100044
60. Chen M, Zhu J-Y, Mu W-J, Guo L. Cysteine dioxygenase type 1 (CDO1): Its functional role in physiologic al and pathophysiological processes. Genes Dis. 2022;10:877-90
61. Yamashita K, Hosoda K, Nishizawa N, Katoh H, Watanabe M. Epigenetic biomarkers of promoter DNA methylation in the new era of ca ncer treatment. Cancer Sci. 2018;109:3695-706
62. Deng P, Chen Y, Ji N. et al. Cysteine dioxygenase type 1 promotes adipogenesis via interaction with peroxisome proliferator-activated receptor gamma. Biochemical and Biophysical Research Communications. 2015;458:123-7
63. Yang J, Sun L, Liu X-Y. et al. Targeted demethylation of the CDO1 promoter based on CRISPR system inh ibits the malignant potential of breast cancer cells. Clin Transl Med. 2023;13:e1423
64. Yokoi K, Harada H, Yokota K. et al. Epigenetic Status of CDO1 Gene May Reflect Chemosensitivity in Colon C ancer with Postoperative Adjuvant Chemotherapy. Ann Surg Oncol. 2018;26:406-14
65. Chen X, Poetsch A. The Role of Cdo1 in Ferroptosis and Apoptosis in Cancer. Biomedicines. 2024;12:918
66. Liu J, Wan Y, Li S. et al. Identification of aberrantly methylated differentially expressed genes and associated pathways in endometrial cancer using integrated bioinf ormatic analysis. Cancer Med. 2020;9:3522-36
67. Kong LH, Xiao XP, Wan R. et al. The role of DNA methylation in the screening of endometrial cancer in postmenopausal women. Zhonghua Yi Xue Za Zhi. 2023;103:907-12
68. Grinshpun A, Chen V, Sandusky ZM, Fanning SW, Jeselsohn R. ESR1 activating mutations: From structure to clinical application. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 2023;1878:188830
69. Betz M, Massard V, Gilson P. et al. ESR1 Gene Mutations and Liquid Biopsy in ER-Positive Breast Can cers: A Small Step Forward, a Giant Leap for Personalization of Endocr ine Therapy? Cancers. 2023;15:5169
70. Li J-W, Deng Q-M, Zhu J-L. et al. Methylation of ESR1 promoter induced by SNAI2-DNMT3B complex pr omotes epithelial-mesenchymal transition and correlates with poor prog nosis in ERα-positive breast cancers. MedComm (2020). 2023;4:e403
71. Bartosch C, Monteiro-Reis S, Vieira R. et al. Endometrial Endometrioid Carcinoma Metastases Show Decreased ER-Alpha and PR-A Expression Compared to Matched Primary Tumors. PLoS One. 2015;10:e0134969
72. Toderow V, Rahmeh M, Hofmann S. et al. Promotor analysis of ESR1 in endometrial cancer cell lines, endometria l and endometriotic tissue. Arch Gynecol Obstet. 2017;296:269-76
73. Li H, Zhou Q, Wu Z, Lu X. Identification of novel key genes associated with uterine corpus endom etrial carcinoma progression and prognosis. Ann Transl Med. 2023;11:100 -
74. He D, Wang X, Zhang Y. et al. DNMT3A/3B overexpression might be correlated with poor patient surviva l, hypermethylation and low expression of ESR1/PGR in endometrioid car cinoma: an analysis of The Cancer Genome Atlas. Chin Med J (Engl). 2019;132:161-70
75. Chen Z, Yang H-J, Lin Q. et al. Estrogen-ERα signaling and DNA hypomethylation co-regulate expression of stem cell protein PIWIL1 in ERα-positive endometrial cancer cells. Cell Commun Signal. 2020;18:84
76. Yurgelun MB, Kulke MH, Fuchs CS. et al. Cancer Susceptibility Gene Mutations in Individuals With Colorectal Ca ncer. J Clin Oncol. 2017;35:1086-95
77. Zhang K, Liu Y, Liu X. et al. Clinicopathological significance of multiple molecular features in und ifferentiated and dedifferentiated endometrial carcinomas. Pathology. 2021;53:179-86
78. Hitchins MP, Alvarez R, Zhou L. et al. MLH1-methylated endometrial cancer under 60 years of age as the "senti nel" cancer in female carriers of high-risk constitutional MLH1 epimut ation. Gynecol Oncol. 2023;171:129-40
79. Pasanen A, Loukovaara M, Bützow R. Clinicopathological significance of deficient DNA mismatch repair and MLH1 promoter methylation in endometrioid endometrial carcinoma. Mod Pathol. 2020;33:1443-52
80. Kahn RM, Gordhandas S, Maddy BP. et al. Universal endometrial cancer tumor typing: How much has immunohistoche mistry, microsatellite instability, and MLH1 methylation improved the diagnosis of Lynch syndrome across the population? Cancer. 2019;125:3172-83
81. Soylemez T, Kir G, Olgun ZC. et al. The Correlation of Histopathologic Parameters With Mismatch Repair Pro tein-deficient Subgroups and MLH1 Methylation in Endometrial Carcinoma s. Int J Gynecol Pathol. 2022;41:484-95
82. Timmerman S, Van Rompuy AS, Van Gorp T. et al. Analysis of 108 patients with endometrial carcinoma using the PROMISE classification and additional genetic analyses for MMR-D. Gynecol Oncol. 2020;157:245-51
83. Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in ge nomic chromatin. Cell. 1997;88:471-81
84. Amir RE, Van den Veyver IB, Wan M. et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methy l-CpG-binding protein 2. Nat Genet. 1999;23:185-8
85. Cohen S, Gabel HW, Hemberg M. et al. Genome-wide activity-dependent MeCP2 phosphorylation regulates nervous system development and function. Neuron. 2011;72:72-85
86. Wang Y, Zhang Y, Wang F. et al. Bioinformatics analysis of prognostic value and immunological role of MeCP2 in pan-cancer. Sci Rep. 2022;12:18518
87. Shen P-C, Wang Y-F, Chang H-C. et al. Developing a novel DNA methylation risk score for survival and identif ication of prognostic gene mutations in endometrial cancer: a study ba sed on TCGA data. Jpn J Clin Oncol. 2022;52:992-1000
88. Xiong Y, Dowdy SC, Eberhardt NL, Podratz KC, Jiang S-W. hMLH1 promoter methylation and silencing in primary endometrial cancer s are associated with specific alterations in MBDs occupancy and histo ne modifications. Gynecol Oncol. 2006;103:321-8
89. Chu Y, Wang Y, Zhang G. et al. Chromatin composition alterations and the critical role of MeCP2 for e pigenetic silencing of progesterone receptor-B gene in endometrial can cers. Cell Mol Life Sci. 2014;71:3393-408
90. Salamon I, Park Y, Miškić T. et al. Celf4 controls mRNA translation underlying synaptic development in the prenatal mammalian neocortex. Nature communications. 2023;14:6025
91. Wang L, Dong L, Xu J. et al. Hypermethylated CDO1 and ZNF454 in Cytological Specimens as Screening Biomarkers for Endometrial Cancer. Front Oncol. 2022;12:714663
92. Cai B, Du J, Wang Y. et al. The endometrial cancer detection using non-invasive hypermethylation o f CDO1 and CELF4 genes in women with postmenopausal blee ding in Northwest China. Cytojournal. 2024;21:15
93. Zhao X, Yang Y, Fu Y, Lv W, Xu D. DNA methylation detection is a significant biomarker for screening end ometrial cancer in premenopausal women with abnormal uterine bleeding. International Journal of Gynecologic Cancer. 2024;34:1165-71
94. Ding H, Wang J, Zhao X. et al. Combination of circulating tumor cells, lncRNAs and DNA methylation fo r the diagnosis of endometrial carcinoma. Oncol Lett. 2024;28:545
95. Dijkhuizen FPHLJ, Mol BWJ, Brölmann HAM, Heintz APM. Cost-effectiveness of the use of transvaginal sonography in the evalua tion of postmenopausal bleeding. Maturitas. 2003;45:275-82
96. Rauf R, Shaheen A, Sadia S. et al. Outpatient endometrial biopsy with Pipelle vs diagnostic dilatation an d curettage. J Ayub Med Coll Abbottabad. 2014;26:145-8
97. Bagaria M, Wentzensen N, Clarke M. et al. Quantifying procedural pain associated with office gynecologic tract s ampling methods. Gynecol Oncol. 2021;162:128-33
98. Woolderink JM, De Bock GH, van Hemel BM. et al. Feasibility of endometrial sampling by vaginal tampons in women with L ynch syndrome. BMC Womens Health. 2020;20:54
99. Bramblet RM, Bakkum-Gamez JN, Slettedahl SW. et al. Methylated DNA Markers for Sporadic Colorectal and Endometrial Cancer Are Strongly Associated with Lynch Syndrome Cancers. Cancer Prev Res (Phila). 2023;16:611-20
100. Bakkum-Gamez JN, Wentzensen N, Maurer MJ. et al. Detection of endometrial cancer via molecular analysis of DNA collecte d with vaginal tampons. Gynecol Oncol. 2015;137:14-22
101. Iavazzo C, Vorgias G, Mastorakos G. et al. Uterobrush method in the detection of endometrial pathology. Anticancer Res. 2011;31:3469-74
102. Raffone A, Raimondo D, Raspollini A. et al. Accuracy of cytological examination of Tao brush endometrial sampling in diagnosing endometrial premalignancy and malignancy. Int J Gynaecol Obstet. 2022;159:615-21
103. Du J, Li Y, Lv S. et al. Endometrial sampling devices for early diagnosis of endometrial lesion s. J Cancer Res Clin Oncol. 2016;142:2515-22
104. DeJong SR, Bakkum-Gamez JN, Clayton AC. et al. Tao brush endometrial cytology is a sensitive diagnostic tool for canc er and hyperplasia among women presenting to clinic with abnormal uter ine bleeding. Cancer Med. 2021;10:7040-7
105. Chen CK, Liao J, Li MS, Khoo BL. Urine biopsy technologies: Cancer and beyond. Theranostics. 2020;10:7872-88
106. Hentschel AE, van den Helder R, van Trommel NE. et al. The Origin of Tumor DNA in Urine of Urogenital Cancer Patients: Local Shedding and Transrenal Excretion. Cancers. 2021;13:535
107. Costas L, Onieva I, Pelegrina B. et al. Evaluation of Somatic Mutations in Urine Samples as a Noninvasive Meth od for the Detection and Molecular Classification of Endometrial Cance r. Clin Cancer Res. 2023;29:3681-90
108. Njoku K, Pierce A, Geary B. et al. Quantitative SWATH-based proteomic profiling of urine for the identifi cation of endometrial cancer biomarkers in symptomatic women. Br J Cancer. 2023;128:1723-32
109. Hanžek A, Siatka C, Duc A-CE. Extracellular urinary microRNAs as non-invasive biomarkers of endometr ial and ovarian cancer. J Cancer Res Clin Oncol. 2023;149:7981-93
110. Wever BMM, van den Helder R, van Splunter AP. et al. DNA methylation testing for endometrial cancer detection in urine, cer vicovaginal self-samples and cervical scrapes. Int J Cancer. 2023;153:341-51
111. van den Helder R, Wever BMM, van Trommel NE. et al. Non-invasive detection of endometrial cancer by DNA methylation analys is in urine. Clin Epigenetics. 2020;12:165
112. Nikanjam M, Kato S, Kurzrock R. Liquid biopsy: current technology and clinical applications. J Hematol Oncol. 2022;15:131
113. Pomerantz T, Brooks R. Circulating Tumor DNA (ctDNA) and Its Role in Gynecologic Malignancies. Curr Treat Options Oncol. 2024;25:510-22
114. Beinse G, Borghese B, Métairie M. et al. Highly Specific Droplet-Digital PCR Detection of Universally Methylate d Circulating Tumor DNA in Endometrial Carcinoma. Clin Chem. 2022;68:782-93
115. S SK, Swamy SN, Premalatha CS, Pallavi VR, Gawari R. Aberrant Promoter Hypermethylation of RASSF1a and BRCA1 in Circulating Cell-Free Tumor DNA Serves as a Biomarker of Ovarian Carcinoma. Asian Pac J Cancer Prev. 2019;20:3001-5
116. Hao W, Yu M, Lin J. et al. The pan-cancer landscape of netrin family reveals potential oncogenic biomarkers. Sci Rep. 2020;10:5224
117. Suehiro Y, Okada T, Okada T. et al. Aneuploidy predicts outcome in patients with endometrial carcinoma and is related to lack of CDH13 hypermethylation. Clin Cancer Res. 2008;14:3354-61
118. Yang J, Li H, Hu S, Zhou Y. ACE2 correlated with immune infiltration serves as a prognostic biomar ker in endometrial carcinoma and renal papillary cell carcinoma: impli cation for COVID-19. Aging (Albany NY). 2020;12:6518-35
119. Hotton J, Gauchotte G, Mougel R. et al. Expressions of HuR, Methyl-HuR and Phospho-HuR in Endometrial Endometr ioid Adenocarcinoma Are Associated with Clinical Features. International journal of molecular sciences. 2024;25:954
120. Hu Y, Jiang N, Wang X. et al. Systematic pan-cancer analysis of RNF186 with potential implications i n progression and prognosis in human cancer. Life Sciences. 2024;338:122389
121. Schuhn A, Tobar TW, Gahlawat AW. et al. Potential of blood-based biomarker approaches in endometrium and breas t cancer: a case-control comparison study. Arch Gynecol Obstet. 2022;306:1623-32
122. Yuan J, Mao Z, Lu Q. et al. Hypermethylated PCDHGB7 as a Biomarker for Early Detection of Endometr ial Cancer in Endometrial Brush Samples and Cervical Scrapings. Front Mol Biosci. 2022;8:774215
123. Wang S-F, Du C-Y, Li M. et al. Endometrial Cancer Detection by DNA Methylation Analysis in Cervical P apanicolaou Brush Samples. Technol Cancer Res Treat. 2024;23:15330338241242637
124. Wentzensen N, Bakkum-Gamez JN, Killian JK. et al. Discovery and validation of methylation markers for endometrial cancer. Int J Cancer. 2014;135:1860-8
125. Wang D, O'Rourke D, Sanchez-Garcia JF. et al. Development of a liquid biopsy based purely quantitative digital dropl et PCR assay for detection of MLH1 promoter methylation in colorectal cancer patients. BMC Cancer. 2021;21:797
Author contact
![]() Corresponding authors: zhoufhedu.cn; liuajcom.cn.
Corresponding authors: zhoufhedu.cn; liuajcom.cn.

 Global reach, higher impact
Global reach, higher impact