Impact Factor
ISSN: 1837-9664
J Cancer 2024; 15(12):3675-3683. doi:10.7150/jca.94202 This issue Cite
Research Paper
Network Pharmacology and Bioinformatics Analysis to Identify the Molecular Targets and its Biological Mechanisms of Sciadopitysin against Glioblastoma
1. Department of Neurosurgery, Renmin Hospital of Wuhan University, Wuhan, Hubei, 430060, P.R. China.
2. Department of Basic Medicine, Medical School, Kunming University of Science and Technology, Kunming, Yunnan, 651701, P.R. China.
3. Guangdong Key Laboratory of Genome Instability and Human Disease Prevention, Department of Biochemistry and Molecular Biology, School of Medicine, Shenzhen University, Shenzhen, Guangdong, 518055, P.R. China.
4. Department of pediatrics, Liyuan Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei, 430077, P.R. China.
5. Department of Gynaecology and Obstetrics, Renmin Hospital of Wuhan University, Wuhan, Hubei, 430060, P.R. China.
Received 2024-1-12; Accepted 2024-4-30; Published 2024-5-13
Abstract
Glioblastoma multiform (GBM) is categorized as the most malignant subtype of gliomas, which comprise nearly 75% of malignant brain tumors in adults. Increasing evidence suggests that network pharmacology will be a novel method for identifying the systemic mechanism of therapeutic compounds in diseases like cancer. The present study aimed to use a network pharmacology approach to establish the predictive targets of sciadopitysin against GBM and elucidate its biological mechanisms. Firstly, targets of sciadopitysin were obtained from the SwissTargetPrediction database, and genes associated with the pathogenesis of GBM were identified from the DiGeNET database. Sixty-four correlative hits were identified as anti-glioblastoma targets of sciadopitysin. Functional enrichment and pathway analysis revealed significant biological mechanisms of the targets. Interaction of protein network and cluster analysis using STRING resulted in two crucial interacting hub genes, namely, HSP90 and AKT1. Additionally, the in vitro cytotoxic potential of sciadopitysin was assessed on GBM U87 cells. The findings indicate that the pharmacological action of sciadopitysin against GBM might be associated with the regulation of two core targets: HSP90 and AKT1. Thus, the network pharmacology undertaken in the current study established the core active targets of sciadopitysin, which may be extensively applied with further validations for treatment in GBM.
Keywords: sciadopitysin, glioblastoma, network pharmacology, HSP90, AKT1
Introduction
Adult glioblastoma (GBM) is the most common malignant primary brain tumor, representing approximately 57% of all gliomas and 48% of all primary malignant central nervous system (CNS) tumors [1]. It is also one of the most deadly and recalcitrant of all malignant solid tumors. Despite recent advances that have been made in multimodality therapy for glioblastoma incorporating surgery, radiotherapy, systemic therapy (chemotherapy, targeted therapy), and supportive care, the overall prognosis is still poor, and long-term survival is rare. The latest research shows that in the United States alone, the annual incidence of glioblastoma is about 34 cases per 1 million people, with a median survival of about 8 months, and more than 7,000 people die from glioblastoma each year [2]. For most patients with GBM, there is no known cause of the disease. It may occur at any age and originate by genetic alterations affecting neuroglial stem or progenitor cells [3]. Incidence increases steadily with age. The therapeutic efficacy of glioblastoma could be improved by figuring out molecular pathways and alterations in the signaling mechanisms of the tumor cells.
Sciadopitysin (SP), a biflavonoid compound that is common in gymnosperms such as Cyperus roxburghii, Cryptomeria fortunei, Podocarpus, Taxus chinensis, and Ginkgo biloba, exhibits various biological properties [4]. In recent years, the biological activity of SP has been gradually investigated. Studies have shown that SP has various pharmacological effects, such as anti-tumor, antioxidation, reducing blood glucose and blood lipid, etc. Bioflavonoids show potential proteins or enzymes related to metabolism, growth, and survival, which are related to tumor growth, tumor metastasis, and angiogenesis [5]. Glioblastoma is a kind of heterogeneous disease, meaning a distinct understanding of its mechanism is required for significant treatment preferences. However, due to limitations in research techniques and economic considerations, it is difficult to reveal the synergy between multiple targets in disease treatment. Currently, the precise antitumor mechanisms of SP are unknown.
Network pharmacology, as an emerging discipline, has been developed very well in recent years by integrating bioinformatics and pharmacology. It is a kind of method that predicts targets against a particular disease by the available biomedical data in system biology and poly-pharmacology [6]. By establishing a pharmaceutical chemistry database, researchers can discover the relationship between drugs and targets, targets and target diseases, systematically mine the existing biological data, abstract these data into a network relation model, and then systematically explain the role of drugs in disease treatment [7]. Network pharmacology can describe the complex relationship between biological systems in targeted therapy and determine the synergistic effect in tumor therapy through network component analysis [8].
So, we used network pharmacology to study multi-target drugs, exploring target sites and action pathways to provide evidence for the clinical application of SP in GBM treatment. The study was divided into the following stages: (1) Identification of the potential targets of SP based on its association with GBM through retrieval from databases; (2) Using gene ontology (GO) terms to study the key role of identified targets through functional enrichment and pathway analysis; (3) Determining the core indicators based on interaction through network analysis; (4) Validation of potential targets by molecular docking verification and in vitro assessment.
Materials and Methods
Identification of potential targets of sciadopitysin and glioblastoma-related targets
SwissTargetPrediction (http://www.swisstargetprediction.ch/, accessed on 20 July 2023) was utilized to screen the potential targets of sciadopitysin [9]. The molecular structure is displayed in Figure S1, and SMILES of sciadopitysin is COC1=CC=C(C=C1)C2OC3C(=C(O)C=C(O)C=3C(=O)C=2)C4=C(OC)C=CC(=C4)C5OC6C(=C(O)C=C(OC)C=6)C(=O)C=5.
The glioblastoma-related targets were screened from the DisGeNET database (http://www.disgenet.org/, accessed on 20 July 2023) using the keyword search “glioblastoma multiforme” [10]. The overlapped targets of sciadopitysin potential targets and glioblastoma-related targets were considered as candidate anti-glioblastoma cancer sciadopitysin targets, which were subjected to further analysis.
Gene ontology and signaling pathway enrichment analysis of the drug-disease targets
To investigate the biological characteristics of drug-disease targets, Gene Ontology (GO) enrichment, and Kyoto Encyclopedia of Genes and Genome (KEGG) pathway analyses were carried out using the clusterProfiler package. The enrichment terms with adjusted p-value <0.05 were deemed to be significantly different. The genes with significant regulatory pathways were chosen for subsequent gene-pathway network analysis.
Establishment of the PPI network and module construction of glioblastoma targets
Based on the Search Tool for the Retrieval of Interacting Genes (STRING), the protein-protein interaction network was constructed by the candidate 64 anti-glioblastoma targets of sciadopitysin, with a lowest confidence score of 0.4.
Network construction and analysis
To elucidate the therapeutic mechanism of sciadopitysin on GBM, the ingredient-target network was built through Cytoscape 3.7.2 software. The drug-disease targets' protein-protein interaction (PPI) network was established by the plugin Bisogenet of Cytoscape version 3.7.2 [11]. Subsequently, topology analysis was conducted using the Cytoscape plugin CytoNCA to calculate the Betweenness Centrality (BC) and Degree Centrality (DC). The nodes with BC and DC were identified as the key nodes. Next, the gene-pathway network was built to identify the key target genes responsible for sciadopitysin-treated GBM. Lastly, the crossover genes between the key nodes and the key target genes were considered the core molecular targets of sciadopitysin for treating GBM and were selected to perform further in vitro experiments.
Molecular docking analysis
To assess the putative targets identified through network analysis, we conducted a molecular docking analysis using the Auto-Dock Vina software [12]. Initially, the three-dimensional (3D) structure of sciadopitysin was retrieved from PubChem (accessed on 9 October 2023) and optimized. Additionally, the 3D structures of the potential targets were downloaded from the Protein Data Bank (PDB) database (accessed on 9 October 2023). We then utilized AutoDockTools to generate a two-dimensional map of sciadopitysin with the potential targets, which enabled us to visualize the direct interactions between the compound and the targets. Finally, PyMol software was used to observe the three-dimensional structure of sciadopitysin-target complex [13].
Cell and cell culture
Human glioblastoma cell line U87 was purchased from the Chinese Academy of Sciences Cell Bank. These cells were incubated at 37°C, 5% CO2 concentration, and high humidity. They were regularly cultured in Dulbecco's modified Eagle's medium (HyClone), which were supplemented with 10 % fetal bovine serum (HyClone).
Active cell apoptosis
Cell apoptosis was measured by the Caspase 3/7Activity Assay Kit (Absin, China). In brief, cells were collected and centrifuged at 600 g at 4°C for 5 minutes, the supernatant was removed, and after washing with PBS, the lysate was added at a ratio of 100 microliters per 2 million cells. The lysate was dissolved in an ice bath for 15 minutes. Then we centrifuged at 16,000 g and 4°C for 10 minutes and transferred the supernatant to a centrifuge tube pre-cooled by an ice bath. The reaction system was as follows: 40 µL buffer, 50 µL experimental cells, 10 µL Ac-DEVD-pNA (2mM). After incubation at 37°C for one hour, A405 was determined when the color change was noticeable.
Flow cytometry
The apoptosis of cells was detected by the Annexin V-fluorescein isothiocyanate/propidium iodide (Annexin V-FITC/PI) apoptosis detection kit (abs50001; Absin Biotechnology Co. Ltd., Shanghai, China) staining. U87 cells were incubated in 6-well plates and treated with 100 μΜ sciadopitysin for 72 h. Cells were collected and washed with cold phosphate-buffered brine, resuspended with 5 µL Annexin V-FITC and 5 µL PI staining solution in 300 µL 1×binding buffer, and incubated at room temperature for 15 min under darkness. Next, we added 300 µL 1×binding buffer and mixed. Finally, cell apoptosis was detected by flow cytometry (Bio-Rad, State of California, USA).
Western blot
Total protein extraction of cell lines was conducted by applying RIPA buffer (Beyotime, Shanghai, China). The proteins were separated on a 10-12% sodium dodecyl sulfate-polyacrylamide gel and then transferred onto polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA). Next, the membrane was incubated with primary antibodies overnight at 4°C. AKT1 (1:1000, rabbit, Cell Signaling Technology, Cat#75692), HSP90α (1:1000, mouse, Abcam, Cat#ab79849), Actin (1:1000, rabbit, Abcam, Cat# ab179467). After the membrane was washed, HRP-labeled Goat Anti-Rabbit IgG (1:1000, Beyotime, Cat# A0208, RRID: AB_2892644) or HRP-labeled Goat anti-mouse IgG (1:1000, Beyotime, A0216, RRID: AB_2860575) were incubated at 25°C for 1 hour. Finally, the enhanced chemiluminescence detection system (Applygen Technology, Beijing, China) was used to detect the signal. The protein expression level was detected by ImageJ software.
Statistical analysis
SPSS version 23.0 was employed to conduct the statistical test. All experiments were performed in triplicate. All data were presented as mean ± SD. Comparisons among different groups were done with one-way ANOVA followed by Dunnett or Bonferroni post hoc analysis. A p-value of <0.01 or <0.05 was deemed statistically significant. The statistical column charts were drawn with Prism 8.0.
Results
Prediction and screening of drug target proteins and glioblastoma-related targets
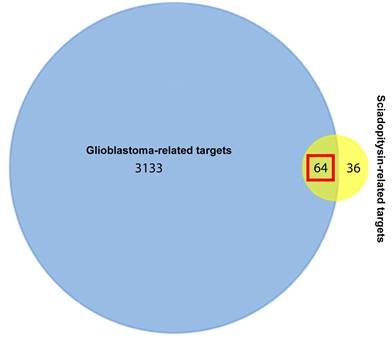
One hundred potential targets of sciadopitysin were screened from the SwissTargetPrediction database. The classes of these potential targets were primarily kinase, enzyme, oxidoreductase (Figure S2). 3197 hits of glioblastoma-related targets were predicted from the DisGeNET database. There were 64 targets overlapped in the drug and disease targets (Figure 1 and Table 1).
Gene ontology analysis of the candidate targets
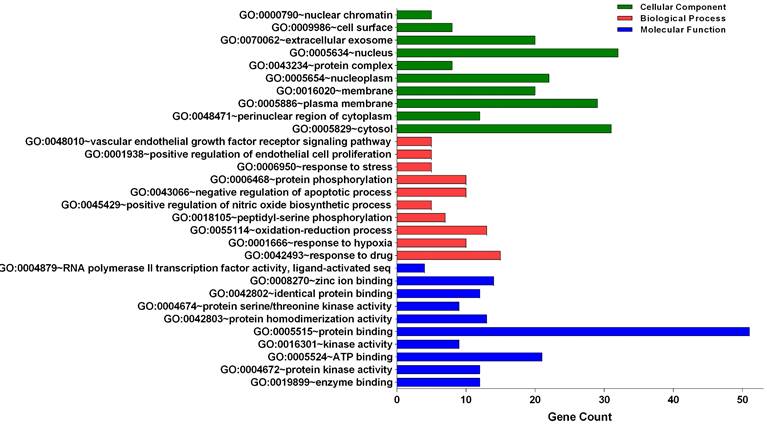
Three categories of anti-glioblastoma targets of sciadopitysin: cellular component, biological process, and molecular function, were classified by gene ontology (GO) analysis (Figure 2). The top 10 significantly enriched GO terms among the 64 core targets were listed in Tables 2-4. Among these GO functions, the enriched GO terms of cellular component were nucleus (GO:0005634), cytosol (GO:0005829), and plasma membrane (GO:0005886). The significant GO terms of the biological process included a response to drug (GO:0042493) and oxidation-reduction process (GO:0055114). The enriched GO terms of molecular function were associated with protein binding (GO:0005515) and ATP binding (GO:0005524).
List of 64 potential anti‐glioblastoma targets of sciadopitysin.
| Gene Name | Uniprot ID | Protein Name |
|---|---|---|
| BACE1 | P56817 | Beta-site APP cleaving enzyme 1 |
| PTPN1 | P18031 | Tyrosine-protein phosphatase non-receptor type 1 |
| VCP | P55072 | Valosin-containing protein |
| PGF | P49763 | Placenta growth factor |
| VEGFA | P15692 | Vascular endothelial growth factor A |
| ABCG2 | Q9UNQ0 | ATP-binding cassette sub-family G member 2 |
| MCL1 | Q07820 | Induced myeloid leukemia cell differentiation protein Mcl-1 |
| ABCB1 | P08183 | ATP-dependent translocase ABCB1 |
| AKR1B1 | P15121 | Aldo-keto reductase family 1 member B1 |
| PRKCB | P05771 | Protein kinase C beta type |
| CYP19A1 | P11511 | Cytochrome P450 19A1 |
| NOX4 | Q9NPH5 | NADPH oxidase 4 |
| KDM1A | O60341 | Lysine-specific histone demethylase 1A |
| RELA | Q04206 | Transcription factor p65 |
| PTGS2 | P35354 | Prostaglandin G/H synthase 2 |
| TNKS | O95271 | Poly [ADP-ribose] polymerase tankyrase-1 |
| ESR1 | P03372 | Estrogen receptor (ER) (ER-alpha) |
| PRKCG | P05129 | Protein kinase C gamma type (PKC-gamma) |
| ABCC1 | P33527 | Multidrug resistance-associated protein 1 |
| ODC1 | P11926 | Ornithine decarboxylase (ODC) |
| LDHA | P00338 | L-lactate dehydrogenase A chain (LDH-A) |
| IDH1 | O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic (IDH) |
| HSP90AA1 | P07900 | Heat shock protein HSP 90-alpha |
| FASN | P49327 | Fatty acid synthase |
| BCHE | P06276 | Cholinesterase |
| MAOA | P21397 | Amine oxidase [flavin-containing] A |
| ESR2 | Q92731 | Estrogen receptor beta (ER-beta) |
| CDK6 | Q00534 | Cyclin-dependent kinase 6 |
| CSNK2A1 | P68400 | Casein kinase II subunit alpha (CK II alpha) |
| AKR1B10 | O60218 | Aldo-keto reductase family 1 member B10 |
| MPG | P29372 | DNA-3-methyladenine glycosylase |
| MELK | Q14680 | Maternal embryonic leucine zipper kinase (hMELK) |
| CA2 | P00918 | Carbonic anhydrase 2 |
| CA12 | O43570 | Carbonic anhydrase 12 |
| KCNH2 | Q12809 | Potassium voltage-gated channel subfamily H member 2 |
| CCR1 | P32246 | C-C chemokine receptor type 1 |
| TYR | P14679 | Tyrosinase |
| AHR | P35869 | Aryl hydrocarbon receptor |
| PDK1 | Q15118 | [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 1, mitochondrial |
| NOS2 | P35228 | Nitric oxide synthase, inducible |
| TRAP1 | Q12931 | Heat shock protein 75 kDa, mitochondrial |
| AKT1 | P31749 | RAC-alpha serine/threonine-protein kinase (Protein kinase B alpha) |
| SRC | P12931 | Proto-oncogene tyrosine-protein kinase Src |
| DAPK1 | P53355 | Death-associated protein kinase 1 |
| MTOR | P42345 | Serine/threonine-protein kinase mTOR |
| PIK3CA | P42336 | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform |
| CHEK1 | O14757 | Serine/threonine-protein kinase Chk1 |
| WEE1 | P30291 | Wee1-like protein kinase |
| CA9 | Q16790 | Carbonic anhydrase 9 |
| PLG | P00747 | Plasminogen |
| HDAC1 | Q13547 | Histone deacetylase 1 |
| AR | P10275 | Androgen receptor |
| CBR1 | P16152 | Carbonyl reductase [NADPH] 1 |
| HSP90B1 | P14625 | Endoplasmin (Heat shock protein 90 kDa beta member 1) |
| CXCR2 | P25025 | C-X-C chemokine receptor type 2 |
| ALOX15 | P16050 | Polyunsaturated fatty acid lipoxygenase ALOX15 |
| PARP1 | P09874 | Poly [ADP-ribose] polymerase 1 |
| MMP9 | P14780 | Matrix metalloproteinase-9 |
| MMP2 | P08253 | 72 kDa type IV collagenase |
| MMP12 | P39900 | Macrophage metalloelastase |
| CD38 | P28907 | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1 |
| TOP1 | P11387 | DNA topoisomerase 1 |
| ARG1 | P05089 | Arginase-1 |
| PRKCI | P41743 | Protein kinase C iota type |
Venn diagram showing the 64 overlapped targets of sciadopitysin potential targets and glioblastoma-related targets.
Signaling pathway enrichment analysis of the candidate targets
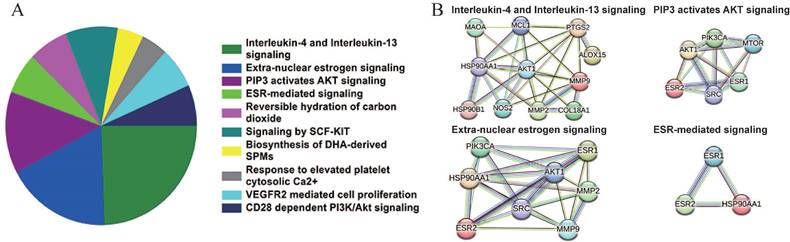
Based on the Reactome database, the top 10 enriched pathways of the 64 candidate targets were found to be Interleukin-4 and Interleukin-13 signaling, Extra-nuclear estrogen signaling, PIP3 activates AKT signaling, ESR-mediated signaling, Reversible hydration of carbon dioxide, Signaling by SCF-KIT, Response to elevated platelet cytosolic Ca2+, Biosynthesis of DHA-derived SPMs, VEGFR2 mediated cell proliferation and CD28 dependent PI3K/Akt signaling (Figure 3A and Table 5). Based on the interaction network constructed by the STRING database, the targets among the pathway enrichment were grouped into four most significant clusters that include ESR-mediated signaling string, extra-nuclear estrogen signaling string, interleukin-4 and Interleukin-13 signaling string, and PIP3 activates AKT signaling string, respectively (Figure 3B).
Top 10 significantly enriched GO terms of cellular component associated with the identified anti‐glioblastoma targets of sciadopitysin
| Term Name | Description | Count | % of Genes | Fold Enrichment | p-value |
|---|---|---|---|---|---|
| GO:0005829 | cytosol | 31 | 48.4375 | 2.66282051 | 8.72E-08 |
| GO:0048471 | perinuclear region of cytoplasm | 12 | 18.75 | 5.50241546 | 8.09E-06 |
| GO:0005886 | plasma membrane | 29 | 45.3125 | 2.00382189 | 9.85E-05 |
| GO:0016020 | membrane | 20 | 31.25 | 2.58863636 | 1.05E-04 |
| GO:0005654 | nucleoplasm | 22 | 34.375 | 2.2501796 | 2.80E-04 |
| GO:0043234 | protein complex | 8 | 12.5 | 5.52912621 | 5.33E-04 |
| GO:0005634 | nucleus | 32 | 50 | 1.68273315 | 8.74E-04 |
| GO:0070062 | extracellular exosome | 20 | 31.25 | 2.02596941 | 0.00232391 |
| GO:0009986 | cell surface | 8 | 12.5 | 4.20295203 | 0.00259475 |
| GO:0000790 | nuclear chromatin | 5 | 7.8125 | 7.37694301 | 0.00446334 |
Gene Ontology analysis of candidate targets. GO analysis classified into 3 groups: molecular function, biological process and cellular component.
Signaling pathway enrichment analysis of the candidate targets of sciadopitysin. (A) Pie chart shows the top 10 enriched pathways of candidate targets of sciadopitysin identified by Reactome database. (B) The four most significant enriched pathways are based on the protein-protein interaction network.
PPI network analysis
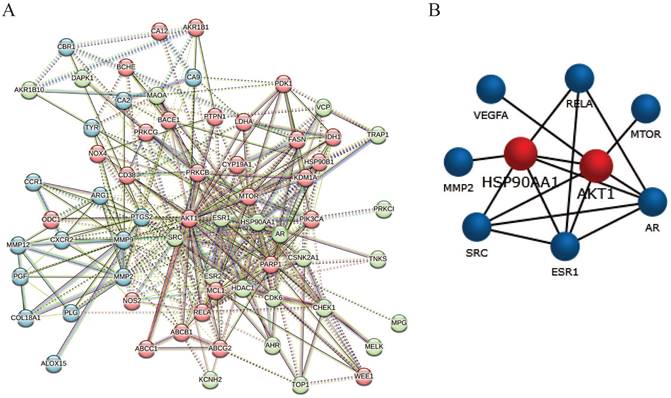
The candidate 64 anti-glioblastoma targets of sciadopitysin were introduced into the STRING database to obtain the PPI network complex. The PPI network contained 64 nodes and 359 edges (Figure 4A), obtained with a medium confidence score of 0.4 and enriched p-value of < 1.0e-16. Using CFinder with a k-cliques value of >12, one module (Figure 4B) was extracted from the constructed PPI network. Based on the key module, HSP90AA1 and AKT1 were selected as the key genes of sciadopitysin targets in glioblastoma.
Top 10 significantly enriched GO terms of biological process associated with the identified anti‐glioblastoma targets of sciadopitysin
| Term Name | Description | Count | % of Genes | Fold Enrichment | p-value |
|---|---|---|---|---|---|
| GO:0042493 | response to drug | 15 | 23.4375 | 12.9461349 | 5.08E-12 |
| GO:0001666 | response to hypoxia | 10 | 15.625 | 15.2543605 | 1.48E-08 |
| GO:0055114 | oxidation-reduction process | 13 | 20.3125 | 5.76161318 | 1.70E-06 |
| GO:0018105 | peptidyl-serine phosphorylation | 7 | 10.9375 | 14.693 | 7.24E-06 |
| GO:0045429 | positive regulation of nitric oxide biosynthetic process | 5 | 7.8125 | 30.5087209 | 1.99E-05 |
| GO:0043066 | negative regulation of apoptotic process | 10 | 15.625 | 5.76648352 | 4.70E-05 |
| GO:0006468 | protein phosphorylation | 10 | 15.625 | 5.75383772 | 4.78E-05 |
| GO:0006950 | response to stress | 5 | 7.8125 | 21.5061475 | 8.00E-05 |
| GO:0001938 | positive regulation of endothelial cell proliferation | 5 | 7.8125 | 19.0126812 | 1.30E-04 |
| GO:0048010 | vascular endothelial growth factor receptor signaling pathway | 5 | 7.8125 | 18.2204861 | 1.53E-04 |
Top 10 significantly enriched GO terms of molecular function associated with the identified anti‐glioblastoma targets of sciadopitysin
| Term Name | Description | Count | % of Genes | Fold Enrichment | p-value |
|---|---|---|---|---|---|
| GO:0019899 | enzyme binding | 12 | 18.75 | 9.505067568 | 3.68E-08 |
| GO:0004672 | protein kinase activity | 12 | 18.75 | 8.816678273 | 7.91E-08 |
| GO:0005524 | ATP binding | 21 | 32.8125 | 3.70506898 | 2.55E-07 |
| GO:0016301 | kinase activity | 9 | 14.0625 | 9.850168568 | 3.02E-06 |
| GO:0005515 | protein binding | 51 | 79.6875 | 1.531251779 | 6.43E-06 |
| GO:0042803 | protein homodimerization activity | 13 | 20.3125 | 4.697196062 | 1.37E-05 |
| GO:0004674 | protein serine/threonine kinase activity | 9 | 14.0625 | 6.313538896 | 7.47E-05 |
| GO:0042802 | identical protein binding | 12 | 18.75 | 4.225884513 | 9.10E-05 |
| GO:0008270 | zinc ion binding | 14 | 21.875 | 3.15886976 | 3.18E-04 |
| GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding | 4 | 6.25 | 29.30729167 | 3.24E-04 |
Top 10 significantly enriched pathways identified by Reactome database and associated anti‐glioblastoma targets of sciadopitysin
| Pathway name | Count | p-value | Genes |
|---|---|---|---|
| Interleukin-4 and Interleukin-13 signaling | 11 | 2.98418E-11 | VEGFA; MCL1; PTGS2; HSP90AA1; MAOA; NOS2; AKT1; HSP90B1; ALOX15; MMP9; MMP2; |
| Extra-nuclear estrogen signaling | 8 | 9.14128E-10 | ESR1; HSP90AA1; ESR2; AKT1; SRC; PIK3CA; MMP9; MMP2; |
| PIP3 activates AKT signaling | 6 | 1.82899E-05 | ESR1; ESR2; AKT1; SRC; MTOR; PIK3CA; |
| ESR-mediated signaling | 3 | 2.95625E-05 | ESR1; HSP90AA1; ESR2; |
| Reversible hydration of carbon dioxide | 3 | 3.92538E-05 | CA2; CA12; CA9; |
| Signaling by SCF-KIT | 4 | 5.84809E-05 | SRC; PIK3CA; CHEK1; MMP9; |
| Response to elevated platelet cytosolic Ca2+ | 2 | 9.86843E-05 | PRKCB; PRKCG; |
| Biosynthesis of DHA-derived SPMs | 2 | 9.86843E-05 | PTGS2; ALOX15; |
| VEGFR2 mediated cell proliferation | 3 | 0.00016796 | VEGFA; PRKCB; SRC; |
| CD28 dependent PI3K/Akt signaling | 3 | 0.000263645 | AKT1; MTOR; PIK3CA; |
Protein-protein interaction network complex and sub-network module construction of candidate targets. (A) Protein-protein interaction network of the anti-glioblastoma targets of sciadopitysin. A total of 64 targets was screened, containing 64 nodes and 359 edges. (B) One key sub-network of the core targets was constructed by module analysis.
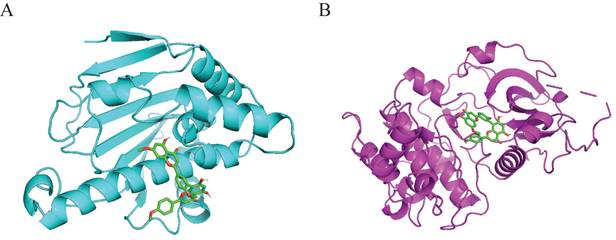
Interaction of sciadopitysin with HSP90α (PDB ID:2VCJ) and AKT1 (PDB ID:4EKL). (A) 3D docking molecule of HSP90α (cyan) and sciadopitysin. (B) 3D docking molecule of AKT1 (magenta) and sciadopitysin.
Molecular docking
Molecular docking was performed to validate the possible binding action mode between sciadopitysin and core targets. Among the two targets, the free binding energy of sciadopitysin with HSP90α -8.1kcal/mol, and with AKT1 was -9.3 kcal/mol, shown in Table 6. In the visualized image, it can be clearly seen that there are docking groups between the cyan labeled HSP90α and sciadopitysin molecules. And magenta-labeled AKT1 and sciadopitysin molecules are identical (Figure 5). The docking diagram shows that HSP90α and AKT1 both interact directly with sciadopitysin.
Sciadopitysin promoted apoptosis of GBM cells by inhibiting HSP90a and AKT1
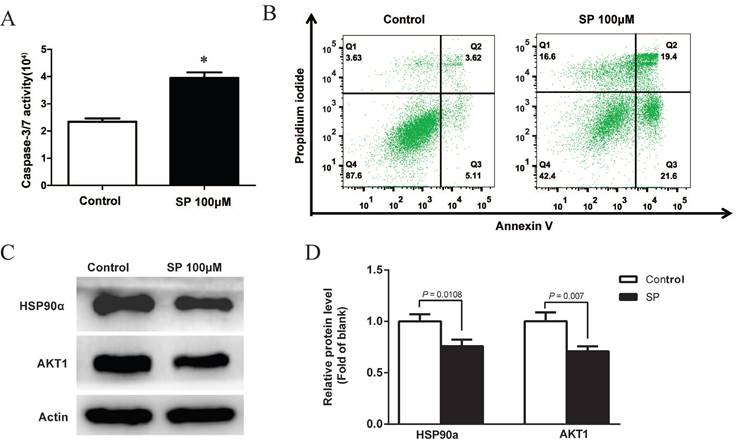
After treatment with 100µM sciadopitysin for 72 h, the apoptosis of GBM cells was determined by flow cytometry, and results showed that the percentage of apoptotic cells was significantly increased after sciadopitysin treatment (Figure 6B). These results strongly suggest that sciadopitysin induces apoptosis of GBM cells. We further verified the molecular target of sciadopitysin involved in inducing apoptosis in U87 cells. According to the above analysis, we detected the HSP90α and AKT1 proteins through western blotting. As shown in Figures 6C and 6D, compared with the control group, the protein expression levels of HSP90α and AKT1 in the treatment group were significantly decreased. The results confirmed that HSP90α and AKT1 were the molecular targets of sciadopitysin in inhibiting glioblastoma.
Discussion
In this study, bioinformatics investigation and network pharmacology were used to investigate the possibility of sciadopitysin in treating glioblastoma. To our knowledge, this is the first study to combine network pharmacology and molecular docking simulations to reveal the anti-glioblastoma effects of sciadopitysin. In the study, firstly, GBM-related genes were predicted from the public database prediction database, the target of sciadopitysin was found, and 64 targets of sciadopitysin anti-glioblastoma were obtained. PPI network and STRING cluster analysis identified HSP90α and AKT1 as two key hubs of action. Finally, cell experiments confirmed that sciadopitysin could regulate GBM apoptosis through HSP90α and AKT1.
HSP90α, encoded by the HSP90AA1 gene, is an important chaperone protein that requires a variety of collaborators to function [14]. And the HSP90 protein plays an important role in basic cellular processes and regulatory pathways such as apoptosis, cell cycle control, and cell signaling [15]. Overexpression of HSP90 is strongly associated with various cancers; for example, in breast cancer, the expression level of HSP90 is closely related to the survival of patients, and the abnormally high expression level of HSP90 reflects the poor treatment effect [16-18]. HSP90 is a promising marker for the diagnosis and prognosis of malignant tumors [14, 19]. In glioblastoma, the expression of HSP90α was abnormally high [20, 21]. Research shows the inhibitor of HSP90, 17-Allylamino-17-deoxykygdanamycin (17-AAG), inhibits the growth of glioma cell lines by targeting intracellular EGFR, AKT, and MAPK proteins [22]. In addition, studies have also shown that inhibiting Hsp90 function in glioblastoma cell lines can reduce the expression level of cell division cycle 2 kinase (cdc2) and cell division cycle 25c (cdc25c), and the proliferation is blocked in G(2)/M [23]. In our study, sciadopitysin promotes apoptosis of U87 cells, which is closely related to the HSP90α protein. In normal tissues, HSP90α is present in the cytoplasm, whereas in glioma cell lines, HSP90α may be abnormally localized to the cell membrane [19, 24]. Sciadopitysin interacts with HSP90α to reduce the activity of intracellular cdc2 and cdc25c and block the cell cycle, thereby inducing apoptosis.
Sciadopitysin promoted apoptosis of U87 cells. (A) Apoptosis of U87 cells was detected by measuring the activity of Caspase 3/7. (B) Apoptosis was analyzed by flow cytometry after Annexin V-FITC/PI staining. (C) Representative Western blots showing the status of HSP90α, AKT1 in U87 cells. Actin was used as an internal control. (D) The expression levels of HSP90α and AKT1 proteins were statistically analyzed.
Molecular docking studies of sciadopitysin with target protein and their binding energies
| Target | drug | Binding energy/(kcal/mol) |
|---|---|---|
| HSP90α | Sciadopitysin | -8.1 |
| AKT1 | Sciadopitysin | -9.3 |
AKT is one of the main downstream effect targets of phosphatidylinositol 3-kinase, which is overexpressed and activated in various cancers, including glioblastoma [25]. The individual functions of the three isomers of AKT, AKT1, AKT2, and AKT3, remain controversial in GBM. AKT2 mRNA and protein levels are elevated in malignant gliomas, while AKT3 expression levels are decreased [26]. In glioma cell lines, AKT2 or AKT3 knockdown inhibited cell growth and induced apoptosis. In contrast, AKT1 knockdown did not affect cell growth and apoptosis, suggesting that AKT2 and AKT3 may be the main contributors to GBM cell growth, and AKT1 may be unnecessary [26]. However, many studies have shown that AKT1 plays an important role in glioblastoma. For example, AKT1 is one of the key hub genes in the gene network of glioblastoma [27]. Overacting insulin receptor substrate 1 may promote GBM cell viability through AKT1 activation [28]. The inhibitory effect of SOX4 on the growth of GBM cells is related to the activation of the p53-p21 signal and the decrease of AKT1 activity [29]. In our experiment, AKT1 is another key target of sciadopitysin to promote apoptosis of glioma cell lines. After sciadopitysin treatment, AKT1 activity is reduced in U87 cells, which may lead to decreased expression levels of Cyclin D1 or p53 in the cells, thus affecting the cell cycle of glioblastoma.
In conclusion, sciadopitysin could be one novel potential targeted medicine for malignant glioblastoma. HSP90α and AKT1 were the key targets that sciadopitysin plays anti-tumor effects. This study provides preliminary experimental evidence of sciadopitysin against glioblastoma to some extent; However, further research is needed to provide more experimental verifications including HSP90α and AKT1 downstream signaling cascades and animal experiments to explore in vivo anti-tumor effects of sciadopitysin.
Supplementary Material
Supplementary figures.
Raw data.
Acknowledgements
Funding
This work was financially supported by the Fundamental Research Funds for the Central Universities (Grant No. 2042018kf0080, 2042022kf1117) and the National Natural Science Foundation of China (Grant No. 82203417).
Data availability statement
The raw data of the study is available in the Supplementary Material.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ostrom QT, Price M, Neff C, Cioffi G, Waite KA, Kruchko C. et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2015-2019. Neuro Oncol. 2022;24:v1-v95
2. Schaff LR, Mellinghoff IK. Glioblastoma and Other Primary Brain Malignancies in Adults: A Review. JAMA. 2023;329:574-87
3. Weller M, Wick W, Aldape K, Brada M, Berger M, Pfister SM. et al. Glioma. Nat Rev Dis Primers. 2015;1:15017
4. Suh KS, Chon S, Jung WW, Choi EM. Protective effects of sciadopitysin against methylglyoxal-induced degeneration in neuronal SK-N-MC cells. J Appl Toxicol. 2022;42:274-84
5. Menezes J, Diederich MF. Bioactivity of natural biflavonoids in metabolism-related disease and cancer therapies. Pharmacol Res. 2021;167:105525
6. Liang JW, Wang MY, Olounfeh KM, Zhao N, Wang S, Meng FH. Network pharmacology-based identifcation of potential targets of the flower of Trollius chinensis Bunge acting on anti-inflammatory effectss. Sci Rep. 2019;9:8109
7. Zhang R, Zhu X, Bai H, Ning K. Network Pharmacology Databases for Traditional Chinese Medicine: Review and Assessment. Front Pharmacol. 2019;10:123
8. Sakle NS, More SA, Mokale SN. A network pharmacology-based approach to explore potential targets of Caesalpinia pulcherima: an updated prototype in drug discovery. Sci Rep. 2020;10:17217
9. Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47:W357-W64
10. Pinero J, Sauch J, Sanz F, Furlong LI. The DisGeNET cytoscape app: Exploring and visualizing disease genomics data. Comput Struct Biotechnol J. 2021;19:2960-7
11. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498-504
12. Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455-61
13. Seeliger D, de Groot BL. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des. 2010;24:417-22
14. Yuan Z, Wang L, Chen C. Analysis of the prognostic, diagnostic and immunological role of HSP90alpha in malignant tumors. Front Oncol. 2022;12:963719
15. Zuehlke AD, Beebe K, Neckers L, Prince T. Regulation and function of the human HSP90AA1 gene. Gene. 2015;570:8-16
16. Barrott JJ, Haystead TA. Hsp90, an unlikely ally in the war on cancer. FEBS J. 2013;280:1381-96
17. Zagouri F, Sergentanis TN, Nonni A, Papadimitriou CA, Michalopoulos NV, Domeyer P. et al. Hsp90 in the continuum of breast ductal carcinogenesis: Evaluation in precursors, preinvasive and ductal carcinoma lesions. BMC Cancer. 2010;10:353
18. Pick E, Kluger Y, Giltnane JM, Moeder C, Camp RL, Rimm DL. et al. High HSP90 expression is associated with decreased survival in breast cancer. Cancer Res. 2007;67:2932-7
19. Birbo B, Madu EE, Madu CO, Jain A, Lu Y. Role of HSP90 in Cancer. Int J Mol Sci. 2021 22
20. Mehta A, Shervington A, Howl J, Jones S, Shervington L. Can RNAi-mediated hsp90alpha knockdown in combination with 17-AAG be a therapy for glioma? FEBS Open Bio. 2013;3:271-8
21. van Ommeren R, Staudt MD, Xu H, Hebb MO. Advances in HSP27 and HSP90-targeting strategies for glioblastoma. J Neurooncol. 2016;127:209-19
22. Sauvageot CM, Weatherbee JL, Kesari S, Winters SE, Barnes J, Dellagatta J. et al. Efficacy of the HSP90 inhibitor 17-AAG in human glioma cell lines and tumorigenic glioma stem cells. Neuro Oncol. 2009;11:109-21
23. Garcia-Morales P, Carrasco-Garcia E, Ruiz-Rico P, Martinez-Mira R, Menendez-Gutierrez MP, Ferragut JA. et al. Inhibition of Hsp90 function by ansamycins causes downregulation of cdc2 and cdc25c and G(2)/M arrest in glioblastoma cell lines. Oncogene. 2007;26:7185-93
24. Hoter A, El-Sabban ME, Naim HY. The HSP90 Family: Structure, Regulation, Function, and Implications in Health and Disease. Int J Mol Sci. 2018 19
25. McDowell KA, Riggins GJ, Gallia GL. Targeting the AKT pathway in glioblastoma. Curr Pharm Des. 2011;17:2411-20
26. Mure H, Matsuzaki K, Kitazato KT, Mizobuchi Y, Kuwayama K, Kageji T. et al. Akt2 and Akt3 play a pivotal role in malignant gliomas. Neuro Oncol. 2010;12:221-32
27. Yang Q, Wang R, Wei B, Peng C, Wang L, Hu G. et al. Gene and microRNA Signatures Are Associated with the Development and Survival of Glioblastoma Patients. DNA Cell Biol. 2019;38:688-99
28. Gorgisen G, Yaren Z. Insulin receptor substrate 1 overexpression promotes survival of glioblastoma cells through AKT1 activation. Folia Neuropathol. 2020;58:38-44
29. Zhang J, Jiang H, Shao J, Mao R, Liu J, Ma Y. et al. SOX4 inhibits GBM cell growth and induces G0/G1 cell cycle arrest through Akt-p53 axis. BMC Neurol. 2014;14:207
Author contact
![]() Corresponding authors: Yun Zhou, E-mail: yunzhouedu.cn; or to Huimin Liu, E-mail: 18040501553com.
Corresponding authors: Yun Zhou, E-mail: yunzhouedu.cn; or to Huimin Liu, E-mail: 18040501553com.

 Global reach, higher impact
Global reach, higher impact